Abstract
Regulated point modification by an RNA editing enzyme occurs at four conserved sites in the Drosophila Shaker potassium channel. Single mRNA molecules can potentially represent any of 24 = 16 permutations (isoforms) of these natural variants. We generated isoform expression profiles to assess sexually dimorphic, spatial, and temporal differences. Striking tissue-specific expression was seen for particular isoforms. Moreover, isoform distributions showed evidence for coupling (linkage) of editing sites. Genetic manipulations of editing enzyme activity demonstrated that a chief determinant of Shaker editing site choice resides not in the editing enzyme, but rather, in unknown factors intrinsic to cells. Characterizing the biophysical properties of currents in nine isoforms revealed an unprecedented feature, functional epistasis; biophysical phenotypes of isoforms cannot be explained simply by the consequences of individual editing effects at the four sites. Our results unmask allosteric communication across disparate regions of the channel protein and between evolved and regulated amino acid changes introduced by RNA editing.
INTRODUCTION
Genomic information unfolds according to a well-established paradigm; the amino acid sequence of a protein is encoded by a literal one-to-one mapping from each genotypic codon to one of 20 amino acids. This central paradigm assumes robust correspondence between DNA and its transcribed RNA copy. A notable interloper in this orderly enterprise is an enzyme that chemically alters individual nucleotides of RNA. Action of the adenosine deaminase acting on RNA (ADAR) enzymes results in the hydrolytic deamination of adenosine-to-inosine (A-to-I) in double-stranded (ds) RNA substrates (Bass, 2002). ADAR modification can affect numerous biological readouts, including alternative RNA splice choices, opposition to RNA interference pathways, and altered microRNA processing (Rueter et al., 1999; Bass, 2006; Nishikura, 2006). One outcome of A-to-I editing, however, has overt consequences for information encoding—inosine is recognized as guanosine (G) by the translation machinery (Basilio et al., 1962), rendering almost half of the codons of the genetic code re-assignable to edited versions encoding different amino acids. Inexplicably, animal genes that encode components of rapid electrical and chemical neurotransmission dominate gene targets of this recoding aspect of editing (Seeburg and Hartner, 2003) and usually require intronic cis elements to form a dsRNA structure that serves as an ADAR substrate (Herbert, 1996). Genetic deficiency for ADAR activity or altered ADAR function can cause behavioral dysfunction, both of which have been implicated in neurological disease (Higuchi et al., 2000; Palladino et al., 2000a, Tonkin et al., 2002; Maas et al., 2006; Mehler and Mattick, 2007). Nevertheless, the functional consequences of A-to-I RNA editing for sites in most ADAR gene targets remain unknown.
Inosine can be detected in mature mRNA from many mammalian tissues, but it reaches peak levels in material isolated from the brain (Paul and Bass, 1998). This simple observation is complicated by certain facts; there are three known editing enzymes (ADAR1-3) in mammals, different isoforms of these ADARs can be produced by alternative processing mechanisms, and ADARs act as a dimer (for review see Keegan et al., 2004). Nevertheless, regulation has been shown to occur at the level of individual editing sites via strong enzyme preference. For instance, the GluR-B AMPA receptor (Q/R) site is edited efficiently only by ADAR2, whereas the paralogous GluR-6 kainate receptor (Q/R) site is edited by ADAR1 (Maas et al., 1996). Even editing sites within several nucleotides of one another can require different ADARs, such as in mammalian serotonin-2C receptor editing (Liu et al., 1999). Conversely, the GluR-B (R/G) site and mammalian GABA receptor transcripts are efficiently edited by either ADAR1 or ADAR2 (Melcher et al., 1996; Ohlson et al., 2007). Both spatial and temporal regulation of specific editing has also been shown to occur. In vertebrates and invertebrates alike, there are marked increases in A-to-I editing for many specific targets throughout development (Bernard and Khrestchatisky, 1994; Lomeli et al., 1994; Palladino et al., 2000b; Keegan et al., 2005; Ohlson et al., 2007). Layering onto this developmental control, spatial regulation of ADAR-mediated recoding produces differing degrees of specific target editing within different regions of the nervous system. In addition, target transcripts with multiple editing sites, like the serotonin-2C receptor, can produce numerous edited isoforms combinatorially (Burns et al., 1997). Neither the temporal nor spatial patterns of specific editing of ADAR targets in mammals have been shown to correlate with known patterns of ADAR gene expression, tacitly implying other unknown factors (Lai et al., 1997; Liu et al., 1999; Paupard et al., 2000).
Voltage-gated potassium channels play crucial roles in determining the firing properties of neurons (Hille, 2001) and are the only common gene target of A-to-I editing among three major animal phyla: chordates, mollusks, and arthropods. In mollusks, extensive editing of the squid channel, sqKv1.1, was shown to regulate functional expression through effects on tetramerization, whereas a subset of the extensive editing sites of sqKv2 affect channel closure and slow inactivation (Patton et al., 1997; Rosenthal and Bezanilla, 2002). In neither case are the RNA structures that direct editing known, nor the reason for such extensive editing. In another invertebrate, Drosophila, RNA editing of Kv2 (Shab) channels has been shown to affect channel biophysics (Ryan et al., 2008). In chordates, the mammalian intronless Kv1.1 gene was shown to undergo spatially regulated editing through the formation of a small RNA hairpin contained within the coding sequence (Hoopengardner et al., 2003; Bhalla et al., 2004). RNA editing of one position within the Kv1.1 potassium channel was shown to dramatically affect the process of channel inactivation.
Here, we describe the in vivo production of editing isoforms for the Shaker potassium channel from the arthropod, Drosophila melanogaster. The Shaker gene possesses four developmentally regulated A-to-I editing sites in highly conserved regions of the channel protein. Expression profiling of the 16 possible isoforms reveals that 15 are expressed. Unexpectedly, we found dramatic tissue-specific differences in Shaker isoform expression levels spanning almost two orders of magnitude. Linkage analyses reveal that the editing of certain sites affects the likelihood that other sites are also edited. ADAR expression studies in transgenic flies revealed that unknown factors, intrinsic to certain locations, predominate in this spatial regulation and that ADAR preference plays a minimal role. The regulatory complexities of Shaker editing extended beyond spatial and temporal scales. Characterization of the biophysical properties of the more abundant Shaker isoforms reveals a functional epistasis; the consequence of an editing mutation, particularly on inactivation rate, depends on whether distant sites are also edited.
MATERIALS AND METHODS
Fly Stocks and Expression Studies
The Drosophila melanogaster wild-type stock used was Canton-S. For rescue experiments, the dADAR5G1-null allele was used. In brief, dADAR5G1/FM7;;elav-pSwitch females were crossed to males containing a rescuing transgene expressing the dADAR-3/4 isoform (TM3∷UAS-dADARwt5/TM6). dADAR5G1/Y;;TM3∷UAS-dADARwt5/elav-pSwitch males were selected and aged for 7 d. Animals were then fed food containing 200 μM RU-486 to induce ADAR expression for 7 d and then harvested for analyses.
RNA Editing Analysis
All RNA extractions were performed using TRIzol (Invitrogen) on whole flies/larvae or various dissected body parts as indicated in Results. Shaker transcripts were amplified by reverse transcription (RT)-PCR using gene-specific primers at all steps. For isoform profiles, cDNAs were cloned from at least three independent RT-PCR reactions for each sample and subjected to automated sequence analysis (see Table S1, available at http://www.jgp.org/cgi/content/full/jgp.200810133/DC1). Levels of editing for individual editing sites determined for developmental and rescue studies were obtained by direct sequencing of RT-PCR products from at least three independent reactions per sample. Areas under the curves were determined from electropherogram traces and editing level expressed as: {% editing} = (area G/total area A+G) * 100. Where editing levels for individual sites were obtained from isoform profiles (Fig. 3), the number of clones edited at a given site was divided by the total number of clones in the sample.
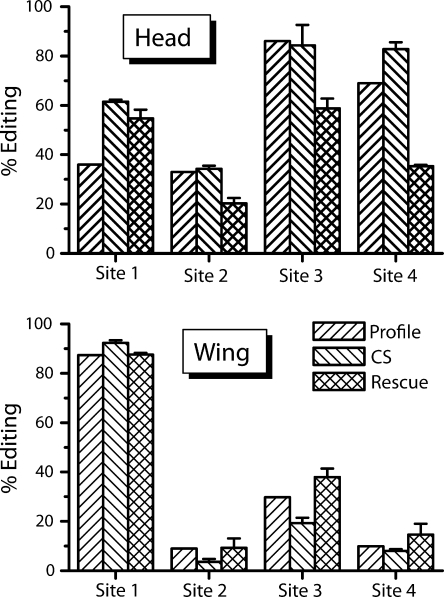
Figure 3.
Contribution of ADAR to tissue-specific editing patterns. The top panel shows quantitation of RNA editing levels in head samples for sites 1–4 from isoform profiling (Profile), direct RT-PCR sequence analysis (see Materials and methods), wild-type (CS), and direct RT-PCR sequence analysis from dADAR-null animals rescued with a single dADAR isoform expressed pan-neuronally. The bottom panel shows quantitation of RNA editing levels in wing samples, as in the top panel.
Expression Clones, Mutagenesis, and Transfection
For functional studies we used chimeras consisting of the N terminus of Shaker B and the C terminus common to Shaker A and C. These cDNA chimeras were generated by using a naturally occurring XbaI restriction enzyme cutting site found in Shaker exon 4. The C-terminal region clones were isolated from Drosophila by RT-PCR and sequence verified. In some experiments fast inactivation was abolished by the deletion of residues 6–46. The point mutations T449V and V463A were constructed to inhibit slow inactivation. All Shaker constructs were inserted into the pGW1 vector for expression in mammalian cells. Editing mutations at the four sites were generated using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) and verified by sequencing. Transfection of tsA201 cells was accomplished using standard calcium phosphate methodology. All isoforms were cotransfected with the auxiliary subunit Hyperkinetic (provided by G. Wilson, University of Michigan, Ann Arbor, MI). Recordings were taken 1–3 d after transfection.
Electrophysiology and Data Analysis
Standard whole cell patch clamp recording methods were used to record ionic currents (Ding and Horn, 2003). Electrode resistance ranged between 0.8 and 1.6 MΩ, and series resistance was compensated so that voltage errors were <3 mV. Patch pipettes contained (in mM): 105 CsF, 35 NaCl, 10 EGTA, and 10 HEPES, pH 7.4. The bath solution contained (in mM): 150 NaCl, 2 KCl, 1.5 CaCl2, 1 MgCl2, and 10 HEPES, pH 7.4. All experiments were performed at room temperature. pCLAMP (MDS Analytical Technologies) software was used for data acquisition and analysis. Further analysis used Origin (Microcal), Microsoft Excel, and in-house FORTRAN programs. We analyzed the isoform distributions (Fig. 2) with the program Mendel 7.0 (http://www.genetics.ucla.edu/software/mendel).
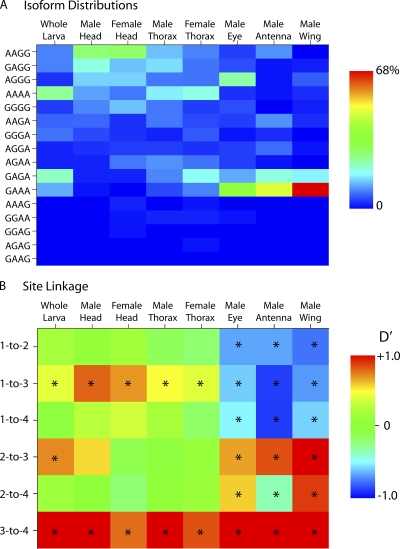
Figure 2.
Isoform distributions. (A) Distributions of 16 possible isoforms in eight populations. Percentage of population accorded to each isoform is color coded. (B) Pairwise linkage between editing at the four sites is color coded as Lewontin's D' statistic, where +1 and −1 represent complete positive or negative coupling, and 0 represents independence. The starred squares indicate significant coupling (P < 0.02; Fisher's exact test).
Online Supplemental Material
The supplemental material includes four tables and three figures. Table S1 summarizes the DNA sequence profiling of 821 Shaker cDNA clones from various tissue samples and their distributions among the various 16 isoforms. Tables S2–S4 summarize the inactivation parameters, conductance-voltage parameters, and deactivation time constants for nine Shaker editing isoforms. Fig. S1 shows the circle diagram depicting the ensemble average base-pairing probabilities for the structure pairing the evolutionarily conserved e1 element with the coding sequence at Shaker editing site 1. Fig. S2 shows the circle diagram depicting base-pairing probabilities with the largest centroid of the structure pairing the evolutionarily conserved e2 and e3 elements with the coding sequences at Shaker editing sites 2–4. Fig. S4 shows the local predicted dsRNA secondary structures pairing conserved intronic editing site complementary sequences (ECSs) with the regions surrounding the edited adenosines. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.200810133/DC1.
RESULTS
RNA Editing of Drosophila Shaker
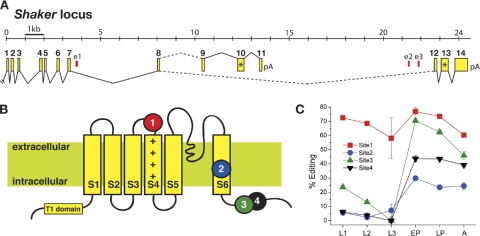
RNA editing of Drosophila Shaker has been reported to occur at six positions (Hoopengardner et al., 2003). Two sites are edited at low levels (<5%) in the T1 domain of the channel and were not considered here. The remaining four editing sites are distributed in two exons (Fig. 1 A). Site 1 is located in exon 7, separated by 19,323 nucleotides in genomic sequence from site 2 in downstream exon 12. Sites 3 and 4 are adjacent in exon 12, 74 nucleotides downstream of site 2, with a separation of only 6 nucleotides. ADAR-mediated recoding requires a dsRNA intermediate, frequently formed by base-pairing interactions with intronic ECSs. We have previously shown that phylogenetic conservation of editing sites is accompanied by a high degree of conservation for the ECS elements that direct dsRNA structure formation (Hanrahan et al., 2000; Reenan et al., 2000). We used comparative genomics of the 12 sequenced Drosophila genomes (http://flybase.org/) to search for conserved ECS elements in Shaker. In the intron downstream of exon 7, we found an invariant conserved element (Fig. 1 A, e1) located 1,289 nt from site 1. Computational folding of the pre-mRNA region encompassing site 1 and e1 was performed using SFOLD, a program that uses sampling of the Boltzmann-weighted ensemble of RNA secondary structures and statistical clustering methods to more effectively characterize structured RNA (Ding et al., 2005, 2006). An ensemble structure was obtained that paired site 1 with e1 generating a structure that, in appearance, is similar to many known ADAR substrates (Figs. S1 and S3). This predicted structure is absolutely conserved in all 12 Drosophila species, whereas all remaining flanking sequences are quite divergent. A similar analysis for the region near editing sites 2–4 revealed two conserved elements (e2 and e3) in the upstream intron flanking exon 12 (Figs. S2 and S3). Thus, it appears that three distinct dsRNA domains control the editing of Shaker sites 1–4. We then compared Shaker editing for five species spanning the phylogenetic distances covered by the “12 genomes” and found editing at sites 1–4 conserved in all cases (unpublished data). Lastly, we assessed editing of the Shaker ortholog in six species of mosquito and honey bee, where we found neither evidence for RNA editing nor any evidence for conservation of intronic elements e1–e3 in species that do not edit (unpublished data). Thus, editing of Shaker at sites 1–4 appears to be Diptera specific and possibly restricted to the family Drosophilidae.
Figure 1.
Editing of the Shaker locus. (A) The Shaker locus transcription unit is shown with numbered exons (yellow), constitutive splice sites (solid lines), alternative splice sites (broken lines), and stop codons (asterisks). The position of conserved elements predicted to direct editing, e1–e3, is shown in red. (B) The position of Shaker editing sites 1–4 is depicted on a generic cartoon of Kv channel topology. (C) Developmental profile of editing in whole animals for sites 1–4 is shown for larval, pupal, and adult stages. Color code as in B.
Editing of Shaker sites 1–4 alters amino acids encoded at positions that are invariant or highly conserved in all vertebrate and invertebrate Kv1 family orthologs (Fig. 1 B). Site 1 is at the top of the voltage-sensing transmembrane segment S4 and results in an isoleucine-to-methionine (I360M) substitution, site 2 is in the S6 segment and is recoded from isoleucine-to-valine (I464V), and sites 3 and 4 are in the S6T cytoplasmic tail separated by one amino acid, resulting in threonine-to-alanine (T489A) and glutamine-to-arginine (Q491R) recoding, respectively.
Because several editing sites have been reported to undergo developmentally regulated modification in vertebrate and invertebrate systems, we investigated the temporal regulation at each editing site in Shaker (Fig. 1 C). Site 1 is edited in both C-terminal alternative splice forms, ShakerA and ShakerB. Determination of the level of editing throughout development revealed that site 1 is largely unregulated in terms of editing level and is edited efficiently (>60%). Despite occurring in the same ion channel transcript, sites 2–4 display highly regulated editing and variable extents of editing. Like numerous other Drosophila editing sites, sites 2–4 are predominantly adult specific (Hanrahan et al., 2000, Palladino et al., 2000b).
Isoform Distributions
Fig. 2 A shows the color-coded distribution of the 24 = 16 possible isoforms in whole larvae and seven different tissues of adult Drosophila. For each tissue, 96–111 individual cDNA clones were sequenced for a total of 821 sequences (Table S1). The editing status at each site is represented by an A (unedited) or a G (edited) in linear order, rather than by amino acid change. We detected 15/16 possible isoforms, spanning a range from 0.1 (1/821 for AGAG and GGAG) to 22.7% (186/821 for GAAA), in our total dataset. Although the male and female populations were statistically indistinguishable (P > 0.3; Fisher's exact test of homogeneity) (Lange, 2003), the distributions were significantly different for all other pairwise comparisons between tissue populations (P < 0.001). The most dramatic difference among the tissues is seen for the isoform GAAA, which comprises, for example, 75 (68%) of the 111 cDNA clones in the male wing and is completely absent in 100 cDNA clones from female heads.
Examination of the distributions of isoforms shows evidence for coupling (linkage) between editing sites. For example, in the adult male wing there are 78 out of 111 clones in which sites 3 and 4 are both unedited (xxAA) and none in which site 3 is unedited and site 4 is edited (xxAG). This is not due simply to the paucity of editing at site 4 because 11 out of 111 clones were of the form xxxG. A similar pattern is seen for all other tissues. For example, there are 14 out of 100 clones of xxAA from male heads, but none of xxAG. In contrast, there are 17 xxGA's and 69 xxGG's. Therefore, if site 3 is unedited, site 4 is almost never edited. Likewise, if site 3 is edited, site 4 is usually edited also. This can be considered a positive cooperativity of editing between sites 3 and 4. Fig. 2 B shows a statistical analysis of the coupling between editing sites in each population (Lange, 2003). The colormap represents Lewontin's D' statistic that ranges between −1 and 1 for sites that show either negative or positive cooperativity, respectively (Lewontin, 1964). At completely independent sites D' = 0. Starred squares show significant coupling between the indicated sites (P < 0.02; Fisher's exact test). An intriguing pattern appears from these data. For all populations, sites 3 and 4 are highly coupled, always in the positive direction. Sites 1 and 3 are also coupled in all tissues, but the sign depends on the tissue: negative cooperativity for eye, antenna, and wing, and positive cooperativity for all other tissues. Moreover, significant coupling occurs among all sites for eye, antenna, and wing, although the sign of D' can be either positive or negative. One of the most surprising observations is the ubiquitous coupling between sites 1 and 3, despite being separated by many thousands of nucleotides in the immature mRNA.
Specificity of ADAR for Particular Editing Sites In Vivo
To seek an explanation for the striking spatial and temporal control of editing seen for Shaker, we examined dADAR's contribution to regulation. The dADAR locus is capable of generating several different isoforms by alternative splicing (Palladino et al., 2000b). dADAR has also been demonstrated to act as a protein dimer on RNA substrates (Gallo et al., 2003). Thus, we reasoned that much of the temporal and spatial regulation of Shaker editing could be attributed to a program of regulated expression and combinatorial action of different dADAR isoforms. To test this hypothesis, we used the pSwitch-GAL4 binary expression system (Roman et al., 2001) to rescue nervous system expression of dADAR in flies genetically deficient for all detectable editing activity of the dADAR locus, including editing of all four Shaker sites studied here (Palladino et al., 2000a, Hoopengardner et al., 2003). We chose one of the most abundant dADAR isoforms produced in adults and constructed transgenic flies expressing its cDNA version, eliminating any possible interaction between alternative ADAR enzymes. Levels of editing were determined for each site individually in transgenic animals from two tissue samples, adult male head and adult male wing, and compared with the levels of editing seen in wild-type controls as well as the isoform expression profile data (Fig. 3). In the adult male head, it is clear that a single isoform of dADAR is capable of editing all four sites in similar ratios as those seen in wild-type controls, albeit at somewhat reduced levels at each site (Fig. 3 A). The site-specific editing levels seen by this method also correlate well with the editing levels determined for each site individually from the profiling data of Fig. 2 A. Contrasting with these data are the levels of editing seen in the adult male wing (Fig. 3 B). Here, site 1 is much more highly edited in the wing than in the adult head, in keeping with control samples and profiling data. We do observe slight differences in the editing levels in rescued animals with respect to controls in both tissues, which we attribute to differences between the artificial expression system (GAL4:UAS) used in these experiments and endogenous dADAR regulation. Nevertheless, there is clear evidence that the efficacy of a single ADAR isoform on particular editing sites can be significantly affected by factors intrinsic to, and predominating in, the cells in which the ADAR is expressed.
Functional Diversity of Shaker Isoforms
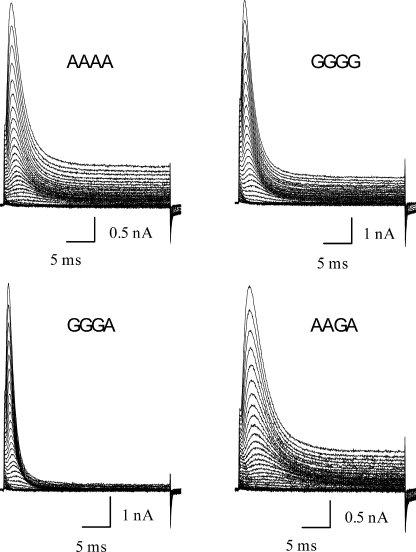
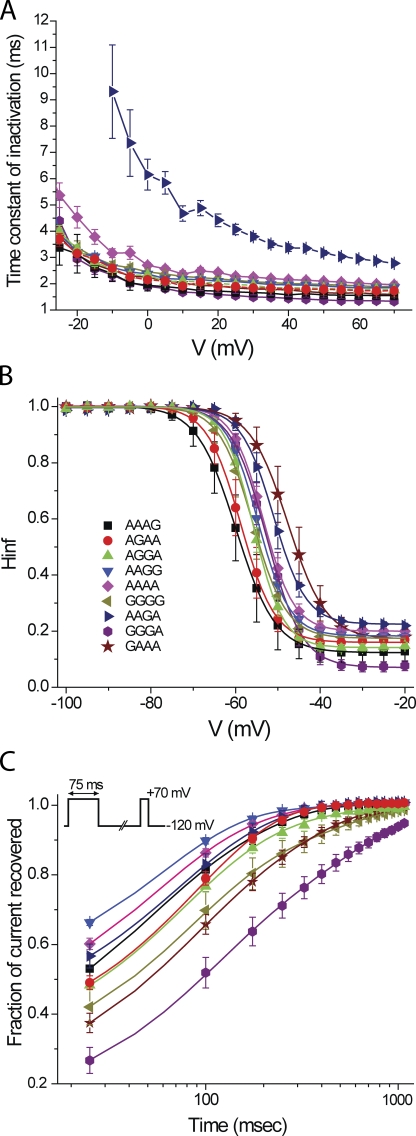
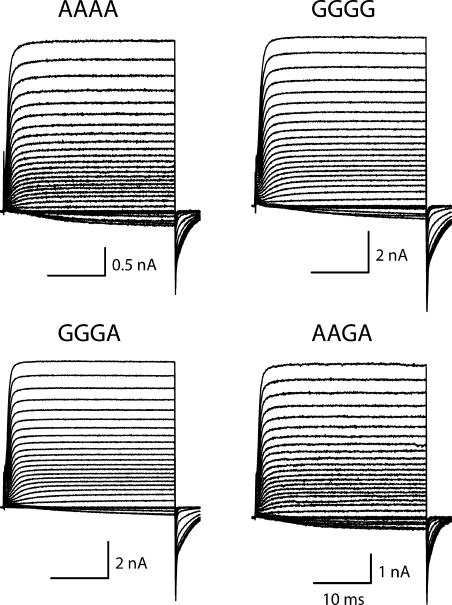
We selected 9 of the 16 possible isoforms for detailed functional characterization. Each was expressed transiently in a mammalian cell line, and whole cell currents were characterized. The channel-forming α-subunits were coexpressed with the auxiliary subunit Hyperkinetic, a cytoplasmic protein that associates with Shaker in Drosophila (Chouinard et al., 1995). Because the expression levels were so high in all of these isoforms, we were able to examine whole cell currents carried by Cs+, which is two orders of magnitude less conductive than K+ (Heginbotham and MacKinnon, 1993). Fig. 4 shows examples of families of currents elicited by depolarizations from a holding potential of −120 up to +70 mV. The top row shows unedited (AAAA) and fully edited (GGGG) isoforms. The bottom row shows the two isoforms with the most extreme functional phenotypes in terms of the kinetics of inactivation. GGGA has the fastest, and AAGA the slowest, rates of inactivation among the nine variants we examined. The rate of inactivation during a depolarization is well fit by a single exponential relaxation. Fig. 5 A shows the time constants for these fits. Although most of these kinetic parameters are comparable among the nine isoforms, AAGA stands out as the slowest over a wide range of voltages, with inactivation time constants approximately threefold slower than those of GGGA. Steady-state inactivation also varied among the nine isoforms, with midpoints differing as much as 11.8 mV (Fig. 5 B and Table S2). The fractional extent of inactivation induced by a 100-ms depolarization to −20 mV (Fig. 5 B) also varied among the isoforms, from 0.92 ± 0.02 for GGGA to 0.78 ± 0.01 for AAGA. This steady-state behavior is consistent with the kinetics of inactivation in that the isoform that inactivates most rapidly also inactivates most completely, and the isoform that inactivates most slowly inactivates least completely. Like entry into the inactivated state, the rate of recovery from inactivation at −120 mV, after a 75-ms depolarization to +70 mV (Fig. 5 C, inset), also had an approximately threefold range, with GGGA the slowest and AAGG the fastest (Table S2). Thus, isoforms that inactivate rapidly tend to recover slowly, and vice versa.
Figure 4.
Whole cell cesium currents of four selected isoforms. Currents were generated from depolarizations between −120 and +70 mV in 5-mV increments. The two isoforms with the fastest and slowest inactivation kinetics are shown in the bottom panels.
Figure 5.
Kinetics and steady-state properties of inactivation. (A) Time constants of inactivation show that AAGA is distinctly slower than the other isoforms. (B) Diversity of steady-state inactivation curves obtained in response to 100-ms prepulses. (C) Kinetics of recovery from inactivation at −120 mV (see inset for voltage protocol). Theory curves are single exponential relaxations (Table S2).
Activation Gating
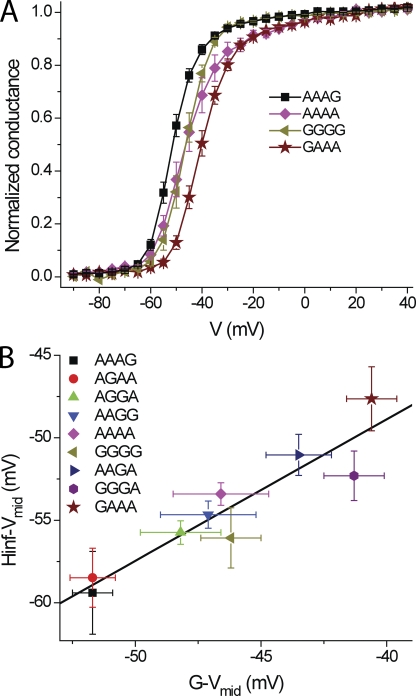
The kinetics and steady-state properties of fast inactivation are coupled to the conformation of the activation gate (Armstrong and Bezanilla, 1974; Bezanilla et al., 1991). Because of this coupling, activation is difficult to characterize in channels that, in response to a depolarization, inactivate on a comparable time scale as they open (e.g., Fig. 4). The differences in fast inactivation among the isoforms could, therefore, be due to differences in activation gating. To test this possibility and analyze the properties of activation gating directly, we examined the same nine isoforms in constructs lacking the N-terminal inactivation ball (Δ6-36) (Hoshi et al., 1990). Fig. 6 shows examples of families of currents, mostly outward currents carried by Cs+, for the same isoforms depicted in Fig. 4. Note the inward current carried by extracellular K+ ions at hyperpolarized voltages and the gating current visible as a shoulder on the rising phase of the activating Cs+ currents. G-V curves were generated from tail currents and had one or two Boltzmann components (Table S3). Fig. 7 plots G-V curves from four isoforms, including the most (GAAA) and least (AAAG) depolarized variants. The midpoints of the major (hyperpolarized) components differed by at most 11 mV among the nine isoforms (Table S3), a similar range as we obtained for steady-state inactivation (Fig. 5 B). The coupling between steady-state activation and inactivation is shown clearly in Fig. 7 B, which plots the midpoints of inactivation against those of activation. The relationship has a slope of 0.86 ± 0.13 with a correlation coefficient of 0.93.
Figure 6.
Cesium currents in selected ball-deleted isoforms. Voltages as in Fig. 4, using the same four isoforms.
Figure 7.
Steady-state activation and inactivation compared. (A) G-V curves for AAAA, GGGG, and the two most shifted isoforms (Table S3). (B) Strong correlation between midpoints of steady-state activation and inactivation.
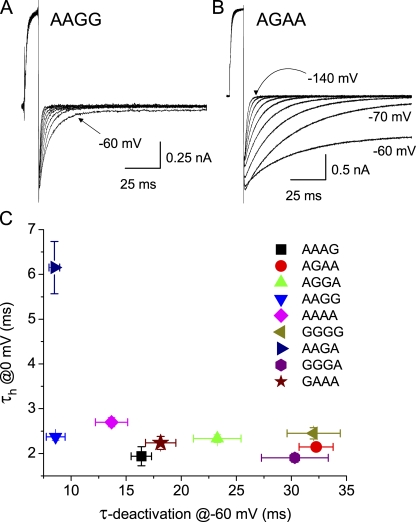
A noticeable difference among the isoforms shown in Fig. 6 is that AAGA has faster kinetics of deactivation at −120 mV than those of the other three isoforms. The rate of deactivation varied widely among the isoforms (Table S4). Fig. 8 (A and B) demonstrates this point with tail currents from the fastest (AAGG) and slowest (AGAA) deactivating isoforms over a voltage range of −60 to −140 mV. Fig. 8 C shows a complete lack of correlation between the kinetics of deactivation and those of inactivation. This suggests that the rate of ball-and-chain inactivation in these isoforms is controlled by factors quite separate from those controlling movement of the activation gate. The kinetics of gate opening were not examined in detail because of the kinetic overlap of gating and ionic current under the conditions of these experiments.
Figure 8.
Kinetics of deactivation and inactivation. (A) Deactivation kinetics in the two most extreme isoforms of inactivation-removed mutants. Tail current kinetics measured over a range from −60 to −140 mV (Table S4). (B) Poor correlation between inactivation and deactivation time constants among the isoforms.
Although the biophysical properties of these nine isoforms manifest the natural functional variability that can be achieved through RNA editing, they also reveal an unprecedented feature, namely a functional epistasis. The isoform AAGA inactivates distinctly slower than any of the others (Figs. 5 A and 8 C). Yet this biophysical phenotype cannot be accounted for by point mutations at any of the four sites. AAGA is edited only at the third site, but the slowing of inactivation occurs only if the other three sites are unedited. For example, the point mutation AAAG-to-AAGG has no effect on inactivation kinetics; nor does AGAA-to-AGGA. Only the point mutation AAAA-to-AAGA produces this slowing. This result unmasks an allosteric communication across disparate regions of the channel protein. Because other biophysical properties of AAGA are unremarkable, it is unlikely that the channel has undergone gross structural changes, leaving the underlying mechanism of this epistasis a matter of speculation. Nevertheless, it suggests that caution is required in interpreting the biophysical consequences of point mutations. A protein's response to mutation at one site may depend in unpredictable ways on seemingly benign or neutral changes at other sites quite removed in primary sequence.
DISCUSSION
RNA modification by ADAR enzymes provides an excellent example of a system for diversifying protein expression from multiple loci, but acting peculiarly, at the behest of neurons. Despite knowledge of editing enzymes and their targets, little information has been gleaned about the regulation of RNA editing in vivo. In mammals, there is clear evidence for spatial and temporal regulation of RNA editing, as well as functional variation between edited and unedited protein isoforms. However, analysis of the regulation of editing in mammals is complicated by the presence of multiple ADAR genes and a paucity of genes targeted for protein recoding. For example, ADAR1 is known to have an RNA editing function in the nervous system, yet ADAR1-deficient mice die early in embryogenesis for reasons that appear to be unrelated to nervous system function (Wang et al., 2000). ADAR1's role in adult editing will need to be addressed in conditional mutants. Animals lacking ADAR2 display profound neuropathological phenotypes, but can be rescued by a copy of the GluR-B subunit pre-edited at the Q/R site, even though substantial residual levels of editing are seen for other known ADAR targets (Higuchi et al., 2000).
In striking contrast with the limitations inherent in mammalian studies, Drosophila melanogaster provides an ideal system for studies of regulation because fruit flies have only one ADAR gene and many (>150) editing sites in nervous system genes. We chose to study RNA editing of the Shaker potassium channel, one of the most well-understood and thoroughly characterized ion channels in biology. Genetic perturbations to Shaker orthologs or their β-subunits have been linked to fly behavioral phenotypes as well as human diseases, such as episodic ataxia, epilepsy, impaired learning, and sleep disorders (Giese et al., 1998; Cirelli et al., 2005; Gasque et al., 2006; Bushey et al., 2007; Douglas et al., 2007; Jen et al., 2007). In addition, Drosophila Shaker has been shown to be subject to posttranscriptional processing by temporally and spatially regulated alternative splicing to generate functionally distinct protein isoforms (Kamb et al., 1987; Hardie et al., 1991; Rogero et al., 1997).
Regulated RNA Editing of Shaker
We show here that Shaker is subject to tightly regulated RNA editing events at four highly conserved sites in two widely separated exons (exon 7 and exon 12) (Fig. 1 A) within the Shaker transcription unit. Comparative genomics reveals conserved RNA editing coupled with ultra-conserved intronic sequences that are the presumptive cis-acting elements proximal to all four editing sites. Computational predictions strongly support three dsRNA structures directed by these elements: e1 pairing with site 1, e3 pairing with site 2, and e2 pairing with sites 3 and 4 (Figs. S1–S3). Corroborative evidence for the independent nature of these structures is seen when addressing the spatial and temporal control of Shaker editing. We observed dramatic developmental regulation for sites 2–4 that reside in exon 12, whereas site 1 in exon 7 is edited at comparable levels at all stages (Fig. 1 C). Spatial regulation suggests an even more fine-grained tuning of this independence (Fig. 2). Male wing tissue, which should contain mostly chemo- and mechanosensory neurons, expresses the GAAA isoform predominantly (68%), whereas this isoform is found at very low levels in the male head (1%). Conversely, the most abundant isoform in heads, AAGG (27%), was not detected in wing tissue. Male antennal tissue also has GAAA as its most abundant isoform, potentially marking this as a Shaker isoform of peripheral sensory neurons. Eye tissue provides an even more stark display of mechanistic independence in editing; the most abundant isoforms here are GAAA (33%) and AGGG (23%). We also observed a near absolute dependence of editing at site 4 on editing at site 3. In 821 cDNAs from our total dataset, 300 were edited at site 4 and 295 of these (98.3%) were also edited at site 3. This dramatic polarized positive cooperativity of editing between two sites may be influenced by the proximity of these sites on the mRNA (Figs. S2 and S3), although the regulatory mechanisms are unknown.
The diverse expression of isoforms seen here could be explained by programmatic expression of different alternative splice forms of the dADAR protein (Palladino et al., 2000b). To test this notion, we genetically eliminated expression of endogenous dADAR and reexpressed, using the pSwitch-GAL4 binary expression system, a single dADAR isoform. All four Shaker editing sites can be edited by this single dADAR isoform in fly brain tissue in a ratio similar to wild-type flies expressing all endogenous dADAR isoforms (Fig. 3). Remarkably, wing tissue expressing a single dADAR isoform also preserves the very skewed pattern of editing seen in the wild-type wing; that is, predominant editing of site 1.
We propose two potential models for how the staging of editing at different sites might be coordinated. In the first, all cells would generate the three predicted structures necessary for editing Shaker sites 1–4 (Figs. S1–S3), but that additional positive or negative factors (we envision RNA-binding proteins) would assist or frustrate ADAR recognition of each RNA structural domain on a site-by-site, cell-specific basis. A second model could invoke RNA chaperones (again, RNA-binding proteins) to act positively or negatively in the formation of the dsRNA structures within each domain. In this model, dADAR passively edits only the Shaker transcripts where appropriate dsRNA structures have formed.
Either of these models could be used to generate the isoform profiles we see. For example, in wing tissue where the GAAA isoform predominates, neurons would express factors that decrease ADAR binding to, or formation of, the structure directing the editing of sites 2–4. In eye tissue, where GAAA and AGGG predominate, two types of neurons could be imagined, a “wing-like” neuron (expressing GAAA) and an additional cell type where factors would eliminate editing at site 1, but promote editing at sites 2–4 (expressing AGGG). Such models do not readily explain the cooperativity of editing seen for the distant sites 1 and 3, positive in some tissues and negative in others. The mechanism of this linkage is speculative but may include a higher-order structure of the dsRNA along with cell-specific factors, bringing these two sites into proximity where ADAR can act cooperatively on them.
Our rescue data also suggest a simple mechanism for the linkage of editing at sites 3 and 4 because clearly one isoform can edit both sites 3 and 4 (Fig. 3 A). The dsRNA structure predicted to form by pairing between e2 and sites 3 and 4 places the editing sites in an imperfect duplex separated by 5 basepairs (Fig. S3). Editing at site 3 would create an A-I mispair, altering the duplex character of this region and changing ADAR binding to the substrate in such as way as to allow site 4 to be modified. Thus, we propose that the “factor” promoting editing at site 4, generating the structure necessary for editing, is the dADAR enzyme itself. Of course, more work at the single-cell level will be necessary to resolve these models. Recently, such single-cell analyses of RNA editing in mammalian cells concluded that, for editing of AMPA and serotonin 2C receptors, additional factors beyond ADAR expression were necessary to explain observed patterns of editing (Sergeeva et al., 2007).
Functional Epistasis
The four conserved editing sites in Shaker are all located in regions associated with channel gating, either on top of the S4 voltage sensor (site 1) or in the transmembrane segment housing the activation gate (sites 2–4). It is not surprising, therefore, that mutations of these residues produce an array of effects on either the voltage dependence or kinetics of gating. The most striking feature of our biophysical interrogation of isoforms is the dramatic slowing of inactivation, seen only in the relatively rare isoform AAGA (Figs. 4, 5 A, and 8 C). Although editing site 3 (residue 489) might be expected to affect ball-and-chain inactivation, because it lies below the cytoplasmic entrance of the open channel where it may encounter either the inactivation ball or its chain (Long et al., 2005), the T489A mutation only has this effect when the other three sites are unedited. We consider this phenotypic interdependence among the sites a type of functional epistasis (Cordell, 2002) that indicates the presence of an allosteric gating network among dispersed residues of the channel protein. Sites 2–4 are in either the helical S6 segment or its cytoplasmic extension, suggesting that mutations may energetically propagate their gating perturbations along the pore-lining S6 segment (Yifrach and MacKinnon, 2002). However, none of these three residues appears to contribute directly to the binding site for the inactivation ball (Zhou et al., 2001). It remains to be seen whether “patterns” of RNA editing, rather than alterations of individual residues, are regulated in specific tissues to produce specific functional consequences. Our results do indicate, nevertheless, that the combinatorics of recoding at multiple sites begets a potpourri of functional phenotypes dependent on context. Moreover, for a multimeric protein like a potassium channel, a further source of diversity is available if heteromers contribute to the functional population of channels in a cell. This will depend on the relative numbers and identities of isoforms expressed in individual cells, a topic that can be addressed by single-cell PCR. Our studies make clear that a thorough appreciation of the organismal significance of editing for ADAR targets will undoubtedly require detailed knowledge of the varied regulatory landscape of editing, even for single gene targets.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants GM079427 (to R. Horn) and Ellison Medical Research Foundation (to R. Reenan).
Edward N. Pugh Jr. served as editor.
L. Ingleby and R. Maloney contributed equally to this work.
Abbreviations used in this paper: ADAR, adenosine deaminase acting on RNA; A-to-I, adenosine-to-inosine; ds, double-stranded; ECS, editing site complementary sequence; RT, reverse transcription.
References
- Armstrong, C.M., and F. Bezanilla. 1974. Charge movement associated with the opening and closing of the activation gates of the Na channels. J. Gen. Physiol. 63:533–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilio, C., A.J. Wahba, P. Lengyel, J.F. Speyer, and S. Ochoa. 1962. Synthetic polynucleotides and the amino acid code. V. Proc. Natl. Acad. Sci. USA. 48:613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. 2002. Annu. Rev. Biochem. 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. How does RNA editing affect dsRNA-mediated gene silencing? 2006. Cold Spring Harb. Symp. Quant. Biol. 71:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, A., and M. Khrestchatisky. 1994. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J. Neurochem. 62:2057–2060. [DOI] [PubMed] [Google Scholar]
- Bezanilla, F., E. Perozo, D.M. Papazian, and E. Stefani. 1991. Molecular basis of gating charge immobilization in Shaker potassium channels. Science. 254:679–683. [DOI] [PubMed] [Google Scholar]
- Bhalla, T., J.J. Rosenthal, M. Holmgren, and R. Reenan. 2004. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 11:950–956. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., H. Chu, S.M. Rueter, L.K. Hutchinson, H. Canton, E. Sanders-Bush, and R.B. Emeson. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 387:303–308. [DOI] [PubMed] [Google Scholar]
- Bushey, D., R. Huber, G. Tononi, and C. Cirelli. 2007. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J. Neurosci. 27:5384–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard, S.W., G.F. Wilson, A.K. Schlimgen, and B. Ganetzky. 1995. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila Hyperkinetic locus. Proc. Natl. Acad. Sci. USA. 92:6763–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli, C., D. Bushey, S. Hill, R. Huber, R. Kreber, B. Ganetzky, and G. Tononi. 2005. Reduced sleep in Drosophila Shaker mutants. Nature. 434:1087–1092. [DOI] [PubMed] [Google Scholar]
- Cordell, H.J. 2002. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 11:2463–2468. [DOI] [PubMed] [Google Scholar]
- Ding, S., and R. Horn. 2003. Effect of S6 tail mutations on charge movement in Shaker potassium channels. Biophys. J. 84:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y., C.Y. Chan, and C.E. Lawrence. 2005. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 11:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y., C.Y. Chan, and C.E. Lawrence. 2006. Clustering of RNA secondary structures with application to messenger RNAs. J. Mol. Biol. 359:554–571. [DOI] [PubMed] [Google Scholar]
- Douglas, C.L., V.V. Vyazovskiy, T.L. Southard, S.Y. Chiu, A. Messing, G. Tononi, and C. Cirelli. 2007. Sleep in Kcna2 knockout mice. BMC Biol. 5:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo, A., L.P. Keegan, G.M. Ring, and M.A. O'Connell. 2003. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22:3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasque, G., P. Labarca, R. Delgado, and A. Darszon. 2006. Bridging behavior and physiology: ion-channel perspective on mushroom body-dependent olfactory learning and memory in Drosophila. J. Cell. Physiol. 209:1046–1053. [DOI] [PubMed] [Google Scholar]
- Giese, K.P., J.F. Storm, D. Reuter, N.B. Fedorov, L.R. Shao, T. Leicher, O. Pongs, and A.J. Silva. 1998. Reduced K+ channel inactivation, spike broadening, and after-hyperpolarization in Kvbeta1.1-deficient mice with impaired learning. Learn. Mem. 5:257–273. [PMC free article] [PubMed] [Google Scholar]
- Hanrahan, C.J., M.J. Palladino, B. Ganetzky, and R.A. Reenan. 2000. RNA editing of the Drosophila para Na(+) channel transcript. Evolutionary conservation and developmental regulation. Genetics. 155:1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie, R.C., D. Voss, O. Pongs, and S.B. Laughlin. 1991. Novel potassium channels encoded by the Shaker locus in Drosophila photoreceptors. Neuron. 6:477–486. [DOI] [PubMed] [Google Scholar]
- Heginbotham, L., and R. MacKinnon. 1993. Conduction properties of the cloned Shaker K+ channel. Biophys. J. 65:2089–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, A. RNA editing, introns and evolution. 1996. Trends Genet. 12:6–9. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., S. Maas, F.N. Single, J. Hartner, A. Rozov, N. Burnashev, D. Feldmeyer, R. Sprengel, and P.H. Seeburg. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 406:78–81. [DOI] [PubMed] [Google Scholar]
- Hille, B. 2001. Ion Channels of Excitable Membranes. Sinauer Associates, Sunderland, MA. 814 pp.
- Hoopengardner, B., T. Bhalla, C. Staber, and R. Reenan. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science. 301:832–836. [DOI] [PubMed] [Google Scholar]
- Hoshi, T., W.N. Zagotta, and R.W. Aldrich. 1990. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 250:533–538. [DOI] [PubMed] [Google Scholar]
- Jen, J.C., T.D. Graves, E.J. Hess, M.G. Hanna, R.C. Griggs, and R.W. Baloh. 2007. Primary episodic ataxias: diagnosis, pathogenesis and treatment. Brain. 130:2484–2493. [DOI] [PubMed] [Google Scholar]
- Kamb, A., L.E. Iverson, and M.A. Tanouye. 1987. Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell. 50:405–413. [DOI] [PubMed] [Google Scholar]
- Keegan, L.P., A. Leroy, D. Sproul, and M.A. O'Connell. 2004. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 5:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan, L.P., J. Brindle, A. Gallo, A. Leroy, R.A. Reenan, and M.A. O'Connell. 2005. Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 24:2183–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, F., C.X. Chen, K.C. Carter, and K. Nishikura. 1997. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 17:2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, K. 2003. Mathematical and Statistical Methods for Genetic Analysis. Springer, NY. 361 pp.
- Lewontin, R.C. 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 49:49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., R.B. Emeson, and C.E. Samuel. 1999. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 274:18351–18358. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., J. Mosbacher, T. Melcher, T. Höger, J.R. Geiger, T. Kuner, H. Monyer, M. Higuchi, A. Bach, and P.H. Seeburg. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 266:1709–1713. [DOI] [PubMed] [Google Scholar]
- Long, S.B., E.B. Campbell, and R. MacKinnon. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. [DOI] [PubMed] [Google Scholar]
- Maas, S., T. Melcher, A. Herb, P.H. Seeburg, W. Keller, S. Krause, M. Higuchi, and M.A. O'Connell. 1996. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 271:12221–12226. [DOI] [PubMed] [Google Scholar]
- Maas, S., Y. Kawahara, K.M. Tamburro, and K. Nishikura. 2006. A-to-I RNA editing and human disease. RNA Biol. 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler, M.F., and J.S. Mattick. 2007. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 87:799–823. [DOI] [PubMed] [Google Scholar]
- Melcher, T., S. Maas, A. Herb, R. Sprengel, P.H. Seeburg, and M. Higuchi. 1996. A mammalian RNA editing enzyme. Nature. 379:460–464. [DOI] [PubMed] [Google Scholar]
- Nishikura, K. Editor meets silencer: crosstalk between RNA editing and RNA interference. 2006. Nat. Rev. Mol. Cell Biol. 7:919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson, J., J.S. Pedersen, D. Haussler, and M. Ohman. 2007. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 13:698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino, M.J., L.P. Keegan, M.A. O'Connell, and R.A. Reenan. 2000. a. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 102:437–449. [DOI] [PubMed] [Google Scholar]
- Palladino, M.J., L.P. Keegan, M.A. O'Connell, and R.A. Reenan. 2000. b. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 6:1004–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, D.E., T. Silva, and F. Bezanilla. 1997. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron. 19:711–722. [DOI] [PubMed] [Google Scholar]
- Paul, M.S., and B.L. Bass. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 17:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupard, M.-C., M.A. O'Connell, A.P. Gerber, and R.S. Zukin. 2000. Patterns of developmental expression of the RNA editing enzyme rADAR2. Neuroscience. 95:869–879. [DOI] [PubMed] [Google Scholar]
- Reenan, R.A., C.J. Hanrahan, and B. Ganetzky. 2000. The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 25:139–149. [DOI] [PubMed] [Google Scholar]
- Rogero, O., B. Hämmerle, and F.J. Tejedor. 1997. Diverse expression and distribution of Shaker potassium channels during the development of the Drosophila nervous system. J. Neurosci. 17:5108–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, G., K. Endo, L. Zong, and R.L. Davis. 2001. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 98:12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, J.J., and F. Bezanilla. 2002. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron. 34:743–757. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., T.R. Dawson, and R.B. Emeson. 1999. Regulation of alternative splicing by RNA editing. Nature. 399:75–80. [DOI] [PubMed] [Google Scholar]
- Ryan, M.Y., R. Maloney, R. Reenan, and R. Horn. 2008. Characterization of five RNA editing sites in Shab potassium channels. Channels. 2-3:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg, P.H., and J. Hartner. 2003. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 13:279–283. [DOI] [PubMed] [Google Scholar]
- Sergeeva, O.A., B.T. Amberger, and H.L. Haas. 2007. Editing of AMPA and serotonin 2C receptors in individual central neurons, controlling wakefulness. Cell. Mol. Neurobiol. 27:669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A., L. Saccomanno, D.P. Morse, T. Brodigan, M. Krause, and B.L. Bass. 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21:6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., J. Khillan, P. Gadue, and K. Nishikura. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 290:1765–1768. [DOI] [PubMed] [Google Scholar]
- Yifrach, O., and R. MacKinnon. 2002. Energetics of pore opening in a voltage-gated K+ channel. Cell. 111:231–239. [DOI] [PubMed] [Google Scholar]
- Zhou, M., J.H. Morais-Cabral, S. Mann, and R. MacKinnon. 2001. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 411:657–661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.