Abstract
The modes of proliferation and differentiation of neural stem cells (NSCs) are coordinately controlled during development, but the underlying mechanisms remain largely unknown. In this study, we show that the protooncoprotein Myc and the tumor suppressor p19ARF regulate both NSC self-renewal and their neuronal and glial fate in a developmental stage–dependent manner. Early-stage NSCs have low p19ARF expression and retain a high self-renewal and neurogenic capacity, whereas late-stage NSCs with higher p19ARF expression possess a lower self-renewal capacity and predominantly generate glia. Overexpression of Myc or inactivation of p19ARF reverts the properties of late-stage NSCs to those of early-stage cells. Conversely, inactivation of Myc or forced p19ARF expression attenuates self-renewal and induces precocious gliogenesis through modulation of the responsiveness to gliogenic signals. These actions of p19ARF in NSCs are mainly mediated by p53. We propose that opposing actions of Myc and the p19ARF–p53 pathway have important functions in coordinated developmental control of self-renewal and cell fate choices in NSCs.
Introduction
Coordinated control of proliferation and differentiation of tissue-specific stem cells is essential for proper morphogenesis of organs and tissues. At early stages of brain development, neuroepithelial progenitors expand by rapid cell divisions and generate a large number and diverse classes of neurons (Caviness et al., 2003). After this early neurogenic period, the growth rate of progenitors gradually decreases, and they begin to preferentially produce glia at late embryonic and early postnatal stages (Sauvageot and Stiles, 2002; Miller and Gauthier, 2007). Recent studies have demonstrated that these developmental changes occur, at least in part, at the level of neural stem cells (NSCs; Martens et al., 2000; Qian et al., 2000; Shen et al., 2006; Fasano et al., 2007). Thus, self-renewal and cell fate choice of NSCs are coordinately controlled in a stage-dependent manner, but the mechanisms underlying such coordination remain poorly understood.
The helix-loop-helix-leucine zipper–type transcription factor family, comprising c-Myc, N-Myc, and L-Myc (herein collectively called Myc), is one of the best-characterized protooncoproteins that plays a vital role in cell cycle control (Grandori et al., 2000). The tumor suppressor p19ARF was originally identified as the protein encoded by an alternative reading frame of the p16INK4a locus (Lowe and Sherr, 2003). Although p16INK4a acts as a cyclin-dependent kinase inhibitor, p19ARF attenuates the cell cycle progression via p53-dependent and -independent pathways (Lowe and Sherr, 2003). In particular, recent studies have shown that p19ARF inhibits Myc activity independently of its action in the p53 pathway (Cleveland and Sherr, 2004; Datta et al., 2004; Qi et al., 2004).
Recent studies have shown that these cell cycle regulators participate in development of multiple organs (for review see Murphy et al., 2005). Brain-specific inactivation of N-Myc and c-Myc in mice has been shown to cause a profound defect in proliferation of granule cell precursors, cells committed to a specific neuronal subtype in the cerebellum (Knoepfler et al., 2002; Hatton et al., 2006; Zindy et al., 2006). Recent studies have also demonstrated the involvement of p19ARF and p53 in the maintenance of NSCs in the adult brain (Bruggeman et al., 2005; Molofsky et al., 2005; Gil-Perotin et al., 2006; Meletis et al., 2006). However, to date, their roles in developmental control of NSCs remain unknown.
In this study, we provide evidence that Myc and the p19ARF–p53 pathway not only regulate NSC self-renewal and proliferation but also control their fate, namely transition from early-stage neurogenesis to late-stage gliogenesis during development. We propose that mutually antagonistic actions of Myc and the p19ARF–p53 pathway are key mechanisms by which the mode of self-renewal and differentiation of NSCs are coordinately controlled in a developmental stage–dependent manner.
Results
Developmental changes in the properties of NSCs
First, we used neurosphere culture to examine developmental changes in the properties of NSCs. In this in vitro assay, cells with a capacity for self-renewal form clonal colonies called neurospheres when seeded at a clonal density (104 cells/ml) in methylcellulose matrix in the presence of FGF2 and EGF (Martens et al., 2000; Nagao et al., 2007). The frequency of primary neurosphere-forming cells represents the content of NSCs in a given tissue, whereas those after subsequent passages reflect the capacity of individual NSCs at a given developmental stage (Martens et al., 2000; Bruggeman et al., 2005; Molofsky et al., 2005).
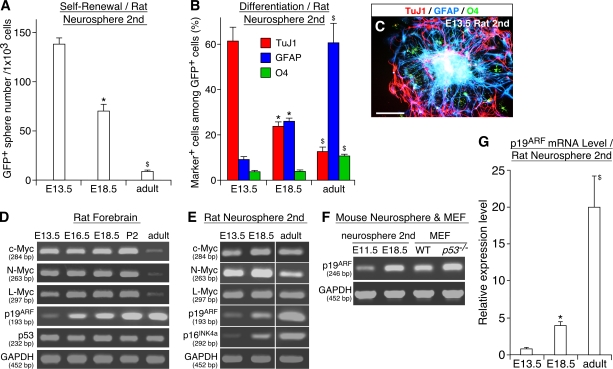
In agreement with previous studies (Martens et al., 2000; Fasano et al., 2007), cells capable of forming clonal neurospheres were detectable in brains at various stages (Fig. 1 A). The vast majority (>95%) of these cells expressed Sox2 and nestin, markers for undifferentiated progenitors, whereas they were negative for markers for more advanced progenitors such as Mash1, Ngn2, and Prox1 (Nagao et al., 2007; unpublished data). Thus, NSCs at different stages, which are selected based on the capacity for self-renewal, share some common properties. Nevertheless, their self-renewing capacity was quantitatively different; cells derived from early-stage (embryonic day [E] 13.5) rat embryos retained a high self-renewal capacity, whereas cells from late-stage (E18.5) forebrains and the subventricular zone of the adult brain showed progressively lower activities. The mode of their differentiation also changed developmentally. The majority (>97%) of neurospheres derived from early-stage (E13.5) embryos retained multipotency (Fig. 1 C; Nagao et al., 2007) and produced many more TuJ1+ neurons than glial fibrillary acidic protein (GFAP+) astrocytes and O4+ oligodendrocytes (Fig. 1 B). In contrast, although late embryonic and adult cells are also multipotent, they preferentially generated astrocytes at the expense of neurons. These changes in vivo can be recapitulated in vitro. When neurospheres were serially passaged at a clonal density, their self-renewal and neurogenic capacities gradually decreased, and conversely, the cells acquired a high gliogenic activity (Fig. S1, C–H, available at http://www.jcb.org/cgi/content/full/jcb.200807130/DC1). These results suggest that the properties of the same progenitor population change over time during development.
Figure 1.
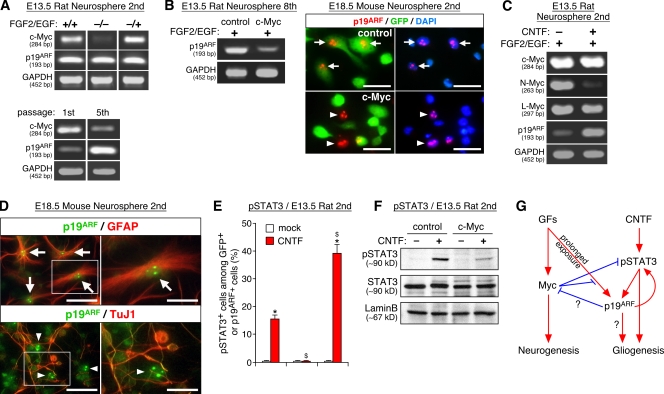
Parallel developmental changes in the properties of NSCs and the expression level of p19ARF. (A–C) Developmental changes in the self-renewal and differentiation potential of NSCs at different stages. NSCs were isolated as neurospheres from rat forebrains at various stages, and their self-renewal activity (A) and differentiation potential (B) were examined. The photograph in C shows clonal neurospheres derived from E13.5 embryos stained for TuJ1 (red), GFAP (blue), and O4 (green). (D–F) The mRNA level of various genes in freshly isolated rat forebrains (D) and neurospheres derived from those tissues (E) is compared by RT-PCR using GAPDH as an internal control. In F, p19ARF expression is compared between E11.5 and E18.5 mouse neurospheres and WT and p53−/− mouse embryo fibroblasts. The sizes of PCR products are shown in base pairs below the gene names. (G) Developmental change in the p19ARF mRNA expression level. *, P < 0.01 compared with values in culture of E13.5 cells; $, P < 0.01 compared with values in culture of E18.5 cells. MEF, mouse embryo fibroblast. Error bars indicate mean + SD. Bar, 100 μm.
Up-regulation of p19ARF in late-stage NSCs
We sought to identify a molecular correlate for this developmental change and found that the level of p19ARF mRNA is gradually up-regulated during development, an ∼20-fold increase in the developing forebrain between E13.5 and postnatal day (P) 2 (Fig. 1 D and Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200807130/DC1). Similarly, the expression of p19ARF was low in neurospheres from early embryos, whereas its level was significantly higher in cells from late-stage and adult brains (4-fold and 20-fold, respectively; Fig. 1, E and G). This up-regulated level of p19ARF was comparable with those detected in p53−/− mouse embryo fibroblasts that have been used widely to study the function of p19ARF (Fig. 1 F and Fig. S2 C). Although similar stage-dependent up-regulation was observed for mRNA for p16INK4a in neurosphere culture, its expression was barely detectable in forebrain tissue in vivo except for very low expression in the adult subventricular zone (Fig. 1 E and not depicted). In contrast, the expression levels of three Myc members and p53 were relatively constant during embryogenesis and in neurosphere culture (Fig. 1, D and E). Like elsewhere in embryos, Myc was highly expressed in proliferating cells in the developing brain (Fig. S1, A–B′). These results demonstrate that the expression level of p19ARF is inversely correlated with the self-renewal and neurogenic activity of NSCs, and its up-regulation parallels gliogenesis at late embryonic stages.
Antagonistic actions of Myc and p19ARF on NSC self-renewal and proliferation
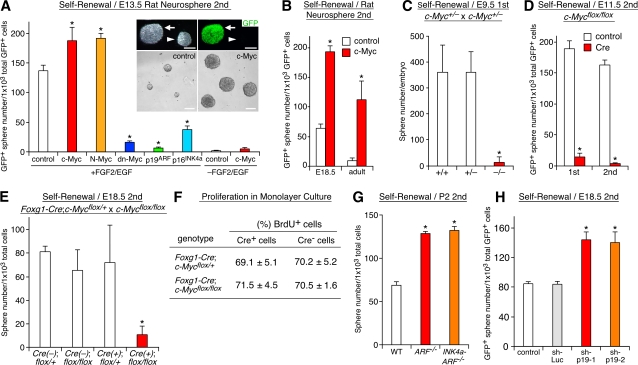
Next, we asked whether Myc and p19ARF regulate NSC self-renewal. We used GFP-expressing retroviruses to modulate their activities in neurosphere culture (Fig. 2 A, inset). GFP+ and GFP− cells in the control virus–infected culture did not show significant difference in terms of the self-renewal or differentiation capacity regardless of the stage of tissue used, indicating that virus-infected cells faithfully recapitulate the properties of the whole cell populations in neurospheres (Nagao et al., 2007; unpublished data). Overexpression of c-Myc and N-Myc significantly enhanced self-renewal of early-stage (E13.5) NSCs, and conversely, a Myc-Mad fusion construct, which acts as a dominant-negative (dn) form of Myc (Berns et al., 1997), markedly attenuated the neurosphere formation (Fig. 2 A). c-Myc also augmented the otherwise very low self-renewal activity of cells derived from late-stage (E18.5) embryos and adults (Fig. 2 B). A similar effect of Myc was observed in repeatedly passaged neurospheres (Fig. S1 C). Conversely, p19ARF and p16INK4a strongly attenuated self-renewal of early-stage cells (Fig. 2 A). No detectable increase in cell death occurred in p19ARF- and p16INK4a-overexpressing culture in the presence of growth factors (Fig. S3 D, available at http://www.jcb.org/cgi/content/full/jcb.200807130/DC1), indicating that the observed decrease was not the result of accelerated cell death.
Figure 2.
Regulation of NSC self-renewal by Myc and p19ARF. (A and B) Effects of retrovirus-mediated overexpression of Myc and p19ARF on self-renewal. The frequency of neurosphere-forming cells among total virus-infected (GFP+) cells is compared using cells derived from different stages. Secondary (2nd) spheres were used for virus infection. Top insets show phase-contrast (left) and fluorescent (right) images of GFP virus–infected (arrows) and uninfected (arrowheads) neurospheres. Bottom insets show larger neurospheres formed by c-Myc–expressing cells (right) compared with those of control cells. (C–E) Effects of inactivation of c-Myc on self-renewal. In C, the frequency of neurosphere-forming cells in the forebrain is compared with c-Myc+/+, c-Myc+/−, and c-Myc−/− mice. In D and E, c-Myc was conditionally inactivated either by infection with Cre viruses in vitro (D) or by crossing with Foxg1-Cre mice using c-Mycflox/flox mice in vivo (E). The formation of neurospheres by cells with different genotypes is compared. (F) Effect of c-Myc inactivation on proliferation in monolayer. Forebrain neuroepithelial cells from Foxg1-Cre;c-Mycflox/+ and Foxg1-Cre;c-Mycflox/flox embryos were cultured in monolayer and labeled with BrdU for 48 h in the presence of FGF2 and EGF. The percentage of BrdU+ cells among Cre+ (c-Myc inactivated) and Cre− (WT) cells was quantified. (G and H) Effects of inactivation in vivo (G) and acute down-regulation in vitro (H) of p19ARF on self-renewal. In G, WT, p19ARF−/−, and p19ARF−/−;p16INK4a−/− mice are compared. In H, short hairpin–p19-1 and -2 are retroviruses expressing shRNAs for p19ARF, whereas short hairpin–Luc expresses shRNA for luciferase. *, P < 0.01 compared with control virus–infected culture (A, B, D, and H) or c-Myc+/+ (C), Foxg1-Cre;c-Mycflox/+ (E and F), and WT (G) mice. Error bars indicate mean + SD. Bars, 100 μm.
We further examined the roles for Myc and p19ARF using several mutant mice. Only 3.2 ± 5.3% (n = 3; P < 0.01) of sphere-forming stem cells were detected in the forebrain of c-Myc−/− mice at E9.5 compared with the wild-type (WT) and heterozygous littermates (Fig. 2 C). Such a severe reduction in the content of NSCs was much more profound than the decrease in the total number of cells in the mutant brain (∼50% of the WT level). Furthermore, we used mice carrying a conditional allele of c-Myc (c-Mycflox/flox) for brain-specific c-Myc deletion (Trumpp et al., 2001). Acute inactivation in vitro by infection with Cre-expressing viruses markedly decreased the self-renewal activity of c-Mycflox/flox cells (Fig. 2 D). Similarly, forebrain-specific inactivation in vivo by crossing c-Mycflox/flox and Foxg1-Cre (Hebert and McConnell, 2000) mice caused a severe reduction of NSCs (Fig. 2 E), demonstrating a cell-autonomous requirement for c-Myc in NSCs.
p19ARF had an opposite effect in late-stage cells. The low neurosphere-forming capacity of cells at P2 was markedly augmented by inactivation of p19ARF (Fig. 2 G; Kamijo et al., 1997). Acute down-regulation of p19ARF by short hairpin RNAs (shRNAs; short hairpin–p19-1 and -2; Sage et al., 2003) also stimulated self-renewal, indicating the direct action of p19ARF on NSCs (Fig. 2 H). However, inactivation of p16INK4a together with p19ARF (Serrano et al., 1996) showed no additional effect compared with p19ARF single deletion (Fig. 2 G). Previous studies have shown the involvement of p19ARF in the maintenance of NSCs in the postnatal and adult brain (Bruggeman et al., 2005; Molofsky et al., 2005). Our data further demonstrate its important role in NSC self-renewal during development.
The formation of neurospheres involves both proliferation and self-renewal. Thus, the observed effect of Myc inactivation could be a secondary consequence of a proliferation defect. To test this possibility, forebrains were isolated from embryos obtained by crossing c-Mycflox/flox with Foxg1-Cre mice, and the cells were cultured in monolayer at a high cell density, the condition in which NSCs and other progenitors can grow in both self-renewing and non–self-renewing modes of divisions. Cell proliferation was evaluated by labeling with BrdU for 48 h. No significant difference was observed between Cre+ (c-Myc−/−) and Cre− (c-Mycf/f) cells (Fig. 2 F), demonstrating that c-Myc inactivation does not result in a general proliferation detect.
We also quantified the overall growth rate of neurospheres and the size of individual spheres. Myc-expressing cells grew faster than control cells and formed larger clonal colonies, whereas p19ARF had an opposite effect (Fig. S3, A–C and not depicted). However, neither Myc nor p19ARF increased cell death in the presence of growth factors (Fig. S3 D). Thus, Myc and p19ARF stimulated and attenuated, respectively, proliferation of NSCs. However, Myc increased not only the proliferation rate but also the frequency of cells capable of forming self-renewing colonies as described above (Fig. 2, A and B). Moreover, although p19ARF-overexpressing cells could not form self-renewing colonies at a clonal density, they could grow as cell aggregates when cultured at a nonclonal high density (105 cells/ml) without methylcellulose matrix. Therefore, simple acceleration or attenuation of the growth rate does not account for the observed effects of Myc and p19ARF in the neurosphere formation assay. It is also unlikely that the action of Myc is attributable to immortalization. c-Myc–expressing cells formed neurospheres only in the presence of growth factors (Fig. 2 A). Moreover, transient (48 h) activation of Myc was sufficient to augment the self-renewing activity (Fig. S3 E). Altogether, these results demonstrate that Myc and p19ARF regulate not only proliferation but also self-renewal of NSCs.
Regulation of NSC differentiation by Myc and p19ARF
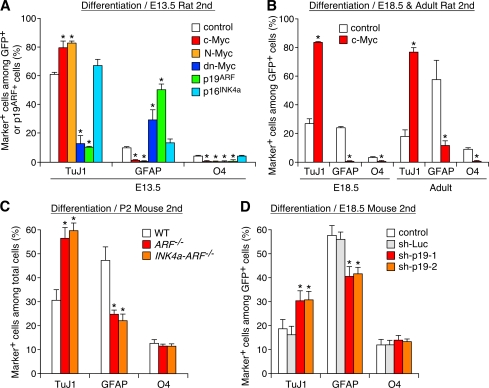
Importantly, Myc and p19ARF also regulated the differentiation potential of NSCs. In culture of early-stage cells, c-Myc and N-Myc significantly augmented differentiation of neurons at the expense of astrocytes and oligodendrocytes, whereas dn-Myc suppressed neurogenesis and stimulated gliogenesis (Fig. 3 A). Moreover, the majority (89.3 ± 5.3%; n = 3) of Myc-expressing neurospheres maintained a high neurogenic capacity after repetitive passages (Fig. S1, D and F–H). Myc also conferred a high neurogenic activity to late-stage and adult NSCs (Fig. 3 B). Conversely, overexpression of p19ARF attenuated neurogenesis and promoted astrogenesis in early-stage cells (Fig. 3 A), whereas inactivation of p19ARF in vivo and acute down-regulation of p19ARF by shRNAs in vitro had opposite effects in late-stage cells (Fig. 3, C and D). Importantly, overexpression of p16INK4a or its inactivation together with p19ARF had no significant effect (Fig. 3, A and C), indicating that the action of p19ARF on differentiation is not a mere consequence of attenuated self-renewal or proliferation.
Figure 3.
Regulation of neurogenesis and gliogenesis by Myc and p19ARF. (A and B) Effects of Myc and p19ARF on differentiation of NSCs. The percentages of neurons and glia among total virus-infected cells are compared. (C and D) Effects of inactivation in vivo (C) and acute down-regulation in vitro (D) of p19ARF on neurogenesis and gliogenesis. *, P < 0.01 compared with control virus–infected culture (A, B, and D) or WT mice (C). Error bars indicate mean + SD.
Myc and p19ARF modulate the response to gliogenic signals
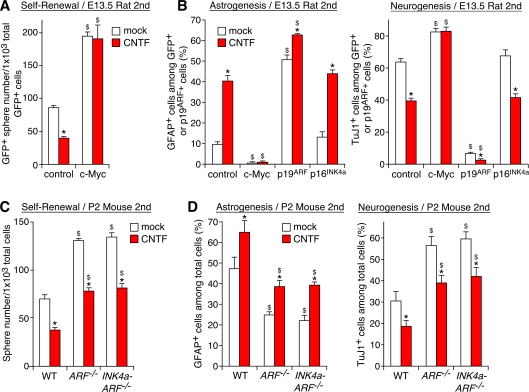
Cytokines of the interleukin-6 family, including ciliary neurotrophic factor (CNTF), act as signals to induce astrocytes late in development (Barnabe-Heider et al., 2005). We asked whether Myc and p19ARF modulate the responsiveness to these gliogenic signals. Myc blocked the action of CNTF to inhibit self-renewal in early-stage cells (Fig. 4 A). Myc also suppressed CNTF-dependent stimulation of astrogenesis and inhibition of neurogenesis (Fig. 4 B). Conversely, overexpression of p19ARF mimicked the action of CNTF, and their combination further shifted the cells toward gliogenesis (Fig. 4 B).
Figure 4.
Modulation of CNTF actions by Myc and p19ARF. (A) Block of CNTF-dependent inhibition of self-renewal by Myc. Control and c-Myc–expressing cells were subjected to the neurosphere formation assay in the presence of CNTF. (B) Effects of c-Myc, p19ARF, and p16INK4a on CNTF-dependent stimulation of astrogenesis (left) and inhibition of neurogenesis (right). Virus-infected cells were induced to differentiate in the presence of CNTF. (C and D) Effects of inactivation of p19ARF and p16INK4a on CNTF-dependent regulation of self-renewal (C) and differentiation (D). *, P < 0.01 compared with mock-treated culture; $, P < 0.01 compared with control virus–infected culture (A and B) or WT mice (C and D). Error bars indicate mean + SD.
We also examined the interactions between CNTF signals and p19ARF in late-stage cells with high p19ARF expression. CNTF inhibited the already low self-renewal activity of cells derived from P2 mouse forebrains (a 46% reduction compared with control cells; Fig. 4 C). However, inactivation of p19ARF significantly attenuated this action. Similarly, CNTF-dependent astrogenesis was significantly attenuated by inactivation of p19ARF (Fig. 4 D). Consequently, although p19ARF−/− cells still responded to CNTF, the self-renewal and gliogenic activity of CNTF-treated p19ARF−/− cells recovered to the level in the untreated WT cells. In contrast, overexpression or inactivation of p16INK4a did not alter the response to CNTF (Fig. 4, B–D). These results demonstrate that Myc and p19ARF modulate the responsiveness to gliogenic signals.
Regulation of Myc and p19ARF expression by growth factors and gliogenic signals
Next, we asked how the expression of Myc and p19ARF is controlled by extracellular signals. Like in many other cell types, c-Myc expression was maintained high in proliferating NSCs (Fig. 5 A, top, +/+), and its level was rapidly down-regulated and up-regulated upon removal (Fig. 5 A, top, −/−) and reexposure (Fig. 5 A, top, −/+), respectively, of growth factors. In contrast, p19ARF expression did not show such a rapid response to growth factors. However, prolonged exposure to growth factors during multiple passages led to marked up-regulation of p19ARF and down-regulation of c-Myc (Fig. 5 A, bottom; and Fig. S2 D). Thus, the expression of c-Myc and p19ARF are regulated by growth factor signals either directly or indirectly (Fig. 5 G). Interestingly, sustained high Myc expression blocked up-regulation of p19ARF during serial passages (Fig. 5 B, left; and Fig. S2 E). c-Myc overexpression in late-stage cells also resulted in acute (<48 h) down-regulation of p19ARF, indicating that the observed suppression was not the result of a genetic lesion of p19ARF (Fig. 5 B, right). Such Myc-dependent inhibition of p19ARF in NSCs is in sharp contrast to oncogenic signal-induced up-regulation of p19ARF during tumorigenesis and cell death in other cell types (Zindy et al., 1998).
Figure 5.
Regulation of Myc and p19ARF expression by growth factors and gliogenic signals. (A) Growth factor–dependent regulation of Myc and p19ARF expression. (top) Neurospheres expanded in the presence of FGF2 and EGF for 3 d (+/+) were starved for 6 h and subsequently reexposed to growth factors for an additional 2 h (−/+) or left unexposed (−/−). (bottom) Neurospheres were serially passaged, and the level of c-Myc and p19ARF mRNAs was compared with the first and fifth spheres. (B) Suppression of p19ARF expression by Myc. The cells examined are control and c-Myc virus–infected neurospheres derived from E13.5 (eighth passage; left) and E18.5 (second passage; right). p19ARF was detected in DAPI+ cell nuclei of control (arrows) but not c-Myc (arrowheads) cells. (C) Regulation of p19ARF and N-Myc by CNTF. Neurospheres were grown in the presence (+) or absence (−) of CNTF for 7 d. (D) Expression of p19ARF in astrocytes. Photographs show the localization of p19ARF in the nucleoli of astrocytes (top, arrows) but not of neurons (bottom, arrowheads). The panels on the right show higher magnification views of the areas indicated by boxes on the left. (E and F) Effects of Myc and p19ARF on CNTF-dependent STAT3 phosphorylation at Y705 (pSTAT3). (G) Model for the relationships of Myc and p19ARF with extracellular signals and neuro/gliogenesis. Arrows and blunt-ended lines indicate stimulation and inhibition, respectively, either at the level of expression or function. The sizes of PCR products are shown in base pairs below the gene names. *, P < 0.01 compared with mock-treated culture; $, P < 0.01 compared with control virus–infected culture. Error bars indicate mean + SD. Bars: (B) 25 μm; (D) 40 μm.
The gliogenic signal CNTF also regulated the expression of Myc and p19ARF. Continuous stimulation by CNTF resulted in marked up-regulation of p19ARF and down-regulation of N-Myc (Fig. 5 C and Fig. S2 F). Thus, CNTF shifts the balance between Myc and p19ARF toward a p19ARF-dominant state during the clonal growth of NSCs. Upon induction of differentiation, p19ARF expression remained in GFAP+ astrocytes but not TuJ1+ neurons, which is consistent with its strong astrogenic action (Fig. 5 D). Accumulation of p19ARF in the nucleolus was observed in these astrocytes as reported for other cell types (Qi et al., 2004). These results demonstrate that Myc and p19ARF are under the control of both growth factor and gliogenic signals, which in turn act as cell-intrinsic modulators of the responsiveness of these extracellular signals. Moreover, given the previous reports that p19ARF can inhibit the activity of Myc (Datta et al., 2004; Qi et al., 2004), the mutual antagonism between Myc and p19ARF could be explained, at least in part, by their cross-regulation at the level of expression and activity (Fig. 5 G).
Myc and p19ARF regulate STAT3 phosphorylation downstream of gliogenic signals
We further examined how Myc and p19ARF regulate differentiation of NSCs. Phosphorylation of signal transducer and activator transcription factor 3 (STAT3) is a crucial step in signal transduction downstream of CNTF (Miller and Gauthier, 2007). We found that a high Myc activity decreased STAT3 phosphorylated at Y705 in response to CNTF, whereas p19ARF overexpression had an opposite effect (Fig. 5, E and F). Such changes were observed 48 h after infection with Myc and p19ARF viruses with no significant change in the level of total STAT3 proteins, indicating that Myc and p19ARF antagonize each other at the level or upstream of STAT3 phosphorylation. These results demonstrate that Myc and p19ARF regulate gliogenesis, at least in part, through modulation of STAT3 signaling.
p53 acts downstream of p19ARF
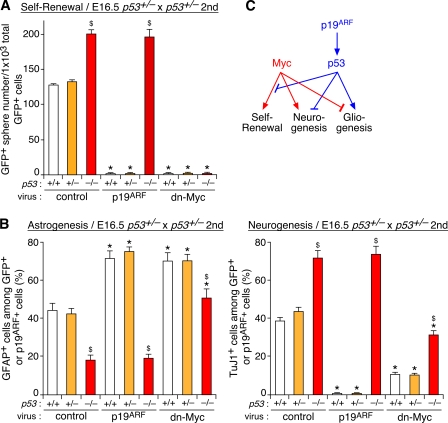
In addition to the direct regulation of Myc, p19ARF exerts its action through p53 in many cell types (Lowe and Sherr, 2003). Thus, we next examined the role of p53 using p53−/− mice (Guo et al., 2003). NSCs derived from late-stage p53−/− mutants showed a higher self-renewal capacity (Fig. 6 A) and produced fewer astrocytes and more neurons (Fig. 6 B) compared with the WT and heterozygous cells. These phenotypes were reminiscent of those of p19ARF−/− cells. Importantly, p19ARF did not attenuate self-renewal or stimulate astrogenesis in p53−/− cells, indicating the essential role of p53 downstream of p19ARF (Fig. 6, A and B).
Figure 6.
Involvement of p53 in Myc- and p19ARF-dependent regulation of NSCs. (A and B) Forebrain neurospheres derived from p53+/+, p53+/−, and p53−/− embryos were infected with control, p19ARF, and dn-Myc viruses and were subsequently subjected to the self-renewal (A) and differentiation (B) assays as described in Figs. 2 and 3. (C) Model for the relationships between Myc, p19ARF, and p53. *, P < 0.01 compared with control virus–infected culture with the same genotypes; $, P < 0.01 compared with WT (+/+) cells infected with the same viruses. Error bars indicate mean + SD.
Next, we examined genetic epistasis between Myc and the p19ARF–p53 pathway. We first tested whether the early lethal phenotype of c-Myc mutants (Davis et al., 1993) can be rescued by concomitant inactivation of p19ARF or p53. However, we could not obtain embryos with a c-Myc−/−;p19ARF−/− or c-Myc−/−;p53−/− genotype that survived beyond E11.5 (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200807130/DC1), probably because of developmental defects in multiple organs other than the brain.
We then used neurosphere culture of late-stage cells that expressed both Myc and p19ARF at substantial levels. As shown in Fig. 6 A, dn-Myc strongly attenuated the neurosphere formation, and its action could not be abrogated by inactivation of p53, suggesting that p53 modulates self-renewal through inhibition of Myc activity or its downstream events. In contrast, both inhibition of neurogenesis and stimulation of astrogenesis by dn-Myc were reversed by p53 inactivation, and conversely, altered cell fates in p53−/− cells were rescued by dn-Myc (Fig. 6 B). Thus, Myc affected the fate choice of NSCs in the absence of p53, and p53 modulated differentiation independently of Myc. These results suggest that Myc and the p19ARF–p53 pathway control neurogenesis and gliogenesis through parallel pathways unlike their hierarchical or linear relationship in regulating self-renewal (Fig. 6 C).
Early neurogenic defect in c-Myc−/− mice
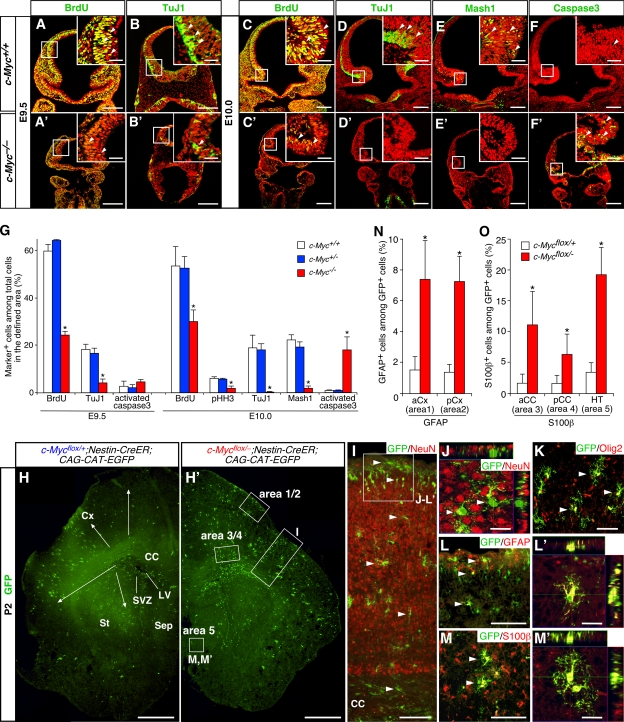
The aforementioned in vitro data suggest that Myc controls NSC fate in a stage-dependent manner. To obtain in vivo evidence supporting this idea, we first examined the consequence of c-Myc inactivation at early stages using c-Myc−/− mice. c-Myc−/− embryos die around E10.5 (Davis et al., 1993), so their phenotypes were examined at E9.5 and E10.0. The mutant had a smaller forebrain and contained a much smaller number of mitotic cells labeled with BrdU and phosphorylated histone H3 compared with the WT (Fig. 7, A, A′, C, C′, and G). TuJ1+ neurons and Mash1+ progenitors were also reduced in the mutant forebrain (Fig. 7, B, B′, D–E′, and G). Although a significant increase of activated caspase3+ dying cells was observed at E10.0 (Fig. 7, F and F′), a substantial decrease of BrdU+ proliferative cells and TuJ1+ neurons was already evident at E9.5, the stage before aberrant cell death began (Fig. 7 G). Given the cell-autonomous effect of c-Myc in NSCs in vitro, a deficiency of NSCs is likely to be part of the reason for the observed defects in proliferation and neurogenesis in c-Myc−/− forebrains.
Figure 7.
Impaired neurogenesis and gliogenesis caused by inactivation of c-Myc in vivo. (A–G) Defects in proliferation and neurogenesis in forebrains of c-Myc−/− mice at E9.5 and E10.0. In A–F′, stained markers are shown in green, and cell nuclei were visualized with PI in red. Insets show higher magnification views of boxed areas. BrdU was administered to pregnant dams 30 min before sampling. G shows a comparison of the percentages of marker-positive cells (arrowheads) among total cells in the areas boxed in A–F′ between c-Myc+/+, c-Myc+/−, and c-Myc−/− embryos. *, P < 0.01 compared with c-Myc+/+ mice. (H–O) Accelerated differentiation of astrocytes by conditional inactivation of c-Myc at late embryonic stages. Tamoxifen was administered to pregnant dams at E16.5 or E17.5, and pups were analyzed at P2 or 3. c-Myc–inactivated cells were visualized as GFP+ cells using CAG-CAT-EGFP Cre reporter. H and H′ show the distribution pattern of GFP+ cells in coronal sections of the forebrain. The arrows indicate migration of GFP+ cells from the subventricular zone (SVZ) toward the outer brain parenchyma. I–M′ show the phenotypes of GFP+ cells (arrowheads) in the neocortex (I–L′) and hypothalamus (M and M′) of c-Mycflox/+;Nestin-CreER;CAG-CAT-EGFP mice. The position of the areas shown in J–L′ is indicated by a box in I. N and O show the comparison of the percentages of GFAP+ and S100β+ cells among GFP+ cells with c-Mycflox/+ and c-Mycflox/− genotypes in the regions indicated by boxes in H′. *, P < 0.01 compared with c-Mycflox/+;Nestin-CreER cells. aCC, anterior corpus callosum; pCC, posterior corpus callosum; aCx, anterior neocortex; pCx, posterior neocortex; CC, corpus callosum; Cx, neocortex; HT, hypothalamus; LV, lateral ventricle; Sep, septum; St, striatum. Error bars indicate mean + SD. Bars: (A–F′) 200 μm; (A–F′, insets) 40 μm; (H and H′) 500 μm; (I, L, and M) 100 μm; (J, L′, and M′) 20 μm; (K) 50 μm.
Conditional inactivation of c-Myc late in development
Next, we examined the effect of late-stage inactivation of c-Myc on gliogenesis. We generated c-Mycflox/flox;Nestin-CreER mice in which c-Myc can be inactivated at a specific developmental stage in a tamoxifen-inducible manner (Burns et al., 2007). We combined CAG-CAT-EGFP Cre reporter (Nakamura et al., 2006) to fate map c-Myc–inactivated cells as GFP+ cells. We first set out the experimental paradigm to study the transition from neurogenesis to gliogenesis. When tamoxifen was administered to the control (c-Mycflox/+;Nestin-CreER;CAG-CAT-EGFP) mice at E15.5, the majority (50–60%) of GFP+ cells became NeuN+ neurons at P3 (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200807130/DC1). In contrast, in animals treated with tamoxifen at E16.5 or 17.5, the fraction of neurons among GFP+ cells was much smaller (7–9% at E16.5 and 0.5–1.0% at E17.5 in the dorsolateral neocortex; Fig. 7, I and J). These late-stage labeled cells were distributed throughout the brain parenchyma (Fig. 7, H and H′). In particular, GFP+ cells located in areas near the lateral ventricle expressed Olig2, a marker for postnatal glial progenitors (Fig. 7 K). It has been shown that these Olig2+ cells migrate to the forming gray and white matters and subsequently differentiate into astrocytes and oligodendrocytes postnatally (Marshall et al., 2005). Thus, tamoxifen treatment at E16.5 and 17.5 allows us to detect the early phase of gliogenesis in vivo.
To compare the timing of astrocyte differentiation between c-Myc+/− and c-Myc−/− cells, we analyzed brains of tamoxifen-treated animals at P2 and 3, the stage when the expression of the astrocyte markers GFAP and S100β begin to be detectable in many areas of the forebrain. At this early postnatal stage, only a small fraction of c-Myc heterozygous (+/−) GFP+ cells expressed GFAP and S100β (Fig. 7, N and O). These cells were confined to specific regions such as the outer margin of the neocortex (Fig. 7, L and L′, arrowheads; and Fig. S5, C′ and D′), the hypothalamus (Fig. 7, M and M′, arrowheads), and the corpus callosum. In contrast, a significantly larger percentage of c-Myc–inactivated (−/−) cells became astrocytes in corresponding regions (Fig. 7, N and O). We observed no noticeable difference in the rate of proliferation, death, or neuronal and oligodendroglial differentiation between c-Myc+/− and c-Myc−/− GFP+ cells (unpublished data), arguing that c-Myc inactivation selectively affects astrocytes. These results demonstrate that conditional inactivation of c-Myc at a late embryonic stage causes precocious differentiation of astrocytes.
Delayed astrogenesis in p19ARF mutant brains
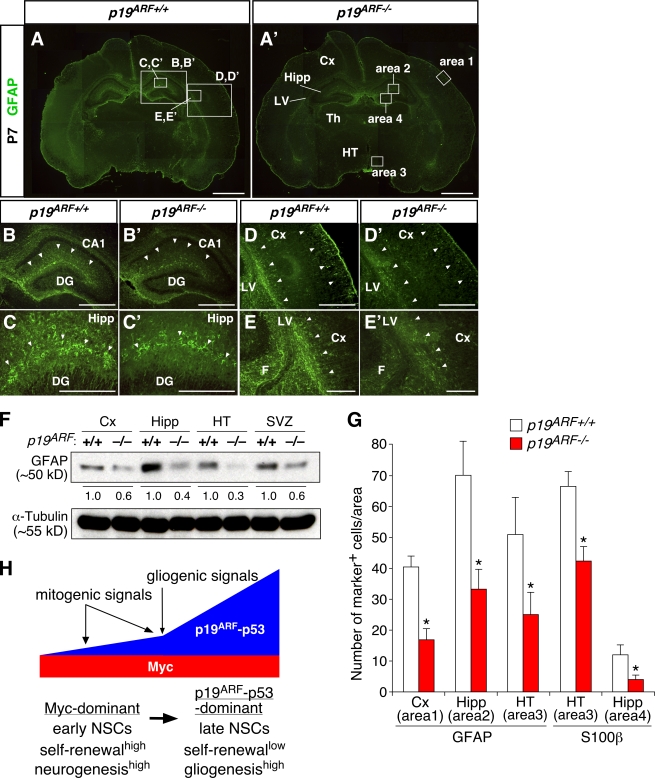
Our in vitro data suggest that p19ARF promotes gliogenesis at postnatal stages. We examined whether inactivation of p19ARF results in delayed gliogenesis. In the WT, differentiation of GFAP+ and S100β+ astrocytes was detectable in widespread regions of the brain at P7. We observed much weaker expression of GFAP and S100β in comparable regions of p19ARF mutants in both immunostaining (Fig. 8, A–E′) and Western blotting (Fig. 8 F). Quantitative analyses demonstrate that the number of astrocytes significantly decreased in many regions of the mutant brain (Fig. 8 G). In contrast, the total cell number in the areas examined was indistinguishable between WT and mutant brains. No noticeable difference in the distribution pattern or density of neurons or dying cells was observed either (unpublished data). Thus, inactivation of p19ARF resulted in attenuated astrocyte differentiation in the postnatal brain as opposed to c-Myc inactivation.
Figure 8.
Attenuated astrocyte differentiation in early postnatal brains of p19ARF mutants. (A–E′) GFAP staining of the WT (p19ARF+/+) and p19ARF−/− brains at P7. B–E′ show higher magnification views of the areas indicated by boxes in A. Differential staining patterns between the WT and mutant are highlighted by arrowheads. (F) Expression level of GFAP in various regions of p19ARF+/+ and p19ARF−/− mice. The level of GFAP proteins in the mutant relative to the WT (designated as 1.0) is indicated. (G) Comparison of the numbers of GFAP+ and S100β+ astrocytes in specific regions of the brain. The locations of areas 1–4 are indicated by boxes in A′. (H) Model for the actions of Myc and the p19ARF–p53 pathway in developmental control of NSCs. Arrows at the top indicate stimulation, and the thick horizontal arrow indicates transition from early- to late-stage cells. *, P < 0.01 compared with the WT. CA1, hippocampal CA1 sector; Cx, neocortex; DG, dentate gyrus; F, fimbria; Hipp, hippocampus; HT, hypothalamus; LV, lateral ventricle; Th, thalamus; SVZ, subventricular zone. Error bars indicate mean + SD. Bars: (A and A′) 1 mm; (B, B′, D, and D′) 500 μm; (C, C′, E, and E′) 200 μm.
Discussion
Coordinated control of NSC self-renewal and differentiation by Myc and p19ARF
Myc and p19ARF have been extensively studied in the context of tumorigenesis, cell death, and senescence of adult tissues and organs (Grandori et al., 2000; Lowe and Sherr, 2003). Recent studies have also begun to reveal their vital roles in normal development (Suzuki et al., 2008; for review see Murphy et al., 2005). In this study, we have demonstrated that Myc and p19ARF exert opposing actions in NSCs during brain development. Given their established roles in cell cycle control, their actions on self-renewal may not be an unexpected result. However, a novel finding in this study is that these molecules also have profound effects on neuronal and glial differentiation. At early embryonic stages, p19ARF expression remains low, whereas Myc expression is maintained high. Our data have shown that this Myc-dominant status links a high self-renewal and neurogenesis capacity in NSCs (Fig. 8 H). As development proceeds, sustained mitogenic signals up-regulate p19ARF, which in turn attenuates self-renewal and neurogenesis. Gliogenic signals further up-regulate p19ARF, thereby promoting the transition from neurogenesis to gliogenesis. This p19ARF-dominant status appears to couple a low self-renewal and a high gliogenic capacity in late-stage NSCs. Thus, we suggest that opposing actions of Myc and p19ARF coordinate the extent of self-renew and the timing of the production of neurons and glia during development.
Antagonistic actions of Myc and the p19ARF–p53 pathway in NSCs
Nervous system–specific inactivation of N-Myc and c-Myc has been shown to cause a defect in proliferation of cerebellar granule cell precursors (Knoepfler et al., 2002; Zindy et al., 2006). Thus, it has been proposed that Myc stimulates proliferation and inhibits differentiation in this specific progenitor subtype that is already committed to a neuronal lineage (Knoepfler et al., 2002). However, we found that Myc not only promotes self-renewal and proliferation but also neurogenesis in multipotent NSCs in the forebrain. Thus, Myc appears to play different roles in distinct progenitor subtypes.
Like in well-studied cell cycle control, p19ARF exerts actions opposite to Myc in NSCs. p19ARF has been shown to inhibit Myc activity independently of p53 (Cleveland and Sherr, 2004; Datta et al., 2004; Qi et al., 2004). However, our data suggest that the action of p19ARF is mainly mediated by p53 in NSCs. Thus, it remains unclear whether direct inhibition of Myc by p19ARF occurs in NSCs. However, we found that Myc suppresses the growth signal–dependent expression of p19ARF. Moreover, they regulated the responsiveness to CNTF at the level of STAT3 phosphorylation in an opposite manner, suggesting reciprocal interactions between Myc and p19ARF at the level of their expression and/or function (Fig. 5 G).
Interestingly, the mode of their antagonism appears to be distinct in controlling self-renewal and differentiation. Our data suggest that the p19ARF–p53 pathway attenuates self-renewal through inhibition of Myc activity or its downstream events similarly to cell cycle control (Seoane et al., 2002; Ho et al., 2005). In contrast, Myc and the p19ARF–p53 pathway appear to regulate differentiation at least in part through independent pathways. Our data have shown that the regulation of STAT3 phosphorylation is part of this mechanism, but other mechanisms are also likely to be involved. Recent studies have demonstrated the role for chromatin remodeling in the temporal control of gliogenesis (Song and Ghosh, 2004; Fan et al., 2005). Myc has been shown to regulate gene expression through chromatin remodeling (Knoepfler et al., 2006). Whether specific epigenetic mechanisms underlie Myc- and p19ARF-dependent control of NSC fate remains to be investigated.
Recent studies have demonstrated important roles of many cell cycle regulators in stem cell self-renewal in adult organs (for review see Bruggeman and van Lohuizen, 2006). For instance, p16INK4a and the polycomb group gene product Bmi-1 have been shown to be involved in the maintenance of NSCs in the adult brain (Bruggeman et al., 2005; Molofsky et al., 2005, 2006). Compared with these molecules, Myc and p19ARF are unique in that it also regulates neuronal and glial fate choices. Interestingly, the expression of p16INK4a was barely detectable in the developing forebrain unlike in the adult brain, whereas robust up-regulation of p19ARF was observed during embryogenesis. Thus, these two closely related cell cycle regulators appear to play distinct roles in NSCs at different stages; i.e., during development and in adults.
Myc and p19ARF in temporal switch from neurogenesis to gliogenesis
Previous studies have shown that cell-intrinsic mechanisms regulate the transition from neurogenesis to gliogenesis by modulating the responsiveness to extracellular signals (Qian et al., 2000; Shen et al., 2006). Our data suggest that Myc and the p19ARF–p53 pathway are integral parts of such cell-intrinsic mechanisms. Myc attenuates the response to gliogenic signals by inhibiting up-regulation of p19ARF and blocking STAT3 phosphorylation. Thus, Myc suppresses precocious gliogenesis in early-stage NSCs by setting a high threshold for gliogenic signals. However, later in development, growth factor–induced p19ARF antagonizes Myc, thereby lowering this threshold. Gliogenic signals further up-regulate p19ARF, which in turn augments gliogenesis via enhanced STAT3 phosphorylation. Thus, the p19ARF–p53 pathway is part of an intracellular positive feedback loop downstream of gliogenic signals, thereby contributing to a sharp transition from neurogenesis to gliogenesis at a specific developmental stage.
Our results also explain apparently puzzling actions of growth factors. It has been shown that growth factors not only stimulate proliferation but also promote gliogenesis (Viti et al., 2003; Song and Ghosh, 2004; Nagao et al., 2007). However, other studies have shown a positive role for growth factor signals in neurogenesis (Menard et al., 2002; Samuels et al., 2008). Our data that both Myc and p19ARF are regulated by growth factors can explain these dual actions. Although growth factors stimulate proliferation in early development, they also maintain a high Myc activity, which in turn supports neurogenesis. As development proceeds, growth factors also up-regulate p19ARF, which in turn promotes gliogenesis. Thus, the outcome of growth factor stimulation changes depending on whether NSCs are at the Myc-dominant early stage or p19ARF-dominant late stage.
In vivo roles for Myc and p19ARF in brain development
Inactivation of N-Myc and c-Myc using Nestin-Cre has been reported to cause no severe morphological defects of the forebrain except for a substantial reduction in its size (Knoepfler et al., 2002; Hatton et al., 2006; Zindy et al., 2006). We observed a similar small brain phenotype in Foxg1-Cre;c-Mycflox/flox mice (our unpublished data). One reason for this mild defect could be that multiple Myc family members share redundant functions. It could also be attributable to incomplete recombination of conditional Myc alleles. Recombined c-Myc−/− cells have a substantial growth disadvantage over nonrecombined cells, and thus, their deficiency could be easily masked by compensatory proliferation of WT cells. In fact, when their self-renewal activity was assessed at the single-cell level, c-Myc–inactivated cells showed a profound deficiency in vitro. Moreover, the forebrain of c-Myc−/− mutants showed a severe neurogenic defect at an early stage. To overcome the problem of incomplete inactivation, we fate mapped Myc-inactivated cells as GFP+ cells in late-stage c-Myc deletion experiments. Such studies have demonstrated that c-Myc inactivation at the perinatal stage results in precocious differentiation of astrocytes.
It has been reported that p19ARF mutants show no obvious defect in brain development or in bulk culture of neurospheres at a high density (Bachoo et al., 2002). No defect in early development is reasonable given the very low expression of p19ARF in the brain at early stages. However, we found that when NSCs were selectively expanded at a clonal density, cells from p19ARF−/− mice at a late stage had a significantly lower gliogenic activity compared with the WT. Moreover, differentiation of astrocytes was significantly delayed or attenuated at the postnatal stage in p19ARF mutants. Notably, this phenotype is reminiscent of that of mice carrying a loss of function mutation of cardiotropin-1, the major STAT3-dependent gliogenic cytokine in the developing forebrain (Barnabe-Heider et al., 2005). Together, these results support the idea that Myc and p19ARF play important roles in controlling neurogenesis and gliogenesis in vivo.
Common mechanisms for stem cell regulation in development, cancer, and aging
Previous studies have established the crucial roles for Myc and the p19ARF–p53 pathway in tumorigenesis and senescence of adult tissues and organs (Grandori et al., 2000; Lowe and Sherr, 2003). It is particularly noteworthy that they are involved in various types of brain tumors (Zhu and Parada, 2002). Our study has now revealed their novel roles in brain development. A recently emerging idea is that deregulation and/or dysfunction of stem cells account for cancer and aging (Clarke and Fuller, 2006). Thus, understanding of the mechanisms underlying the control of stem cells in normal development has become increasingly important. We suggest that mutually antagonistic actions of Myc and the p19ARF–p53 pathway are key mechanisms common for development, cancer, and aging of the brain.
Materials and methods
Animals
All animal procedures were performed according to the guidelines and regulations of the Institutional Animal Care and Use Committee and the National Institutes of Health. The generation and genotyping of c-Myc−/− and c-Mycflox/flox (Trumpp et al., 2001; maintained in an FVB/N background) and Nestin-CreER and CAG-CAT-EGFP mice (Nakamura et al., 2006; Burns et al., 2007) were described previously. p53−/− and Foxg1-Cre mice (Hebert and McConnell, 2000; Guo et al., 2003) were provided by Y. Zheng and F. Guo (Cincinnati Children's Hospital Research Foundation, Cincinnati, OH) and S. McConnell (Stanford University, Stanford, CA), respectively. p19ARF−/− and p19ARF−/−;p16INK4a−/− mutant mice (Serrano et al., 1996; Kamijo et al., 1997) were obtained from the National Cancer Institute Mouse Models of Human Cancers Consortium and were maintained in a mixed background (129X1/SvJ × C57BL/6 and 129/Sv × C57BL/6, respectively). To induce Cre activity in c-Mycflox/−;Nestin-CreER;CAG-CAT-EGFP triple transgenic mice, a single dose of tamoxifen (Sigma-Aldrich) dissolved in maize oil (5 mg/40 g of body weight) was administered to pregnant dams on the indicated embryonic days. Embryos and pups of these mice and WT Sprague-Dawley rats were collected from timed pregnant females. Adult brains were obtained from 7–8-wk-old animals.
Cell culture
Neurosphere culture was performed as described previously (Nagao et al., 2007; Sugimori et al., 2007) with some modifications. Embryos and pups were collected from timed pregnant rats or mice and placed in an artificial cerebrospinal fluid (124 mM NaCl, 5 mM KCl, 1.3 mM MgCl2, 2 mM CaCl2, 26 mM NaHCO3, and 10 mM d-glucose). The forebrain was removed from the rest of the embryos under a dissection microscope (SV-11; Carl Zeiss, Inc.). For samples derived from E18.5 embryos or later stages, brains were removed from the skull, and coronal sections were prepared either manually or using rodent brain matrix (ASI Instruments). Thin tissue strips near the lateral ventricles (∼100-μm thick from the adult brain) were further obtained from these coronal slices. The resultant tissue was dissociated by incubation in a low Ca2+ and high Mg2+ artificial cerebrospinal fluid (124 mM NaCl, 5 mM KCl, 3.2 mM MgCl2, 0.1 mM CaCl2, 26 mM NaHCO3, 10 mM d-glucose, 100 U/ml penicillin, and 100 μg/ml streptomycin [Mediatech]) containing 0.05% (wt/vol) trypsin (Sigma-Aldrich), 0.67 mg/ml hyaluronidase (Sigma-Aldrich), and 0.1 mg/ml deoxyribonuclease I (Roche) at 37°C for 10 min. Subsequently, trypsin was neutralized with 0.7 mg/ml ovamucoid (Sigma-Aldrich), and the resultant tissue suspension was triturated mechanically to yield a single-cell suspension. The cells were filtered through a 40-μm sterile nylon mesh (BD) and washed twice with a basal medium (1:1 mixture of DME and F-12 medium [Invitrogen] containing 100 U/ml penicillin and 100 μg/ml streptomycin). Numbers of viable cells were determined by staining with trypan blue (Sigma-Aldrich).
In experiments using monolayer culture, cells isolated from embryos with specific genotypes were directly seeded on 100 μg/ml poly-d-lysine (Sigma-Aldrich)–coated glass chambers at a density of 6 × 104 cells/cm2 in the basal medium supplemented with the B-27 Supplement (Invitrogen), 20 ng/ml bovine FGF2 (R&D Systems), 20 ng/ml mouse EGF (Roche), 2 μg/ml heparin (molecular weight of 3,000; Sigma-Aldrich), and 1 mg/ml bovine serum albumin (Sigma-Aldrich). Dividing cells were labeled with 1 μM BrdU (Sigma-Aldrich) for 48 h.
To determine the frequency of self-renewing NSCs in a given tissue, dissociated single cells were directly seeded in a medium containing methylcellulose matrix at a density of 104 viable cells/ml. To manipulate cells by virus infection, primary neurospheres were dissociated at day 2 after seeding and were replated to yield secondary spheres. The cells were harvested at day 4 and subjected to retrovirus infection by incubating with high titer virus stock to yield 30–60% infection efficiency. These virus-infected secondary spheres were used for self-renewal and differentiation assays unless otherwise stated. For multiple passages of neurospheres, clonal colonies formed by virus-infected cells in methylcellulose matrix were recovered, dissociated to single cells by trypsin, and seeded again in the medium containing methylcellulose. The same procedure was repeated to yield serially passaged spheres. The number of passage used in a specific set of experiments is indicated in the figure legends. We confirmed the clonal nature of neurospheres formed under the aforementioned conditions using culture infected with GFP-expressing retroviruses; all clones were composed entirely of either GFP+ or GFP− cells but not both.
To induce differentiation of neurons and glia, cells were seeded onto poly-d-lysine–coated chambers as either individual clonal colonies (at a density of ∼100 colonies/cm2) or dissociated single cells (at a density of 6 × 104 cells/cm2) and were subsequently cultured for 6 d without FGF2 and EGF. In some experiments, cells were treated with 50 ng/ml human CNTF (Sigma-Aldrich) as described previously (Nagao et al., 2007). The quantitative results are expressed as mean ± SD of three to five independent culture experiments, and statistical analyses were performed with a two-tailed unpaired t test.
Retroviral vectors
Recombinant retroviruses expressing GFP together with human c-Myc, human dn-Myc (Berns et al., 1997), human MycER (provided by M. Eilers, Philipps University Marburg, Marburg, Germany; Eilers et al., 1989), and mouse N-Myc (provided by H. Kondoh, Osaka University, Osaka, Japan) were prepared using the retrovirus vector pMXIG (provided by T. Kitamura, University of Tokyo, Tokyo, Japan) as described previously (Nagao et al., 2007; Sugimori et al., 2007). The retrovirus vectors pMSCV expressing the full-length cDNAs for mouse p19ARF and p16INK4a and pMIEG3 expressing Cre recombinase were provided by Y. Zheng (Guo et al., 2003). p19ARF cDNA was also provided by S.R. Hann (Vanderbilt University School of Medicine, Nashville, TN; Qi et al., 2004). Viral vectors expressing shRNAs for p19ARF (pSIRIPP-p19-1 and pSIRIPP-p19-2), which were originally generated by the laboratory of T. Jacks (Massachusetts Institute of Technology, Cambridge, MA; Sage et al., 2003), were purchased from Addgene. Their control vectors pSIRIPP and p(SI)2-Luc (short hairpin–Luc) were provided by T.R. Golub (Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA) and S.L. Lessnick (University of Utah, Salt Lake City, UT). The fragment containing the puromycin-resistant gene in the original pSIRIPP vector was replaced with the coding sequence of GFP. All of these viral vectors were transfected into the packaging cell line Platinum-E to produce GFP retroviruses as described previously (Morita et al., 2000).
Immunostaining and histological analysis
To examine the brain phenotypes of various mutant mice, 12–16-μm-thick cryosections were prepared from embryonic and postnatal forebrains. Immunostaining of brain sections and cultured cells was performed as described previously (Nagao et al., 2007; Sugimori et al., 2007) using the following antibodies: TuJ1 (mouse [1:2,000]; Babco), NeuN (mouse [1:200]; Millipore), GFAP (mouse [1:1,000] and rabbit [1:1,000]; Millipore), S100β (mouse [1:1,000]; Sigma-Aldrich), O4 (mouse IgM [1:1,000]; Millipore), GFP (mouse [1:500] and rabbit [1:5,000]; Invitrogen; rat [1:5,000]; Nacalai Tesque), BrdU (mouse [1:1,000]; GE Healthcare), p19ARF (rat [1:200]; Santa Cruz Biotechnology, Inc.), p16INK4a (rabbit [1:200]; Santa Cruz Biotechnology, Inc.), phospho–c-Myc (T58/S62; rabbit [1:1,000]; Cell Signaling Technology), phosphorylated STAT3 (pSTAT3-Y705; rabbit [1:1,000]; Cell Signaling Technology), activated caspase3 (rabbit [1:1,000]; BD), phospho–histone H3 (S10; rabbit [1:1,000]; Millipore), and Cre (rabbit [1:10,000]; EMD). Affinity-purified rabbit polyclonal antibodies for Mash1 (1:2,000) and Olig2 (1:3,000) were described previously (Sugimori et al., 2007).
Immunoreactive cells were visualized by staining with secondary antibodies conjugated with Alexa Fluor 350, 488, 568, and 594 (1:400; Invitrogen). Cell nuclei in cultured samples were stained with 1 μg/ml DAPI (Sigma-Aldrich), whereas 2.5 μg/ml propidium iodide (PI; Invitrogen) was used for tissue sections. Specimens were mounted in Prolong Antifade (Invitrogen) and viewed at room temperature. Phase-contrast and fluorescent images of cultured cells and low magnification montage images of brain sections were captured using 10× Plan Apochromat NA 0.32, 20× Plan Apochromat NA 0.60, and 40× Plan Neofluar NA 0.75 objective lenses attached to a microscope (Axiophoto2; Carl Zeiss, Inc.) in combination with a digital charge-coupled device camera and attached software (C5810 version 1.1.4; Hamamatsu Photonics). High magnification fluorescent images of tissue sections were collected on a laser-scanning confocal microscope (LSM 510; Carl Zeiss, Inc.) with 40× Plan Neofluar NA 0.75 and 63× C Apochromat NA 1.2 W Korr objective lenses using imaging software (version 4.0 SP2; Carl Zeiss, Inc.). Images were imported into Photoshop (version 7.0; Adobe) for cropping and linear contrast adjustment.
To examine c-Myc−/− embryos, the percentage of marker-positive cells among neuroepithelial cells with PI+ nuclei in defined areas was measured. To examine differentiation of astrocytes in postnatal brains of c-Mycflox/−;Nestin-CreER;CAG-CAT-EGFP mice, the percentage of GFP+ cells costained with GFAP and S100β was quantified in defined areas. Likewise, astrocyte differentiation in p19ARF−/− mice was examined by quantifying the total number of DAPI+ nuclei and those surrounded by GFAP+ or S100β+ cytoplasm in defined areas (400 μm × 280 μm = 0.112 mm2). From each animal, two to three (c-Myc mutant mice) or two to five (p19ARF mice) coronal sections were chosen to cover comparable regions of the anterior and posterior parts of the forebrain. To maintain the consistency of staining results and avoid possible variability of developmental stages among litters, three to five pairs of the WT and mutant embryos or pups derived from independent crossings were examined.
Western blot analysis
The expression of pSTAT3-Y705 and GFAP was examined by Western blot analyses as described previously (Nagao et al., 2007) using the following antibodies: pSTAT3-Y705 (rabbit [1:1,000]; Cell Signaling Technology), STAT3 (mouse [1:2,500]; BD), and GFAP (mouse [1:1,000]; Millipore). α-Tubulin (mouse [1:1,000]; Sigma-Aldrich) and lamin B (goat [1:200]; Santa Cruz Biotechnology, Inc.) were used as internal controls. The relative expression level among samples was compared by analyzing digital images of blots using ImageJ software (National Institutes of Health).
RT-PCR
RNAs were isolated using the RNeasy kit (QIAGEN). The mRNA expression level was quantified using Opticon DNA Engine (Bio-Rad Laboratories), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The following sense and antisense primers were used for RT-PCR and gel imaging (amplified PCR product size is given in parentheses): rat c-Myc, 5′-TCCTGTACCTCGTCCGATTC-3′ and 5′-ACATGGCACCTCTTGAGGAC-3′ (284 bp); rat N-Myc, 5′-ACCAGCGCTGTGGTCACTA-3′ and 5′-GTCTTGGGGCGTACAGTGAT-3′ (263 bp); rat L-Myc, 5′-AAGCTTGGGGCAGGAATTAT-3′ and 5′-GTCTCTTCTCCACCGTCACC-3′ (297 bp); rat p19ARF, 5′-ACCCCAAGTGAGGGTTTTCT-3′ and 5′-GCACCATAGGAGAGCAGGAG-3′ (193 bp); mouse p19ARF, 5′-GTCGCAGGTTCTTGGTCACT-3′ and 5′-CGAATCTGCACCGTAGTTGA-3′ (246 bp); rat p16INK4a, 5′-GAACACTTTCGGTCGTACCC-3′ and 5′-CCCAGCGGAGGAGAGTAGAT-3′ (292 bp); rat p53, 5′-TTTGAGGTTCGTGTTTGTGC-3′ and 5′-CCTCATTCAGCTCTCGGAAC-3′ (232 bp); and rat and mouse GAPDH, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ (452 bp).
For quantitative RT-PCR using Opticon DNA Engine, the following sense and antisense primers were used: rat c-Myc, 5′-AAAGGCCCCCAAGGTAGTTA-3′ and 5′-CTCGCCGTTTCCTCAGTAAG-3′; rat N-Myc, 5′-AGGCAGTCACCACTTTCACC-3′ and 5′-GTTGTGTTGCTGATGGATGG-3′; rat p19ARF, 5′-ACCCCAAGTGAGGGTTTTCT-3′ and 5′-GATCCTCTCTGGCCTCAACA-3′; mouse p19ARF, 5′-GCTCTGGCTTTCGTGAACAT-3′ and 5′-TGAGCAGAAGAGCTGCTACG-3′; and rat and mouse GAPDH, 5′-AAGGGCTCATGACCACAGTC-3′ and 5′-GGATGCAGGGATGATGTTCT-3′.
Online supplemental material
Fig. S1 shows Myc-dependent regulation of self-renewal and differentiation during serial passages of neurospheres. Fig. S2 shows developmental and extracellular signal-dependent changes in the expression level of Myc and p19ARF. Fig. S3 shows modulation of cell growth and death by c-Myc and p19ARF. Fig. S4 shows failure of rescue of the early lethality of c-Myc−/− mutation by inactivation of p19ARF and p53. Fig. S5 shows stage-dependent genetic labeling of neural progenitors using Nestin-CreER;CAG-CAT-EGFP mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200807130/DC1.
Supplementary Material
Acknowledgments
We are grateful to Drs. M. Eilers, H. Kondoh, S.R. Hann, Y. Zheng, F. Guo, S. McConnell, T. Kitamura, T. Jacks, T.R. Golub, and S.L. Lessnick for reagents and R. Lang, Y. Zheng, and K. Tago for critical reading of the manuscript and helpful discussion.
This work was supported in part by the Ohio Eminent Scholar Award of the State of Ohio.
Abbreviations used in this paper: CNTF, ciliary neurotrophic factor; dn, dominant negative; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; NSC, neural stem cell; PI, propidium iodide; shRNA, short hairpin RNA; STAT3, signal transducer and activator transcription factor 3; WT, wild type.
References
- Bachoo, R.M., E.A. Maher, K.L. Ligon, N.E. Sharpless, S.S. Chan, M.J. You, Y. Tang, J. DeFrances, E. Stover, R. Weissleder, et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 1:269–277. [DOI] [PubMed] [Google Scholar]
- Barnabe-Heider, F., J.A. Wasylnka, K.J. Fernandes, C. Porsche, M. Sendtner, D.R. Kaplan, and F.D. Miller. 2005. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 48:253–265. [DOI] [PubMed] [Google Scholar]
- Berns, K., E.M. Hijmans, and R. Bernards. 1997. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene. 15:1347–1356. [DOI] [PubMed] [Google Scholar]
- Bruggeman, S.W., and M. van Lohuizen. 2006. Controlling stem cell proliferation: CKIs at work. Cell Cycle. 5:1281–1285. [DOI] [PubMed] [Google Scholar]
- Bruggeman, S.W., M.E. Valk-Lingbeek, P.P. van der Stoop, J.J. Jacobs, K. Kieboom, E. Tanger, D. Hulsman, C. Leung, Y. Arsenijevic, S. Marino, and M. van Lohuizen. 2005. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 19:1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, K.A., A.E. Ayoub, J.J. Breunig, F. Adhami, W.L. Weng, M.C. Colbert, P. Rakic, and C.Y. Kuan. 2007. Nestin-CreER mice reveal DNA synthesis by nonapoptotic neurons following cerebral ischemia hypoxia. Cereb. Cortex. 17:2585–2592. [DOI] [PubMed] [Google Scholar]
- Caviness, V.S. Jr., T. Goto, T. Tarui, T. Takahashi, P.G. Bhide, and R.S. Nowakowski. 2003. Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb. Cortex. 13:592–598. [DOI] [PubMed] [Google Scholar]
- Clarke, M.F., and M. Fuller. 2006. Stem cells and cancer: two faces of eve. Cell. 124:1111–1115. [DOI] [PubMed] [Google Scholar]
- Cleveland, J.L., and C.J. Sherr. 2004. Antagonism of Myc functions by Arf. Cancer Cell. 6:309–311. [DOI] [PubMed] [Google Scholar]
- Datta, A., A. Nag, W. Pan, N. Hay, A.L. Gartel, O. Colamonici, Y. Mori, and P. Raychaudhuri. 2004. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J. Biol. Chem. 279:36698–36707. [DOI] [PubMed] [Google Scholar]
- Davis, A.C., M. Wims, G.D. Spotts, S.R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671–682. [DOI] [PubMed] [Google Scholar]
- Eilers, M., D. Picard, K.R. Yamamoto, and J.M. Bishop. 1989. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 340:66–68. [DOI] [PubMed] [Google Scholar]
- Fan, G., K. Martinowich, M.H. Chin, F. He, S.D. Fouse, L. Hutnick, D. Hattori, W. Ge, Y. Shen, H. Wu, et al. 2005. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 132:3345–3356. [DOI] [PubMed] [Google Scholar]
- Fasano, C.A., J.T. Dimos, N.B. Ivanova, N. Lowry, I.R. Lemischka, and S. Temple. 2007. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 1:87–99. [DOI] [PubMed] [Google Scholar]
- Gil-Perotin, S., M. Marin-Husstege, J. Li, M. Soriano-Navarro, F. Zindy, M.F. Roussel, J.M. Garcia-Verdugo, and P. Casaccia-Bonnefil. 2006. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J. Neurosci. 26:1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori, C., S.M. Cowley, L.P. James, and R.N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653–699. [DOI] [PubMed] [Google Scholar]
- Guo, F., Y. Gao, L. Wang, and Y. Zheng. 2003. p19Arf-p53 tumor suppressor pathway regulates cell motility by suppression of phosphoinositide 3-kinase and Rac1 GTPase activities. J. Biol. Chem. 278:14414–14419. [DOI] [PubMed] [Google Scholar]
- Hatton, B.A., P.S. Knoepfler, A.M. Kenney, D.H. Rowitch, I.M. de Alboran, J.M. Olson, and R.N. Eisenman. 2006. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 66:8655–8661. [DOI] [PubMed] [Google Scholar]
- Hebert, J.M., and S.K. McConnell. 2000. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222:296–306. [DOI] [PubMed] [Google Scholar]
- Ho, J.S., W. Ma, D.Y. Mao, and S. Benchimol. 2005. p53-dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 25:7423–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo, T., F. Zindy, M.F. Roussel, D.E. Quelle, J.R. Downing, R.A. Ashmun, G. Grosveld, and C.J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 91:649–659. [DOI] [PubMed] [Google Scholar]
- Knoepfler, P.S., P.F. Cheng, and R.N. Eisenman. 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 16:2699–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler, P.S., X.Y. Zhang, P.F. Cheng, P.R. Gafken, S.B. McMahon, and R.N. Eisenman. 2006. Myc influences global chromatin structure. EMBO J. 25:2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, S.W., and C.J. Sherr. 2003. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13:77–83. [DOI] [PubMed] [Google Scholar]
- Marshall, C.A., B.G. Novitch, and J.E. Goldman. 2005. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J. Neurosci. 25:7289–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, D.J., V. Tropepe, and D. van Der Kooy. 2000. Separate proliferation kinetics of fibroblast growth factor-responsive and epidermal growth factor-responsive neural stem cells within the embryonic forebrain germinal zone. J. Neurosci. 20:1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis, K., V. Wirta, S.M. Hede, M. Nister, J. Lundeberg, and J. Frisen. 2006. p53 suppresses the self-renewal of adult neural stem cells. Development. 133:363–369. [DOI] [PubMed] [Google Scholar]
- Menard, C., P. Hein, A. Paquin, A. Savelson, X.M. Yang, D. Lederfein, F. Barnabe-Heider, A.A. Mir, E. Sterneck, A.C. Peterson, et al. 2002. An essential role for a MEK-C/EBP pathway during growth factor-regulated cortical neurogenesis. Neuron. 36:597–610. [DOI] [PubMed] [Google Scholar]
- Miller, F.D., and A.S. Gauthier. 2007. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 54:357–369. [DOI] [PubMed] [Google Scholar]
- Molofsky, A.V., S. He, M. Bydon, S.J. Morrison, and R. Pardal. 2005. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 19:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky, A.V., S.G. Slutsky, N.M. Joseph, S. He, R. Pardal, J. Krishnamurthy, N.E. Sharpless, and S.J. Morrison. 2006. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 443:448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. [DOI] [PubMed] [Google Scholar]
- Murphy, M.J., A. Wilson, and A. Trumpp. 2005. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 15:128–137. [DOI] [PubMed] [Google Scholar]
- Nagao, M., M. Sugimori, and M. Nakafuku. 2007. Cross talk between Notch and growth factor/cytokine signaling pathways in neural stem cells. Mol. Cell. Biol. 27:3982–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., M.C. Colbert, and J. Robbins. 2006. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 98:1547–1554. [DOI] [PubMed] [Google Scholar]
- Qi, Y., M.A. Gregory, Z. Li, J.P. Brousal, K. West, and S.R. Hann. 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 431:712–717. [DOI] [PubMed] [Google Scholar]
- Qian, X., Q. Shen, S.K. Goderie, W. He, A. Capela, A.A. Davis, and S. Temple. 2000. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 28:69–80. [DOI] [PubMed] [Google Scholar]
- Sage, J., A.L. Miller, P.A. Perez-Mancera, J.M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 424:223–228. [DOI] [PubMed] [Google Scholar]
- Samuels, I.S., J.C. Karlo, A.N. Faruzzi, K. Pickering, K. Herrup, J.D. Sweatt, S.C. Saitta, and G.E. Landreth. 2008. Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J. Neurosci. 28:6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageot, C.M., and C.D. Stiles. 2002. Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 12:244–249. [DOI] [PubMed] [Google Scholar]
- Seoane, J., H.V. Le, and J. Massague. 2002. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 419:729–734. [DOI] [PubMed] [Google Scholar]
- Serrano, M., H. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R.A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 85:27–37. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Y. Wang, J.T. Dimos, C.A. Fasano, T.N. Phoenix, I.R. Lemischka, N.B. Ivanova, S. Stifani, E.E. Morrisey, and S. Temple. 2006. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 9:743–751. [DOI] [PubMed] [Google Scholar]
- Song, M.R., and A. Ghosh. 2004. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 7:229–235. [DOI] [PubMed] [Google Scholar]
- Sugimori, M., M. Nagao, N. Bertrand, C.M. Parras, F. Guillemot, and M. Nakafuku. 2007. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development. 134:1617–1629. [DOI] [PubMed] [Google Scholar]
- Suzuki, A., S. Sekiya, D. Buscher, J.C. Izpisua Belmonte, and H. Taniguchi. 2008. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development. 135:1589–1595. [DOI] [PubMed] [Google Scholar]
- Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G.R. Martin, and J.M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 414:768–773. [DOI] [PubMed] [Google Scholar]
- Viti, J., A. Feathers, J. Phillips, and L. Lillien. 2003. Epidermal growth factor receptors control competence to interpret leukemia inhibitory factor as an astrocyte inducer in developing cortex. J. Neurosci. 23:3385–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., and L.F. Parada. 2002. The molecular and genetic basis of neurological tumours. Nat. Rev. Cancer. 2:616–626. [DOI] [PubMed] [Google Scholar]
- Zindy, F., C.M. Eischen, D.H. Randle, T. Kamijo, J.L. Cleveland, C.J. Sherr, and M.F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy, F., P.S. Knoepfler, S. Xie, C.J. Sherr, R.N. Eisenman, and M.F. Roussel. 2006. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc. Natl. Acad. Sci. USA. 103:11579–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.