Abstract
The microtubule cytoskeleton is crucial for the internal organization of eukaryotic cells. Several microtubule-associated proteins link microtubules to subcellular structures. A subclass of these proteins, the plus end–binding proteins (+TIPs), selectively binds to the growing plus ends of microtubules. Here, we reconstitute a vertebrate plus end tracking system composed of the most prominent +TIPs, end-binding protein 1 (EB1) and CLIP-170, in vitro and dissect their end-tracking mechanism. We find that EB1 autonomously recognizes specific binding sites present at growing microtubule ends. In contrast, CLIP-170 does not end-track by itself but requires EB1. CLIP-170 recognizes and turns over rapidly on composite binding sites constituted by end-accumulated EB1 and tyrosinated α-tubulin. In contrast to its fission yeast orthologue Tip1, dynamic end tracking of CLIP-170 does not require the activity of a molecular motor. Our results demonstrate evolutionary diversity of the plus end recognition mechanism of CLIP-170 family members, whereas the autonomous end-tracking mechanism of EB family members is conserved.
Introduction
Live-cell fluorescence imaging has revealed that a large and diverse subclass of microtubule-associated proteins, the +TIPs, associate dynamically with the growing ends of microtubules (Carvalho et al., 2003; Galjart and Perez, 2003; Lansbergen and Akhmanova, 2006; Akhmanova and Steinmetz, 2008). They link microtubules to subcellular structures like organelles (Perez et al., 1999), actin filaments (Tsvetkov et al., 2007), or the cell cortex (Miller et al., 2000). The mechanism by which +TIPs end-track is intimately linked to the dynamic state of the microtubule end. The elucidation of the end-tracking mechanism has proven to be challenging. Because plus end tracking of the large majority of +TIPs has until now only been observed in living cells, it remained unclear if end tracking of a given +TIP is a direct or indirect ability (Schuyler and Pellman, 2001). Furthermore, the multitude of interactions between +TIPs opened the possibility that redundant mechanisms of end accumulation might exist (Akhmanova and Steinmetz, 2008).
The most prominent plus end–tracking proteins conserved in all eukaryotes are members of the end-binding protein (EB) and CLIP-170 family (Perez et al., 1999; Mimori-Kiyosue et al., 2000; Tirnauer et al., 2002; Galjart, 2005; Akhmanova and Steinmetz, 2008). Recently, the in vitro reconstitution of plus end–tracking of fission yeast EB and CLIP-170 family members revealed how, on a molecular level, these yeast +TIPs track growing microtubule ends (Bieling et al., 2007). The molecular mechanism of plus end tracking of vertebrate EB and, in particular, of CLIP-170 proteins is, however, still under debate. The observation that fragments of vertebrate CLIP-170 containing the N-terminal tandem microtubule binding (cytoskeleton-associated protein glycine-rich [CAP-Gly]) domain bind to unpolymerized tubulin suggested that CLIP-170 autonomously tracks dynamic ends by a copolymerization mechanism (Diamantopoulos et al., 1999; Arnal et al., 2004; Folker et al., 2005; Ligon et al., 2006; Slep and Vale, 2007), although it is clear that CLIP-170 orthologues in yeast require a molecular motor for end tracking (Busch et al., 2004; Carvalho et al., 2004; Bieling et al., 2007).
Here, we reconstituted microtubule end tracking of vertebrate EB1 and CLIP-170 (Fig. 1 A) in vitro. We established the minimal requirements and elucidate the molecular mechanism underlying their ability to end-track. We find that this mechanism differs from previously suggested models and demonstrate evolutionary diversity of part of the plus end–tracking mechanism.
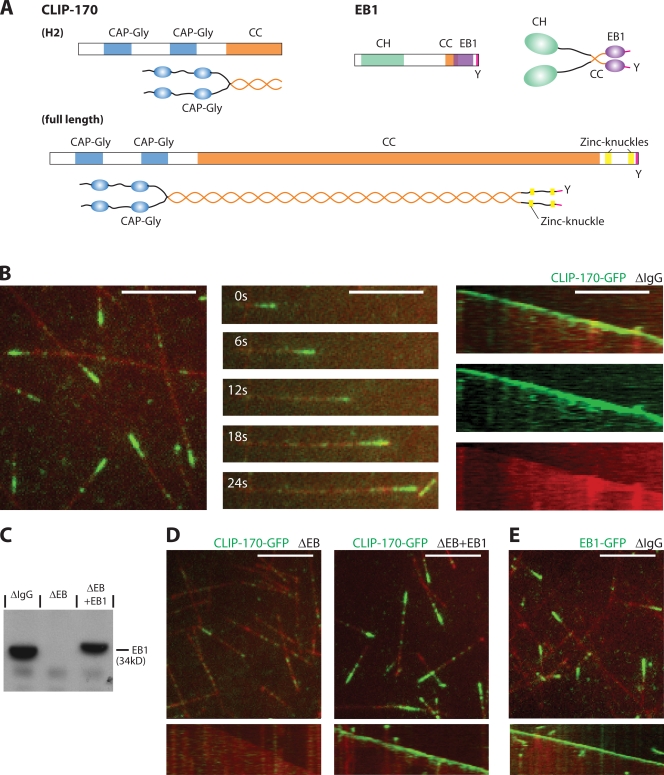
Figure 1.
CLIP-170 tracks growing microtubule ends in X. laevis egg extract in an EB1-dependent manner. (A) Scheme of the domain architecture of CLIP-170 and EB1. (B) TIRF microscopy of CLIP-170–GFP (green) on dynamic Alexa Fluor 568–labeled microtubules (red) in mock-depleted interphasic egg extract: an image of several microtubules (left), a time sequence (middle), and the corresponding kymograph (space-time plot) as overlay and separate channels (right) of a single microtubule are shown. (C) Western blot of mock-depleted (ΔIgG), EB-depleted (ΔEB), and EB-depleted extract with added recombinant EB1 (ΔEB+EB1), probed with an anti-EB1 antibody. (D) Images (top) and kymographs (bottom) of CLIP-170–GFP and dynamic microtubules in EB-depleted interphasic extract (left) and in extract with added recombinant EB1 (right). (E) Image (top) and kymograph (bottom) of EB1-GFP and microtubules in mock-depleted extract. Recombinant CLIP-170–GFP or EB1-GFP was added to a final concentration of 125 nM. Kymographs display a period of 46 s. Bars, 5 μm.
Results and discussion
CLIP-170 tracks growing microtubule ends in Xenopus laevis egg extract in an EB-dependent manner
We prepared recombinant, full-length CLIP-170 fused to GFP (CLIP-170–GFP), and we first tested for its functionality by examining its behavior in interphasic X. laevis egg extracts by time-lapse total internal reflection fluorescence (TIRF) microscopy. As in living cells (Perez et al., 1999), we observed selective accumulation of CLIP-170–GFP at growing microtubule ends (Fig. 1 B and Video 1, left, available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1). Although we detected a weak signal of a lattice-associated fraction of CLIP-170–GFP, we did not observe any evidence of processive transport toward the plus end, which argues against the involvement of a motor protein in end tracking of vertebrate CLIP-170 (Fig. 1 B and Video 1, left). A multitude of studies have shown that vertebrate CLIP-170 members interact with a large number of different +TIPs (Lansbergen et al., 2004; Watson and Stephens, 2006; Niethammer et al., 2007; Akhmanova and Steinmetz, 2008). In particular, EB proteins have been implicated in CLIP-170 end tracking, but a strict hierarchy has not firmly been established (Komarova et al., 2005).
We found that quantitative immunodepletion of all EB family proteins from egg extract (Fig. 1 C; see Materials and methods) completely abrogated plus end tracking by CLIP-170–GFP (Fig. 1 D, left; and Video 1, middle), which clearly demonstrates that one or several EB proteins are strictly required for end tracking of vertebrate CLIP-170. Interestingly, add-back of only recombinant EB1 (Fig. 1 C) fully restored the end accumulation of CLIP-170 (Fig. 1 D, right; and Video 1, right). Recombinant EB1 fused to GFP (EB1-GFP) also selectively accumulated at growing microtubule ends in egg extracts (Fig. 1 E), as expected from in vivo observations (Mimori-Kiyosue et al., 2000). To determine whether CLIP-170 is directly recruited to growing microtubule ends by end-associated EB1 or if other additional proteins present in the extract are also necessary, we identified the minimal requirements for plus end tracking of CLIP-170 by reconstitution experiments with purified proteins in buffer.
CLIP-170, in contrast to EB1, is not an autonomous microtubule end–binding protein
In buffer, dynamic microtubules extending from short surface-immobilized microtubules in the presence of purified tubulin and GTP were imaged using time-lapse TIRF microscopy. Recombinant, full-length CLIP-170–GFP failed to localize efficiently to dynamic microtubules in vitro (Fig. 2 A and Video 2, left, available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1). To test whether the lack of microtubule binding resulted from a proposed inhibitory intramolecular head-to-tail association of its N-terminal microtubule-binding domains with its C-terminal tail region (Lansbergen et al., 2004; Hayashi et al., 2007), we generated a fragment of CLIP-170 previously called H2 (Fig. 1 A; Scheel et al., 1999; Arnal et al., 2004), which contains the N-terminal CAP-Gly domains but lacks the C-terminal tail. This fragment is able to accumulate at growing microtubule ends inside living cells (Perez et al., 1999). In the absence of other proteins, purified GFP-tagged fragments (H2-GFP) became bound along the microtubule lattice in buffer (Fig. 2 A and Video 3, left; Scheel et al., 1999; Arnal et al., 2004), which demonstrates that full-length CLIP-170 is autoinhibited in the absence of potentially required interaction partners. Strikingly, however, H2-GFP did not track growing microtubule ends in vitro (Fig. 2 A and Video 3, left), despite binding weakly to soluble tubulin, as demonstrated by analytical gel filtration (Fig. 2 D). Increasing the concentration of H2-GFP or decreasing the ionic strength of the buffer lead to more binding of H2-GFP along the microtubule lattice, which is similar to previous observations (Diamantopoulos et al., 1999; Folker et al., 2005), but not to a selective accumulation at growing microtubule ends (unpublished data). H2-GFP also did not strongly promote microtubule growth (Fig. S1 A; Arnal et al., 2004). In conclusion, CLIP-170 is not an autonomous end-tracking protein.
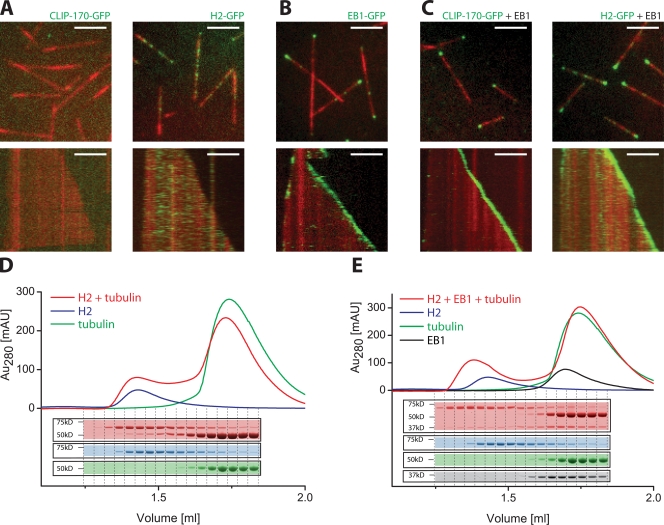
Figure 2.
EB1 is necessary and sufficient for end tracking of CLIP-170 in buffer. (A–C) TIRF microscopy images (top) and kymographs (bottom) of dynamic Alexa Fluor 568–labeled microtubules (red) in buffer in the presence of the following purified GFP-labeled +TIPs (green): 50 nM CLIP-170–GFP (A, left) or 50 nM H2-GFP (A, right); 150 nM EB1-GFP (B); and 35 nM CLIP-170–GFP (C, left) or 35 nM H2-GFP (C, right) in the presence of 70 nM of unlabeled EB1 in the presence of 15.5 μM tubulin. Kymographs display a period of 5 min. Bars, 5 μm. (D and E) Analytical gel filtrations: UV absorbance profiles and the corresponding Coomassie-stained SDS gel fractions of runs of a mixture of 20 nM H2 and 40 nM GTP-tubulin (D, red), and of 20 nM H2, 20 nM EB1, and 40 nM GTP-tubulin (E, red). Runs of H2 alone (blue), GTP-tubulin alone (green), and EB1 alone (black) at concentrations as in the mixtures are shown for comparison.
Next, we tested whether recombinant EB1 is an autonomous end-tracking protein. We found that EB1-GFP alone indeed selectively accumulated at growing, but not shrinking, microtubule ends in buffer (Fig. 2 B and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1). This was previously also observed for the fission yeast orthologue Mal3 (Bieling et al., 2007), which indicates that autonomous end tracking is an evolutionary conserved property of this protein family. Increasing the concentration of EB1 or decreasing the ionic strength of the buffer also revealed weak binding of EB1 to the microtubule lattice (unpublished data), which is in agreement with earlier observations (Sandblad et al., 2006; Manna et al., 2008). Within the concentration range of selective end tracking (up to ∼500 nM), EB1-GFP had only a mild effect on the microtubule growth velocity (Fig. S1 A). Furthermore, EB1 did not form a complex with soluble tubulin (Fig. S1 B, left). This is similar to its fission yeast orthologue Mal3 (Bieling et al., 2007) and agrees with the observation that EB1 only has a strong growth promoting activity at much higher concentrations (Vitre et al., 2008).
EB1 is necessary and sufficient for promoting the end tracking of CLIP-170
Because EB proteins were strictly necessary for localization of CLIP-170 in X. laevis egg extract (Fig. 1 D), we next tested if EB1 was able to recruit CLIP-170 to growing microtubule ends in the absence of other proteins in vitro. Strikingly, we found that in the presence of unlabeled EB1, full-length CLIP-170–GFP (Fig. 2 C and Video 2, right) and H2-GFP (Fig. 2 C and Video 3, right) strongly accumulated at growing, but not at depolymerizing microtubule ends, which demonstrates that end tracking of CLIP-170 can be reconstituted in vitro with three proteins only. Therefore, EB1 is both necessary and sufficient for recruiting CLIP-170 to growing microtubule ends. Our finding that EB1-dependent end tracking does not require the C-terminal tail of CLIP-170 in vitro is in agreement with biochemical and structural data that has demonstrated a weak but highly specific interaction between EB1 and CAP-Gly domains (Honnappa et al., 2006). Accordingly, analytical gel filtration demonstrated that EB1 and H2 indeed interact under the conditions of our experiments both in the absence (Fig. S1 B, right) and presence (Fig. 2 E) of excess soluble tubulin. Our identification of a minimal vertebrate end-tracking system consisting of only EB1 and CLIP-170 contrasts the previously described fission yeast system that additionally requires a motor protein for recruitment of the CLIP-170 orthologue (Busch et al., 2004; Carvalho et al., 2004; Bieling et al., 2007).
CLIP-170 and EB1 decorate growing microtubule ends for several seconds
Most previous studies favored a copolymerization mechanism for plus end tracking of CLIP-170 (Diamantopoulos et al., 1999; Arnal et al., 2004; Folker et al., 2005; Ligon et al., 2006; Slep and Vale, 2007) that requires tight binding of CLIP-170 to soluble tubulin. However, we found by analytical gel filtration that the interaction between H2 and tubulin under our in vitro conditions is rather weak (Fig. 2 D), as compared with the interaction between EB1 and H2 (Fig. 2 E). In the simplest copolymerization scenario, CLIP-170 bound to soluble tubulin would be loaded exclusively from solution to the very distal end of the growing microtubule, and the dissociation rate of CLIP-170 from the microtubule end region would directly determine the length of the CLIP-170 comets. We tested this prediction by measuring the comet tail length of the end-associated CLIP-170 ensemble and compared it to the dissociation kinetics of individual end-associated CLIP-170 molecules.
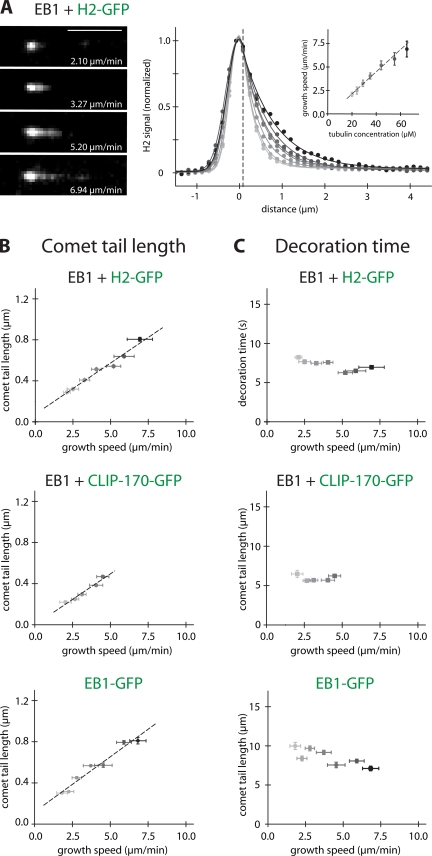
The decay length of the comet-like signal of H2-GFP and CLIP-170–GFP at microtubule ends in the presence of unlabeled EB1 (Fig. 3 A, left; and Fig. S2 A, left, available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1) increased with the microtubule growth velocity, which was varied by changing the tubulin concentration (Fig. 3 A, right, inset; and Fig. S2 A, right, inset). Averaged fluorescence intensity profiles from 50 individual growing microtubule ends revealed that the intensity of the comet tails decayed exponentially almost immediately after the maximum at the microtubule end that was toward the region of the microtubule lattice (Fig. 3 A, right; and Fig. S2 A right). Quantitative analysis showed that the comet tail lengths of both H2-GFP (Fig. 3 B, top) and of CLIP-170–GFP (Fig. 3 B, middle) in the presence of unlabeled EB1 increased roughly linearly with increasing growth velocity. The mean decoration times (comet tail length divided by growth velocity) of ∼7.2 and 5.9 s, respectively, were almost independent of the growth velocity (Fig. 3 C, top, middle). The corresponding analysis for EB1 alone (Fig. 3 B, bottom; and Fig. S2 B) showed a similar end decoration time (Fig. 3 C, bottom), which indicates that the distribution of CLIP-170 is largely determined by the distribution of end-associated EB1. The differences between the decoration times might be a consequence of the potentially cooperative nature of the binding events between the various domains of the +TIPs and tubulin.
Figure 3.
Analysis of the comet-shaped accumulation of +TIPs at microtubule ends. (A, left) TIRF microscopy images of comet-like accumulations of H2-GFP (added at 75 nM) at the ends of individual microtubules growing with the indicated velocities in the presence of 150 nM of unlabeled EB1. Bar, 5 μm. (A, right) Averaged fluorescence intensity profiles of H2-GFP comets at different tubulin concentrations (from 50 individual comets per tubulin concentration; dots) were fitted (lines) using Gaussian (to the left of the vertical broken line) and single exponential (to the right of the vertical broken line) functions. (A, inset) Microtubule growth velocities as a function of the used tubulin concentrations (error bars indicate SD). (B) Comet tail lengths of H2-GFP (top) and CLIP-170–GFP (middle) in the presence of unlabeled EB1, and of EB1-GFP alone (bottom) as a function of the microtubule growth speed. The comet tail lengths were obtained from the single exponential fits to the averaged intensity profiles. H2-GFP and CLIP-170–GFP were added at 75 nM, and EB1 and EB1-GFP at 150 nM. (C) Characteristic end-decoration times of the +TIPs as indicated corresponding to the comet tail lengths in B. The characteristic decoration time in the comet tail was obtained by dividing the comet tail length by the microtubule growth speed. Vertical error bars in B and C represent the standard error, and horizontal error bars represent the SD of the growth velocity.
CLIP-170 and EB1 turn over fast on binding sites at microtubule ends that exist for several seconds
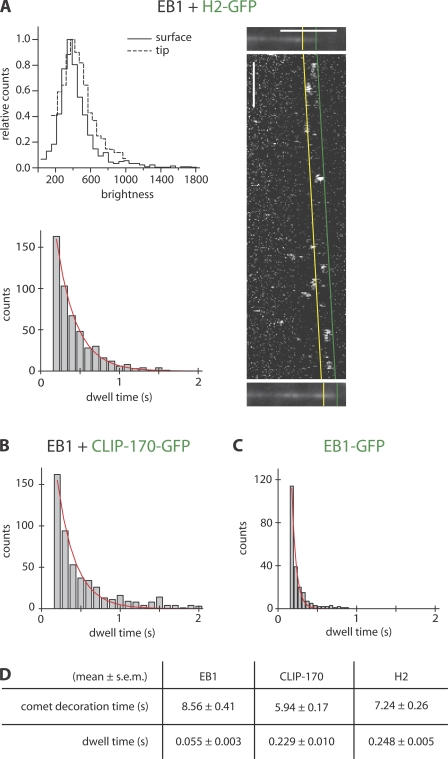
We next tested if the measured decoration times represent the mean dwell time of individual +TIPs at the microtubule end, as a copolymerization mechanism would predict, or, alternatively, the lifetime of binding sites existing at the microtubule end for the +TIPs. To distinguish between these possibilities, we measured the turnover of single, end-associated H2-GFP or CLIP-170–GFP molecules by fast single-molecule imaging using low concentrations of GFP-labeled protein in the presence of excess unlabeled EB1 and excess unlabeled H2 or CLIP-170, respectively. Brightness analysis of binding/unbinding events (Fig. 4 A, top left) established that individual H2-GFP molecules could be visualized at growing microtubule ends. To obtain single molecule dwell times, we analyzed binding events of individual +TIP molecules at growing microtubule ends using kymographs (e.g., Fig. 4 A, right). The analysis of 506 individual binding/unbinding events of H2-GFP at growing microtubule ends yielded an exponential dwell time distribution with a mean value of only 0.25 s (Fig. 4 A, bottom left). The behavior of full-length CLIP-170–GFP was very similar, with a mean end-dwell time of 0.23 s (541 events; Fig. 4 B and Fig. S3 B). These rapid unbinding kinetics are in agreement with recent turnover measurements in living cells (Dragestein et al., 2008). Together with the large difference in concentrations between the CLIP-170 (nM range) and soluble tubulin (μM range) under our conditions of selective end-tracking, the large difference between the ensemble decoration time and the single molecule dwell time excludes a copolymerization mechanism for CLIP-170. Therefore, the shape of CLIP-170 comets at growing microtubule ends is a signature of the distribution of binding sites that exist there in the presence of EB1 for 6–7 s, and on which CLIP-170 turns over rapidly.
Figure 4.
Single-molecule imaging: fast turnover of CLIP-170 and EB1 at growing microtubule ends. (A, top left) Brightness distribution of the fluorescence of single H2-GFP molecules as measured when attached to a surface and bound to the growing microtubule end, as indicated. The uniformity of the distributions and their similarity indicates that the large majority of the observed H2-GFP molecules at microtubule ends are individual dimers (see also Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1). (A, right) Individual H2-GFP molecules (added at 1 nM) binding at growing microtubule ends in the presence of an excess 74 nM of unlabeled H2, 150 nM of unlabeled EB1, and 35 μM tubulin. Images of a growing Alexa Fluor 568–labeled microtubule directly before (top) and after (bottom) a fast H2-GFP time lapse (shown as a kymograph between the microtubule images) were used to identify the growth trajectory of the microtubule end region. Binding events within the end region reaching from the distal end of the microtubule (green) to the end of the comet tail (yellow) as obtained from the averaged intensity profile (Fig. 4 A, right) were analyzed. Horizontal bar, 5 μm; vertical bar, 2 s. (A, bottom left) Histogram of the dwell times of end-associated single H2-GFP molecules. (B) Histogram of single CLIP-170–GFP molecules added at 5 nM in the presence of 70 nM of unlabeled CLIP-170, 150 nM of unlabeled EB1, and 35 μM tubulin. (C) Histogram of single EB1-GFP molecules added at 0.75 nM in the presence of 149 nM of unlabeled EB1 and 35 μM tubulin. Red lines in the histograms are single exponential fits to the data, providing a mean (single molecule) dwell time for each +TIP that is approximately two orders of magnitude lower than the mean bleaching time of the fluorescent label (Fig. S3). (D) Summary of the mean +TIP comet decoration times as determined from data shown in Fig. 3 C and the mean single molecule dwell times from histogram fits. Comet decoration times are significantly different (Mann-Whitney U test, P < 0.05), even if in a similar range. Dwell times differ from comet decoration times by more than an order of magnitude.
The corresponding analysis for EB1-GFP at growing microtubule ends also showed a large difference between the mean decoration time of 8.5 s, as obtained from the comet shape analysis (Fig. 3 C, bottom; and Fig. S2 B), and the mean single molecule dwell time of 0.05 s measured under single-molecule imaging conditions (217 events; Fig. 4 C and Fig. S3 B). This indicates that EB1, like its fission yeast orthologue Mal3 (Bieling et al., 2007), turns over rapidly on binding sites at microtubule ends that it can recognize autonomously. This is in agreement with EB1 not binding efficiently to soluble GTP tubulin (Fig. S1 B, left).
Strikingly, the characteristic mean decoration times at microtubule ends were quite similar among all +TIPs studied so far in vitro (Fig. 4 D; Bieling et al., 2007). This suggests that the lifetime of end binding sites is determined by the kinetics of a characteristic structural transition at the microtubule end because it is largely independent of the identity of the specific molecular probe used for its measurement. This transition could be related to the closure of microtubule sheets that are observed by cryo-electron microscopy at the growing end (Chretien et al., 1995) and/or to GTP hydrolysis in the GTP cap (Schek et al., 2007).
CLIP-170 binds at microtubule ends to composite binding sites consisting of tyrosinated α-tubulin and tyrosinated EB1
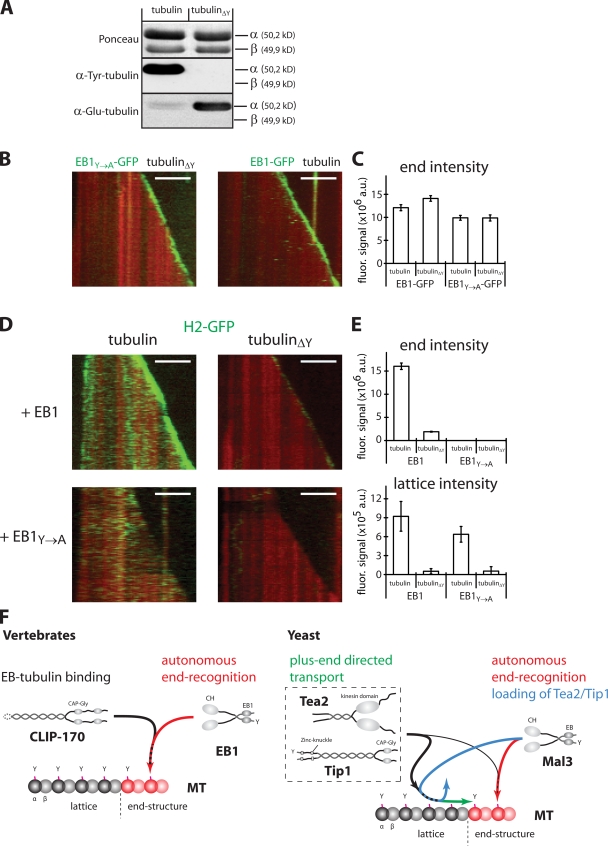
Finally, we asked if CLIP-170 tracks growing microtubule ends by purely hitchhiking on EB1 or if it also interacts with the tubulin polymer on microtubule ends. Both α-tubulin and EB1 carry a characteristic C-terminal EEY motif, of which the tyrosine is important for plus end tracking of CLIP-170, but not of EB1, in vivo (Badin-Larcon et al., 2004; Erck et al., 2005; Peris et al., 2006). Structural studies have shown that CAP-Gly domains can bind to the C-terminal regions of EB proteins (Honnappa et al., 2005; Weisbrich et al., 2007). To determine whether CLIP-170 interacts with both tubulin and EB1 while bound to the microtubule end, we prepared detyrosinated tubulin and an EB1 mutant with a C-terminal tyrosine-to-alanine substitution (EB1Y→A; Fig. 5 A). First, we showed that EB1 and EB1Y→A bound similarly to the growing ends of microtubules, irrespective of the tyrosination state of tubulin in vitro (Fig. 5, B and C). In contrast, both lattice association and end binding of H2-GFP in the presence of EB1 (Fig. 5 E, bottom) were drastically reduced (by 94 and 87%, respectively) when microtubules assembled from detyrosinated tubulin (Fig. 5 D, top right; and Video 5, top, available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1) instead of from normal, tyrosinated tubulin (Fig. 5 D, top left). Additionally, end tracking of H2-GFP on normal microtubules was abolished in the presence of EB1Y→A, whereas lattice binding was not strongly affected (Fig. 5 D, bottom left; Fig. 5 E, bottom; and Video 5, left). Finally, the use of detyrosinated microtubules and EB1Y→A completely abolished both end tracking and lattice binding of H2-GFP (Fig. 5 D, bottom right; Fig. 5 E; and Video 5, bottom). These measurements on dynamic microtubules agree with crystallographic data that lead to the proposal that CAP-Gly domain–containing proteins act as EEY motif–recognizing proteins (Honnappa et al., 2006; Weisbrich et al., 2007). Our results suggest that end tracking of CLIP-170 requires direct interactions with composite binding sites consisting of both tyrosinated α-tubulin and, most importantly, of end-associated, tyrosinated EB1, which explains why end accumulation of CLIP-170 is dependent on tyrosinated tubulin in vivo (Peris et al., 2006).
Figure 5.
CLIP-170 recognizes composite EB1/tubulin-binding sites at the microtubule end. (A) Western blot of mock-treated and detyrosinated tubulin (tubulinΔY) either Ponceau-stained or probed with an anti–Tyr-tubulin or anti–Glu-tubulin antibody. (B) Kymographs of a growing TAMRA-labeled microtubule (red) in the presence of 75 nM EB1Y→A-GFP (left, green) or EB1-GFP (right, green) in buffer. (C) The peak signal of the EB1 comets obtained from averaged intensity profiles at the indicated conditions. Error bars indicate standard error.(D) Kymographs of a growing TAMRA-labeled microtubule (red) in the presence of 35 nM H2-GFP (green) growing with either mock-treated (left) or detyrosinated tubulin (right) in the presence of unlabeled EB1 (top) or EB1Y→A (bottom). Bars, 5 μm. (E) The peak signal of the H2 comets (top) obtained from averaged intensity profiles at the indicated conditions. Signal of H2-GFP bound to the microtubule lattice (bottom) as averaged from intensity line scans. Error bars indicate the standard error (top) or the standard deviation of the mean lattice intensity from the line scans (bottom). (F) Schematic illustration of the mechanisms of end tracking by vertebrate (left) and fission yeast (right) +TIPs. See text for details.
Conclusion
We have reconstituted selective tracking of growing microtubule ends by the two major vertebrate +TIPs, CLIP-170 and EB1, in vitro, revealing the molecular mechanism of plus end tracking of these vertebrate +TIPs. Members of the EB1 family autonomously recognize a structure that is associated with the growing microtubule end on which they turn over rapidly. Together with binding motifs contributed by the C terminus of α-tubulin, EB1 establishes highly dynamic, composite binding sites for CLIP-170, which is by itself not able to track growing microtubule ends (Fig. 5 F). EB proteins might in fact be unique in their ability to autonomously end track, and could also create end-associated binding sites in a similar manner for other EB1-dependent +TIPs. Although the mechanism of autonomous plus end tracking by EB1 family members appears to be evolutionary conserved, the interaction of CLIP-170 family members with the composite EB1/end tubulin–binding sites is not. In contrast to vertebrate CLIP-170, orthologues from yeasts additionally require a motor protein for plus end tracking (Fig. 5 F; Busch et al., 2004; Carvalho et al., 2004; Bieling et al., 2007). This might be the result of differences in the modular composition between the members of the CLIP-170 or EB protein family and could reflect differing intracellular constraints in different species.
Materials and methods
Protein biochemistry
Full length, human CLIP-170 and CLIP-170–GFP (cytoplasmic linker protein 170 α-2, GenBank/EMBL/DDBJ accession no. AAA35693; 59% amino acid identity to the putative CLIP-170 homologue from X. laevis) were expressed in baculovirus-infected insect cells using the BactoBac system according to the manufacturer's instructions (Invitrogen). Harvested cells were resuspended in ice-cold buffer A (50 mM KPi, pH 7.5, 500 mM NaCl, 2 mM MgCl2, 10 mM β-mercaptoethanol, and 0.25% [wt/wt] Brij-35) containing protease inhibitors (Roche) and lysed by being passed through a high-pressure homogenizer twice. Clarified lysates were batch-incubated for 1.5 h at 4°C with Ni-Proteino resin (Macherey-Nagel). After binding of the protein, the resin was transferred into an empty column, which was washed with buffer A containing 3 mM imidazole, 50 mM arginine, and 50 mM glutamate. Proteins were eluted in buffer A containing 300 mM imidazole, 50 mM arginine, and 50 mM glutamate, then directly gel filtered over a HiLoad Superdex 200 (GE Healthcare) column equilibrated against buffer A containing 50 mM arginine and 50 mM glutamate. Peak fractions were pooled and either directly used for experiments for up to 2 d or supplemented with glycerol to a final concentration of 20% (vol/vol) and flash frozen in liquid ethane.
Full-length EB1 and EB1-GFP from X. laevis (XEB1A, GenBank/EMBL/DDBJ accession no. BAB84522; 75% amino acid identity to human EB1) were expressed in Escherichia coli (BL21(DE3) CodonPlus-RIL) induced with 0.5 mM IPTG for 8 h at 25°C). Harvested cells were resuspended in ice-cold buffer B (50 mM KPi, pH 7.2, 400 mM NaCl, 2 mM MgCl2, and 10 mM β-mercaptoethanol) containing protease inhibitors (Roche) and lysed by being passed through a high-pressure homogenizer twice. Clarified lysates were batch incubated for 1.5 h at 4°C with Ni-Proteino resin. After binding of the protein, the resin was transferred into an empty column, which was washed with buffer B containing 2.5 mM imidazole. Proteins were eluted in buffer B containing 300 mM imidazole and directly gel filtered over a HiLoad Superdex 200 column equilibrated against buffer B. Peak fractions were pooled, supplemented with glycerol to a final concentration of 20% (vol/vol), and flash frozen in liquid nitrogen.
H2 and H2-GFP were expressed in E. coli (BL21(DE3) CodonPlus-RIL) and induced with 1 mM IPTG for 5 h at 30°C. Harvested cells were resuspended in ice-cold buffer C (50 mM KPi, pH 7.5, 500 mM NaCl, 1 mM MgCl2, and 1 mM β-mercaptoethanol) containing protease inhibitors (Roche) and lysed by passing through a high-pressure homogenizer twice. Clarified lysates were loaded onto a Co2+-loaded HiTrap chelating column. After binding of the protein, the column was washed with buffer C containing 8.5 mM imidazole followed by elution with buffer C containing 300 mM imidazole. H2 was then directly gel filtered over a HiLoad Superdex 200 column equilibrated against buffer C, whereas H2-GFP was cleaved overnight at 4°C by adding His-tagged tobacco etch virus (TEV) protease, then desalted over a PD-10 column (GE Healthcare) equilibrated against buffer C and passed over a Co2+-loaded HiTrap chelating column to remove the cleaved z tag as well as the TEV protease. The flow-through containing H2-GFP was then gel filtered over a HiLoad Superdex 200 column equilibrated against buffer C. Peak fractions after gel filtration were pooled, supplemented with glycerol to a final concentration of 20% (vol/vol), and flash frozen in liquid nitrogen.
Labeling of tubulin with Alexa Fluor 568 carboxylic acid succinimidyl ester (Invitrogen), with 5-(and-6)-carboxytetramethylrhodamine succinimidyl ester, or with 6((biotinoyl)amino)hexanoic acid, succinimidyl ester (Invitrogen) was performed as described previously (Hyman et al., 1991).
Polyclonal antibodies
Rabbit polyclonal antibodies recognizing X. laevis EB1 and EB3 were a gift of I. Kronja, S. Rybina, and E. Karsenti (European Molecular Biology Laboratory, Heidelberg, Germany).
Preparation of egg extracts
Preparation of cytostatic factor (CSF)-arrested egg extracts and immunodepletions were performed as described previously (Desai et al., 1999). Recombinant CLIP-170–GFP or EB1-GFP were added 10 min before the experiment to a final concentration of 125 nM of recombinant protein.
End-tracking assay in interphasic egg extract
Flow chambers consisting of two poly-l-lysine–polyethylene glycol (PLL-PEG) passivated glass surfaces separated by double-sided tape (Tesa) were prepared in a temperature-controlled room (18 ± 1°C). The chambers were washed twice with CSF-XB buffer, and the experiment was initiated by flowing interphasic egg extract supplemented with 230 μg/ml glucose oxidase (Serva), 35 μg/μl catalase (Sigma-Aldrich), and 1 μM Alexa Fluor 568–labeled tubulin into the flow chamber. As the extract quickly heats up to room temperature, microtubule nucleation and growths occurs very rapidly. Therefore, the flow chamber was directly sealed with nail polish and immediately transferred to the microscope for imaging.
Analytical gel filtration
50 μl of 20 μM H2, EB1, and/or 40 μM tubulin was incubated in gel filtration buffer (80 mM K-Pipes, pH 6.8, 85 mM KCl, 85 mM potassium-acetate, 4 mM MgCl2, 0.2 mM GTP, 1 mM EGTA, 10 mM β-mercaptoethanol, and 0.25% [wt/wt] Brij-35) either individually or in combinations, as indicated, on ice for 15 min before loading on a Superose 6 PC 3.2/30 (GE Healthcare) equilibrated in gel filtration buffer. The absorbance of the eluted protein was measured at 280 nm. Fractions of 36 μl were collected, supplemented with SDS sample buffer, and separated on 4–12% Bis-Tris acrylamide gels (Invitrogen). Proteins were stained with Coomassie brilliant blue.
Detyrosination of tubulin
A mixture of pig brain tubulin and tetramethyl-6-carboxyrhodamine (TAMRA)-labeled tubulin (at 25 mg/ml and a labeling ratio of 1:15) was detyrosinated at 37°C by the addition of carboxypeptidase A (Sigma-Aldrich) at 6 U/mg of tubulin. After 5 min, 1 mM GTP and 2 mM MgCl2 were added to the reaction to promote tubulin polymerization. After a 30-min incubation at 37°C, microtubules were centrifuged at 60,000 g, then resuspended and depolymerized in ice-cold buffer X (100 mM MOPS, pH 6.9, 2 mM EGTA, 1 mM MgSO4, and 2 mM DTT). Detyrosinated tubulin was recovered from the supernatant after centrifugation for 20 min at 60,000 g, subjected to another round of polymerization/depolymerization in BRB80 buffer (80 mM K-Pipes, pH 6.8, 2 mM MgCl2, and 1 mM EGTA), and frozen again in liquid nitrogen. For control experiments, tubulin was treated in the same way but without carboxypeptidase A (yielding “mock-treated” tubulin). To test for the efficiency of detyrosination, mock-treated or detyrosinated tubulin were separated by 10% SDS-PAGE, blotted onto nitrocellulose, and stained with Ponceau S. Western blots were performed with anti–Tyr-tubulin antibody (YL1/2; Millipore) or anti–Glu-tubulin (Millipore).
Glass surface treatment
Biotin-PEG functionalized and PLL-PEG passivated glass slides were prepared as described previously (Bieling et al., 2007).
End-tracking assay in buffer
Flow chambers were assembled in a heated room (30 ± 1°C) by sticking a biotin-PEG functionalized coverslip to a PLL-PEG passivated microscope slide with double-sided tape. The flow cell was then washed with assay buffer (80 mM K-Pipes, pH 6.8, 85 mM KCl, 85 mM potassium-acetate, 4 mM MgCl2, 1 mM GTP, 1 mM EGTA, 10 mM β-mercaptoethanol, and 0.25% [wt/wt] Brij-35), and potential residual unspecific binding sites were blocked by flowing in 1% Pluronic F-127 and 50 μg/ml κ-casein in assay buffer. The flow cell was then incubated on ice with 50 μg/ml NeutrAvidin (Invitrogen) and 50 μg/ml κ-casein in assay buffer for 5 min, transferred to room temperature, and washed with 15 chamber volumes of assay buffer. Then, the chambers were incubated with brightly labeled, short GMP-CPP (guanosine-5′-[(α,β)]-methylene)triphosphate) microtubules (containing 26% Alexa Fluor 568–labeled tubulin and 26% biotinylated tubulin) in assay buffer at 30°C for 5 min. Microtubule growth was initiated by flowing in 15 μM of dimly labeled tubulin (containing 10% Alexa Fluor 568–labeled tubulin and no biotinylated tubulin) together with +TIP proteins at the indicated concentrations in assay buffer containing 0.1% methyl cellulose (4,000 cP; Sigma-Aldrich), 50 μg/ml β-casein (Sigma-Aldrich), and oxygen scavengers (20 mM glucose, 320 μg/ml glucose oxidase [Serva], and 55 μg/ml catalase [Sigma-Aldrich]). For the determination of the comet tail length and the dwell time of individual +TIP molecules at the microtubule end, the concentration of tubulin was elevated as indicated. The flow chamber was then sealed with nail polish and transferred to the microscope for imaging. The temperature was maintained at 30 ± 1°C. For experiments in Fig. 5, normal tubulin was replaced by either enzymatically detyrosinated tubulin or tubulin treated in the same manner in the absence of the enzyme (mock tubulin).
TIRF microscopy
Imaging was performed on a custom TIRF microscope equipped with a Cascade II, cooled charge-coupled device camera (Photometrics) as described previously (Bieling et al., 2007). Two-color time-lapse imaging for the localization of +TIPs on dynamic microtubules was performed at 1 frame/3 s for experiments in buffer and at 1 frame/s in egg extract with a 100-ms exposure time. For the dwell time analysis of single +TIP molecules at the microtubule plus end, one-color imaging was performed at an increased frame rate of 20 frames/s, with a 50-ms exposure time.
Data analysis of +TIP comets
The fluorescence signal of +TIP “comets” at growing microtubule plus ends was analyzed as described previously (Bieling et al., 2007). The error of the comet tail length represents the standard error of the mean length as a fit parameter. The error of the decoration time was calculated by error propagation from the standard errors of the growth rates and the fitted comet tail length.
Analysis of single molecule dwell times at the microtubule end
First, continuously growing microtubule plus ends were selected from the images of growing microtubules taken directly (1 s) before and after a fast time-lapse sequence, as described previously (Bieling et al., 2007). Dwell times of individual binding events within the tip region were then measured by kymograph analysis. The tip region was defined to begin at the distal end of the microtubule and to extend for 0.9 μm (for CLIP-170–GFP), 1.1 μm (for H2-GFP), or 1.2 μm (for EB1-GFP), as obtained from the corresponding averaged intensity profile. The mean dwell time and the standard error were obtained from a single exponential fit to the dwell time histogram from three independent experiments for each +TIP protein analyzed.
Single molecule brightness and bleaching analysis
To determine the brightness of individual surface-bound molecules, GFP-tagged +TIPs at a final concentration of 0.2–2 nM in assay buffer were flowed into a flow chamber consisting of PLL-PEG–coated glass. After 3-min incubation at room temperature, unbound molecules were washed out, and imaging was performed at the same conditions as in end-tracking experiments. Single fluorescent spots seen in fast time-lapse movies either from surface-adhered or end-tracking GFP-tagged +TIPs were automatically identified using commercially available software (Kalaimoscope; TransInsight GmbH). To compare the brightness of the three constructs, intensity histograms were generated and maximum-normalized. For bleaching analysis, the number of spots was plotted against time and fitted with a single exponential function with characteristic time T.
Online supplemental material
Fig. S1 illustrates that microtubule growth speed is only weakly dependent on +TIP concentration, and shows results from two analytical gel filtrations. Fig. S2 shows the comet shape analysis of CLIP-170 and EB1. Fig. S3 provides information about the brightness distribution and the bleaching characteristics of all three +TIPs. Video 1 shows CLIP-170–GFP in interphasic X. laevis egg extract. Video 2 shows CLIP-170–GFP on dynamic microtubules in buffer. Video 3 shows H2-170–GFP on dynamic microtubules in buffer. Video 4 shows EB1-GFP on dynamic microtubules in buffer. Video 5 shows the importance of C-terminal tyrosine of α-tubulin and EB1 for end tracking of CLIP-170. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200809190/DC1.
Acknowledgments
We thank Iva Kronja, Sofia Rybina, and Eric Karsenti for reagents, Krzysztof Rogowski for insightful discussions, and Anamarija Kruljac-Letunic for advice on EB1 purification.
P. Bieling and T. Surrey acknowledge financial support from the Deutsche Forschungsgemeinschaft, and I.A. Telley acknowledges support from the European Union STREP Active Biomics and the Swiss National Science Foundation (SNSF). C. Janke was supported by the Centre National de la Recherche Scientifique, Association de la Recherche contre le Cancer (ARC) award 3140, and French National Research Agency (ANR) award JC05_42022.
Abbreviations used in this paper: CAP-gly, cytoskeleton-associated protein glycine-rich; EB, end-binding protein; PEG, polyethylene glycol; PLL, poly-l-lysine; TAMRA, tetramethyl-6-carboxyrhodamine; TIRF, total internal reflection fluorescence.
References
- Akhmanova, A., and M.O. Steinmetz. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. [DOI] [PubMed] [Google Scholar]
- Arnal, I., C. Heichette, G.S. Diamantopoulos, and D. Chretien. 2004. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 14:2086–2095. [DOI] [PubMed] [Google Scholar]
- Badin-Larcon, A.C., C. Boscheron, J.M. Soleilhac, M. Piel, C. Mann, E. Denarier, A. Fourest-Lieuvin, L. Lafanechere, M. Bornens, and D. Job. 2004. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc. Natl. Acad. Sci. USA. 101:5577–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling, P., L. Laan, H. Schek, E.L. Munteanu, L. Sandblad, M. Dogterom, D. Brunner, and T. Surrey. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105. [DOI] [PubMed] [Google Scholar]
- Busch, K.E., J. Hayles, P. Nurse, and D. Brunner. 2004. Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell. 6:831–843. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., J.S. Tirnauer, and D. Pellman. 2003. Surfing on microtubule ends. Trends Cell Biol. 13:229–237. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., M.L. Gupta Jr., M.A. Hoyt, and D. Pellman. 2004. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 6:815–829. [DOI] [PubMed] [Google Scholar]
- Chretien, D., S.D. Fuller, and E. Karsenti. 1995. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 129:1311–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., A. Murray, T.J. Mitchison, and C.E. Walczak. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. [DOI] [PubMed] [Google Scholar]
- Diamantopoulos, G.S., F. Perez, H.V. Goodson, G. Batelier, R. Melki, T.E. Kreis, and J.E. Rickard. 1999. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragestein, K.A., W.A. van Cappellen, J. van Haren, G.D. Tsibidis, A. Akhmanova, T.A. Knoch, F. Grosveld, and N. Galjart. 2008. Dynamic behavior of GFP-CLIP-170 reveals fast protein turnover on microtubule plus ends. J. Cell Biol. 180:729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erck, C., L. Peris, A. Andrieux, C. Meissirel, A.D. Gruber, M. Vernet, A. Schweitzer, Y. Saoudi, H. Pointu, C. Bosc, et al. 2005. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl. Acad. Sci. USA. 102:7853–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker, E.S., B.M. Baker, and H.V. Goodson. 2005. Interactions between CLIP-170, tubulin, and microtubules: implications for the mechanism of Clip-170 plus-end tracking behavior. Mol. Biol. Cell. 16:5373–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart, N. 2005. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 6:487–498. [DOI] [PubMed] [Google Scholar]
- Galjart, N., and F. Perez. 2003. A plus-end raft to control microtubule dynamics and function. Curr. Opin. Cell Biol. 15:48–53. [DOI] [PubMed] [Google Scholar]
- Hayashi, I., M.J. Plevin, and M. Ikura. 2007. CLIP170 autoinhibition mimics intermolecular interactions with p150Glued or EB1. Nat. Struct. Mol. Biol. 14:980–981. [DOI] [PubMed] [Google Scholar]
- Honnappa, S., C.M. John, D. Kostrewa, F.K. Winkler, and M.O. Steinmetz. 2005. Structural insights into the EB1-APC interaction. EMBO J. 24:261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa, S., O. Okhrimenko, R. Jaussi, H. Jawhari, I. Jelesarov, F.K. Winkler, and M.O. Steinmetz. 2006. Key interaction modes of dynamic +TIP networks. Mol. Cell. 23:663–671. [DOI] [PubMed] [Google Scholar]
- Hyman, A., D. Drechsel, D. Kellogg, S. Salser, K. Sawin, P. Steffen, L. Wordeman, and T. Mitchison. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485. [DOI] [PubMed] [Google Scholar]
- Komarova, Y., G. Lansbergen, N. Galjart, F. Grosveld, G.G. Borisy, and A. Akhmanova. 2005. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell. 16:5334–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen, G., Y. Komarova, M. Modesti, C. Wyman, C.C. Hoogenraad, H.V. Goodson, R.P. Lemaitre, D.N. Drechsel, E. van Munster, T.W. Gadella Jr., et al. 2004. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 166:1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen, G., and A. Akhmanova. 2006. Microtubule plus end: a hub of cellular activities. Traffic. 7:499–507. [DOI] [PubMed] [Google Scholar]
- Ligon, L.A., S.S. Shelly, M.K. Tokito, and E.L. Holzbaur. 2006. Microtubule binding proteins CLIP-170, EB1, and p150Glued form distinct plus-end complexes. FEBS Lett. 580:1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, T., S. Honnappa, M.O. Steinmetz, and L. Wilson. 2008. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry. 47:779–786. [DOI] [PubMed] [Google Scholar]
- Miller, R.K., S.C. Cheng, and M.D. Rose. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 11:2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868. [DOI] [PubMed] [Google Scholar]
- Niethammer, P., I. Kronja, S. Kandels-Lewis, S. Rybina, P. Bastiaens, and E. Karsenti. 2007. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 5:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, F., G.S. Diamantopoulos, R. Stalder, and T.E. Kreis. 1999. CLIP-170 highlights growing microtubule ends in vivo. Cell. 96:517–527. [DOI] [PubMed] [Google Scholar]
- Peris, L., M. Thery, J. Faure, Y. Saoudi, L. Lafanechere, J.K. Chilton, P. Gordon-Weeks, N. Galjart, M. Bornens, L. Wordeman, et al. 2006. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 174:839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandblad, L., K.E. Busch, P. Tittmann, H. Gross, D. Brunner, and A. Hoenger. 2006. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 127:1415–1424. [DOI] [PubMed] [Google Scholar]
- Scheel, J., P. Pierre, J.E. Rickard, G.S. Diamantopoulos, C. Valetti, F.G. van der Goot, M. Haner, U. Aebi, and T.E. Kreis. 1999. Purification and analysis of authentic CLIP-170 and recombinant fragments. J. Biol. Chem. 274:25883–25891. [DOI] [PubMed] [Google Scholar]
- Schek, H.T. III, M.K. Gardner, J. Cheng, D.J. Odde, and A.J. Hunt. 2007. Microtubule assembly dynamics at the nanoscale. Curr. Biol. 17:1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., and D. Pellman. 2001. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 105:421–424. [DOI] [PubMed] [Google Scholar]
- Slep, K.C., and R.D. Vale. 2007. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell. 27:976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J.S., S. Grego, E.D. Salmon, and T.J. Mitchison. 2002. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov, A.S., A. Samsonov, A. Akhmanova, N. Galjart, and S.V. Popov. 2007. Microtubule-binding proteins CLASP1 and CLASP2 interact with actin filaments. Cell Motil. Cytoskeleton. 64:519–530. [DOI] [PubMed] [Google Scholar]
- Vitre, B., F.M. Coquelle, C. Heichette, C. Garnier, D. Chretien, and I. Arnal. 2008. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 10:415–421. [DOI] [PubMed] [Google Scholar]
- Watson, P., and D.J. Stephens. 2006. Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J. Cell Sci. 119:2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrich, A., S. Honnappa, R. Jaussi, O. Okhrimenko, D. Frey, I. Jelesarov, A. Akhmanova, and M.O. Steinmetz. 2007. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 14:959–967. [DOI] [PubMed] [Google Scholar]