Abstract
Poly-ADP ribose polymerase 1 (PARP-1) is activated by DNA damage and has been implicated in the repair of single-strand breaks (SSBs). Involvement of PARP-1 in other DNA damage responses remains controversial. In this study, we show that PARP-1 is required for replication fork slowing on damaged DNA. Fork progression in PARP-1−/− DT40 cells is not slowed down even in the presence of DNA damage induced by the topoisomerase I inhibitor camptothecin (CPT). Mammalian cells treated with a PARP inhibitor or PARP-1–specific small interfering RNAs show similar results. The expression of human PARP-1 restores fork slowing in PARP-1−/− DT40 cells. PARP-1 affects SSB repair, homologous recombination (HR), and nonhomologous end joining; therefore, we analyzed the effect of CPT on DT40 clones deficient in these pathways. We find that fork slowing is correlated with the proficiency of HR-mediated repair. Our data support the presence of a novel checkpoint pathway in which the initiation of HR but not DNA damage delays the fork progression.
Introduction
A DNA double-strand break (DSB) can be induced by ionizing radiation, chemicals, and single-strand breaks (SSBs) in replication. Camptothecin (CPT) inhibits topoisomerase I (Topo I) and is one of the DSB-inducing reagents for which the action mechanism has been characterized in detail. The drug reversibly abolishes the religation activity of Topo I to generate SSBs to which the protein is covalently linked. DSBs arise when replication forks collide with the SSBs and run off (Pommier et al., 2003). Thus, CPT-induced DSBs are largely replication dependent. Eukaryotes have two pathways for repairing DSBs: homologous recombination (HR) and nonhomologous end joining (NHEJ). The relative contribution of these two DSB repair pathways seems to differ depending on the cell cycle; HR occurs more frequently in the S and G2 phases, and NHEJ is most efficient in the G1 phase (Takata et al., 1998; Essers et al., 2000). For these reasons, CPT-induced replication-dependent DSBs are usually repaired by the HR pathway (Arnaudeau et al., 2001). The choice of these two DSB repair pathways is likely to be decided by a molecular mechanism activated immediately after DSB formation.
Poly-ADP ribosylation is a posttranslational modification catalyzed by poly-ADP ribose polymerase 1 (PARP-1) and PARP-2 and is one of the earliest cellular responses to DNA damage. PARP-1 and -2 bind to the damage sites and activate themselves by automodification. This process causes chromatin decondensation around damage sites, recruitment of repair machineries such as DNA ligase III–XRCC-1 base excision repair complexes, and accelerates DNA damage repair, especially in the case of SSBs (Caldecott et al., 1996; Masson et al., 1998; Leppard et al., 2003). PARP-1 also seems to affect DSB repair because PARP-1–deficient cells are hypersensitive to DSB-inducing agents, especially to CPT (Chatterjee et al., 1989; Bowman et al., 2001; Pommier et al., 2003). Biochemical studies showed that NHEJ proteins such as Ku and DNA-PK are poly-ADP ribosylated by PARP-1, and the affinity of Ku to DSBs was decreased (Ariumi et al., 1999; Galande and Kohwi-Shigematsu, 1999; Li et al., 2004). Moreover, a genetic study showed that PARP-1 could protect the HR pathway from toxic interference by Ku70 (in DT40 and mammalian cells) in response to CPT-induced DSBs (Hochegger et al., 2006). These studies suggest that PARP-1 can interfere with NHEJ, which might be unfavorable for DSB repair in S phase and provide access for the HR machineries. These findings have also suggested the possibility that PARP-1 is involved in the regulation of replication fork progression coupled with HR-dependent DSB repair via its poly-ADP ribosylation activity. However, the role of PARP-1 in HR-coupled replication fork modulation in vivo has not been fully investigated.
In this study, we investigated the contributions of PARP-1 to the progression of replication forks on CPT-damaged DNA using an in vivo replication labeling assay and a dynamic molecular combing technique that allowed us to analyze the progression of individual replication forks in vivo. Interestingly, the addition of CPT significantly slowed down fork progression when PARP-1 was active, whereas inhibition or deficiency of PARP-1 resulted in unchanged fork progression rates in the presence of CPT. We also found that this slowing down of the replication fork in wild-type cells depended on the HR pathway, which was diminished in PARP-1 knockout cells as a result of negative interference by Ku.
Results and discussion
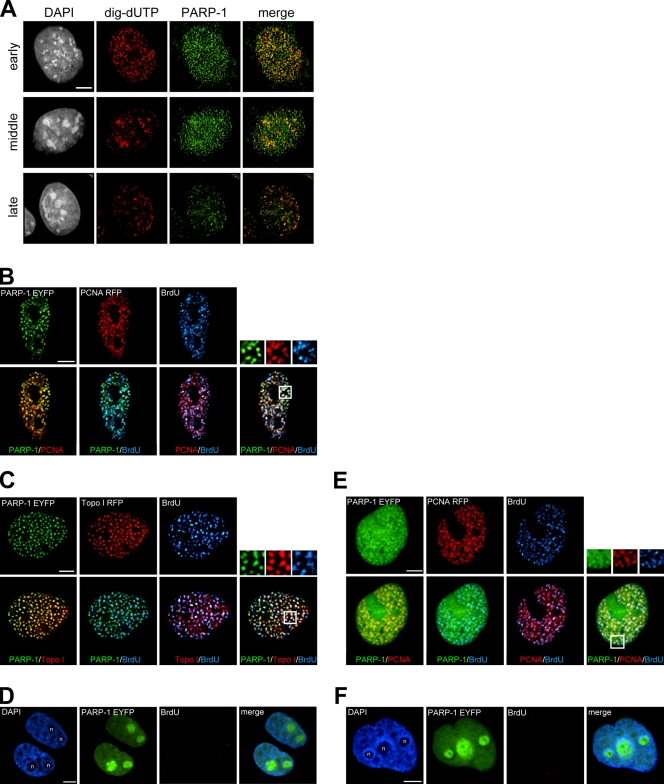
Previous studies showed that PARP-1 interacted with some replication fork machineries (Dantzer et al., 1998; Frouin et al., 2003). To test for a possible association between PARP-1 and replication forks in our experimental system, we examined the immunofluorescence of PARP-1 and replicating DNA in mouse fibroblast cells, m5S. We found that PARP-1 is likely to interact with replication forks because some of the PARP-1 foci in the nucleus colocalized with replication foci throughout S phase (Fig. 1 A). Next, PARP-1–enhanced YFP (EYFP) and proliferating cell nuclear antigen (PCNA)–RFP were transiently expressed in COS-7 cells, and BrdU pulse labeling was performed. We found that in S phase, PARP-1–EYFP was adjacent or colocalized to PCNA-RFP and BrdU foci (Fig. 1 B). Similar results were also observed when we transiently expressed Topo I–DsRed instead of PCNA-RFP (Fig. 1 C). In BrdU-negative cells, PARP-1–EYFP was enriched in nucleoli as previously reported (Yung et al., 2004) and did not form any foci as seen in S-phase cells (Fig. 1 D). We confirmed that a fraction of PARP-1–EYFP seemed to colocalize to PCNA- and BrdU-containing foci in HeLa cells. Again, PARP-1–EYFP appeared to be concentrated in nucleoli in nonreplicating HeLa cells (Fig. 1 F).
Figure 1.
Localization of PARP-1 to the replicating genomic region. (A) Digoxigenin–deoxy-UTP was introduced into m5S cells to label replicating cells. PARP-1 and digoxigenin–deoxy-UTP (dig-dUTP) were visualized by immunofluorescence. DNA was counterstained with DAPI. (B and C) Localization of PARP-1–EYFP and PCNA-RFP or PARP-1–EYFP and Topo I–DsRed during S phase in COS-7 cells. After pulse labeling with BrdU, cells were fixed and BrdU was immunodetected. (D) Typical localization of PARP-1–EYFP out of S phase in COS-7 cells. (E) Localization of PARP-1–EYFP and PCNA-RFP during S phase in HeLa cells. (F) Typical localization of PARP-1–EYFP out of S phase in HeLa cells. Overlaid images and magnified views of the boxed areas are shown. n, nucleoli. Bars, 5 μm.
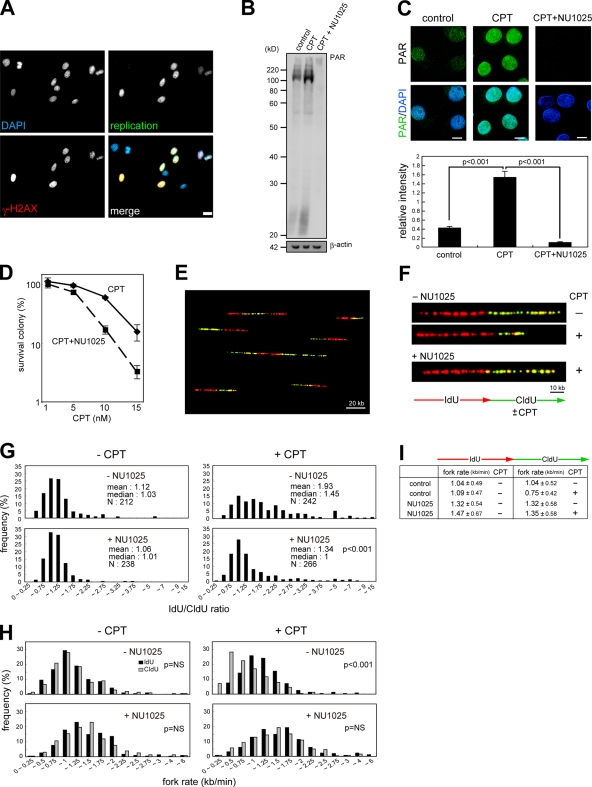
To investigate the involvement of PARP-1 in modulation of fork progression on DNA damaged with CPT, we treated cells with CPT in combination with the PARP inhibitor NU1025. Before analysis of replication fork kinetics, we evaluated poly-ADP ribosylation activity after CPT treatment and the inhibitory effect of NU1025. CPT-induced DNA damage is largely replication dependent. In fact, γ-H2AX foci were detected only in the presence of CPT in S-phase nuclei (Fig. 2 A). First, we performed Western blotting to detect poly-ADP ribosylated proteins using FITC-NAD+ as a substrate. Compared with untreated cells, the poly-ADP ribosylation level in CPT-treated cells was higher over a wide size range of proteins (Fig. 2 B). NU1025 treatment led to the disappearance of poly-ADP ribosylated proteins. Using fluorescence microscope analysis, we confirmed that poly-ADP ribosylation was inhibited by NU1025 in the presence of CPT and found that NU1025 sensitized cells to CPT (Fig. 2, C and D).
Figure 2.
A PARP inhibitor abrogates CPT-induced replication fork slowing. (A) m5S cells were labeled with digoxigenin–deoxy-UTP and release cultured in the presence of CPT. Merged images show that γ-H2AX are detected only in S-phase nuclei. (B) Cells were pretreated with NU1025 before CPT treatment and incubated with FITC-NAD+ and CPT for 15 min as described in Materials and methods. β-Actin was used as a loading control. (C) Fluorescent microscope images of poly-ADP ribose in cells were treated as indicated. Fluorescence intensities of each nucleus (>50 cells) were quantified. (D) Sensitivity of NU1025-treated cells to CPT. The mean of three independent experiments and standard deviation are shown. (E) A typical global image of DNA fibers. HeLa cells were pulse labeled with IdU (red) and sequentially with CldU (green). After molecular combing, replicated DNA was immunodetected as described in Materials and methods. (F) The images show the typical DNA fibers in cells treated with the indicated inhibitors. Cells were pretreated with NU1025 and pulse labeled with IdU for 20 min. After washing in PBS, cells were treated with CldU in the absence or presence of CPT for 20 min. (G) Distribution of the ratio of the rate of fork progression in cells treated as indicated. The p-value of the Kolmogorov-Smirnov test for the ratio distribution of NU1025-treated cells for CPT treatment compared with NU1025-untreated cells is shown. (H) Distribution of the rate of fork progression during IdU and CldU pulse labeling in each cell. The p-value of the Wilcoxon signed-rank test for the distribution of fork rate in each cell is shown. (I) The tabular data are mean fork rates for each cell treated as indicated. The mean rates were calculated from the data described in H. Bars, 10 kb.
We hypothesized that the modulation of replication fork progression is one of the critical steps of DNA damage repair for replication-coupled DSBs. To measure the rate of fork progression, HeLa cells were pulse labeled for 20 min with IdU followed by CldU for 20 min in the presence or absence of CPT. After genomic DNA preparation and dynamic molecular combing, IdU- and CldU-labeled DNA were immunostained with each appropriate antibody. This allows use of the first label, which is always incorporated under normal circumstances, to normalize the second-label fork rate for differences in fork rates that are intrinsic to different forks. Fig. 2 (E and F) shows typical images of replicating DNA in cells treated as indicated. Fig. 2 G shows the histogram of the distribution of the ratio of fork rates during first and second pulse label. Saturating amounts of CPT were added during CldU labeling. The mean ratio in CPT-treated HeLa cells was about twofold higher compared with untreated cells. If CPT-induced DSBs decrease the rates of replication fork progression in a PARP-1–dependent manner, a PARP inhibitor should affect the replication fork progression kinetics. Interestingly, a similar mean ratio was observed in NU1025-treated cells in spite of CPT treatment during CldU pulse label. These results indicated that NU1025 counteracted the fork slowing induced by CPT. This result was confirmed in another cell line, m5S (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200806068/DC1). These results were also evident when the distribution of fork rates during IdU and CldU labeling was quantified and plotted. Compared with the fork rates during IdU labeling, the entire distribution of fork rates shifted to slower fork rates during CldU labeling after CPT addition, and this decrease was reversed by NU1025 (Fig. 2 H). These results suggest that PARP activity could be responsible for lower rates of fork progression in response to CPT-induced DNA damage in mammalian cells.
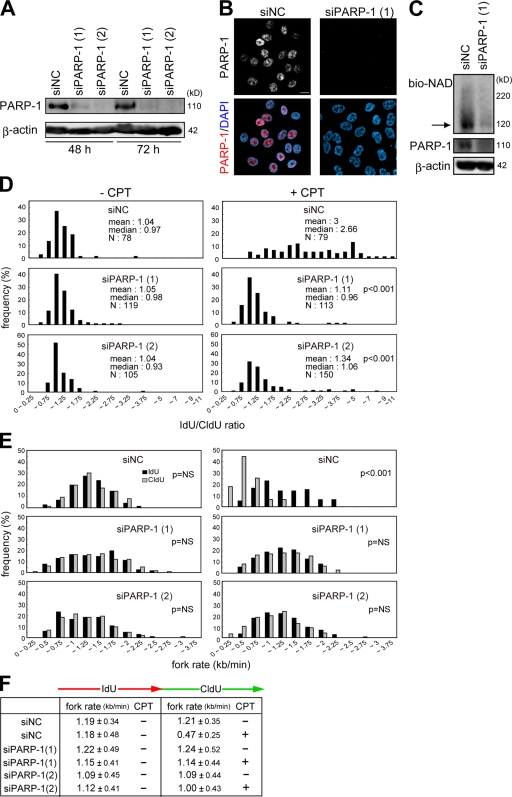
To further evaluate the effect of PARP on replication forks, PARP-1 was reduced with a PARP-1–specific siRNA. PARP-1 was effectively decreased in HeLa cells at 48 and 72 h after transfection of two individual siRNAs (Fig. 3, A and B). Compared with cells transfected with the negative control siRNA, the poly-ADP ribosylation level was decreased to the background level (Fig. 3 C). The ratio of fork rates during IdU and CldU labeling was about twofold higher in cells transfected with PARP-1–specific siRNA compared with those with the negative control siRNA in the presence of CPT (Fig. 3 D). Consistent with this result, the entire distribution of fork rates during CldU labeling shifted to faster fork rates in the presence of CPT in those cells transfected with the PARP-1 siRNAs compared with cells transfected with negative control siRNA (Fig. 3 E). These results indicate that PARP-1 can affect replication fork progression on a damaged chromosome caused by CPT.
Figure 3.
Effects of PARP-1 knockdown on kinetics of replication forks. (A and B) Evaluation of PARP-1 knockdown in HeLa cells. The amount of PARP-1 in cells transfected with two different siRNA duplexes against PARP-1 and negative control siRNA (siNC) was evaluated by Western blotting (A) and immunofluorescence (B). β-Actin was used as a loading control. (C) Poly-ADP ribosylated proteins were detected as shown in Fig. 1 B. The arrow indicates the molecular mass of PARP-1. (D) Distribution of the ratio of the rate of fork progression in cells transfected with each siRNA. (E) Distribution of the rate of fork progression during IdU and CldU pulse labeling in cells transfected with each siRNA. (F) The tabular data are mean fork rates for each cell treated as indicated. The mean rates were calculated from the data described in E. Bar, 10 μm.
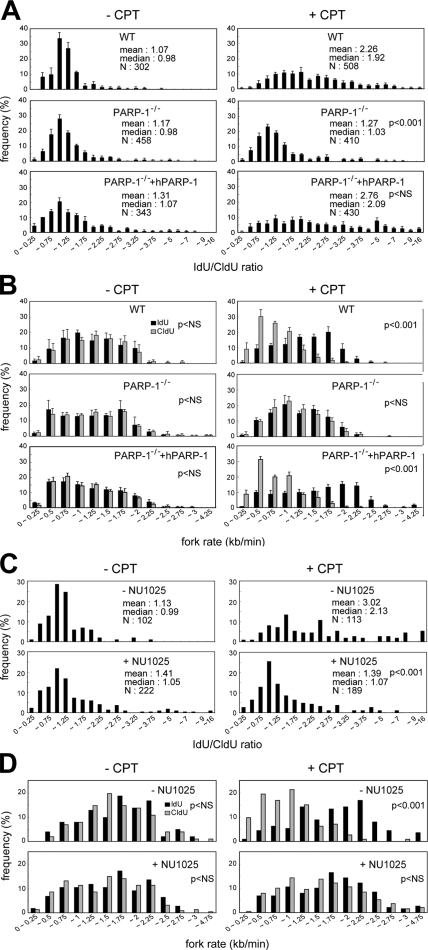
Chicken DT40 cells are useful for the analysis of gene functions caused by the abundant stocks of mutants. Chicken cells appear to lack a PARP-2 homologue, which is another repair-related PARP, so PARP-1−/− DT40 cells could have no repair-associated poly-ADP ribosylation activity and, therefore, are hypersensitive to CPT as well as other DSB-inducible reagents (Hochegger et al., 2006). Thus, we analyzed the involvement of PARP-1 in fork progression on damaged DNA using PARP-1−/− DT40 cells. The mean ratio of fork rates in CPT-treated wild-type cells was about twofold higher compared with untreated cells, whereas a similar mean ratio was observed in PARP-1−/− cells in spite of CPT treatment during CldU pulse labeling (Fig. 4 A). Quantification of the mean fork rate revealed that CPT treatment did not induce fork slowing in PARP-1−/− cells, and the rate of fork progression was similar in the absence of CPT (Fig. 4 B). NU1025 treatment also recovered the CPT-induced slowing of fork progression in DT40 cells (Fig. 4, C and D). If CPT-induced replication fork slowing is dependent on PARP-1 activity, additional expression of PARP-1 should restore the impact of CPT on replication fork progression. Therefore, we measured the rate of fork progression in PARP-1−/− DT40 cells stably expressing human PARP-1. Strikingly, human PARP-1 completely restored the kinetics of fork progression in the presence of CPT in PARP-1−/− cells (Fig. 4, A and B). These results clearly indicate that PARP-1 can affect the replication fork progression in response to CPT-induced DNA damage.
Figure 4.
Replication fork progression in PARP-1−/− DT40 cells. (A) Distribution of the ratio of the rate of fork progression in wild-type (WT), PARP-1−/− DT40, and PARP-1−/− + human PARP-1 (hPARP-1) DT40 cells. (B) Distribution of the rate of fork progression during IdU and CldU pulse labeling in each cell. (C and D) Distribution of the ratio of the rate of fork progression and the rate of fork progression during IdU and CldU pulse labeling in wild-type DT40 cells pretreated with NU1025. Data bars are the means of three independent experiments, and error bars represent SEM.
It is possible that the CPT-induced fork slowing is caused by PARP-1–induced repair activities because PARP plays a role in damage response to CPT in the following two manners: first, CPT stabilizes Topo I–DNA covalent complexes, and resulting SSBs are repaired by PARP, XRCC1, and DNA polymerase β (Plo et al., 2003; El-Khamisy et al., 2005). Second, Topo I–DNA covalent complexes that encounter replication forks lead to the formation of DSBs. PARP may suppress the toxic effect of NHEJ-mediated repair, which may result in chromosomal translocation and thereby indirectly facilitate accurate HR-mediated DSB repair (Hochegger et al., 2006).
To address a role of PARP-1 in SSB repair (SSBR), we analyzed the rate of fork progression in polymerase β–deficient DT40 cells, but CPT-induced fork slowing was not reversed in polymerase β−/− cells (Fig. S2, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200806068/DC1). To confirm these results in mammalian cells, HeLa cells in which another SSBR factor, XRCC1, was knocked down by siRNA were used. The protein level of XRCC1 was significantly reduced at 48 or 72 h after siRNA transfection (Fig. S2 C). Similar to polymerase β−/− DT40 cells, CPT-induced fork slowing was not recovered in XRCC1-depleted HeLa cells (Fig. S2, D–F), suggesting that PARP-1 regulates replication fork progression in response to CPT-induced DNA damage independently of its SSBR-related activity.
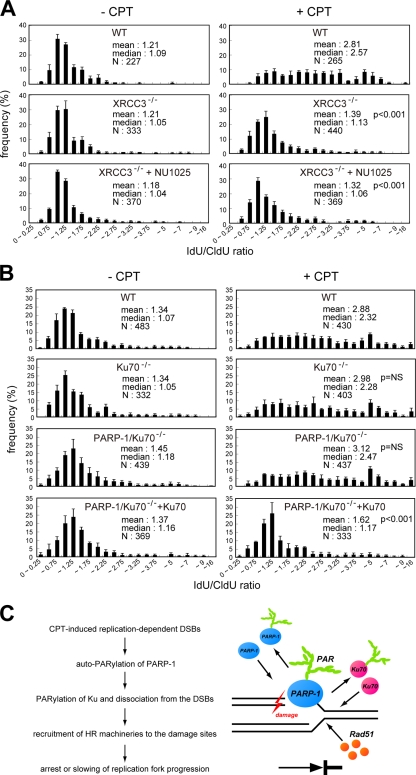
We hypothesized that a PARP-1–related DSB repair pathway is responsible for CPT-induced fork slowing. To address this hypothesis, we tested two DT40 mutants in DNA DSB repair pathways, XRCC3−/− mutant cells that are defective in HR and Ku70−/− mutants that are defective in NHEJ. Interestingly, slowing of replication fork progression occurred at the same levels as wild type in the Ku70−/− single mutant but was clearly reversed in XRCC3−/− cells (Fig. 5 and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200806068/DC1). No effect of NU1025 was observed in XRCC3−/− cells. Similar results were observed in other HR-defective BRCA2tr/− DT40 cells (Fig. S3, C and D; Hatanaka et al., 2005). These results suggest that replication forks slow down in response to CPT as a result of the activation of the HR pathway.
Figure 5.
Recovery of the lower rates of fork progression in the presence of CPT in PARP-1 mutant is not observed under the deficiency of NHEJ capacity. (A and B) Distribution of the ratio of the rate of fork progression during IdU and CldU pulse labeling in each indicated DT40 cell. The total number of the forks analyzed in each cell is also indicated. (C) Model illustrating how PARP-1 affects replication fork progression on damaged DNA. Data bars are the means of three independent experiments, and error bars represent SEM. WT, wild type.
Previously, Hochegger et al. (2006) showed that PARP-1−/− DT40 cells had reduced levels of HR, and concomitant deletion of Ku70 in these PARP-1−/− cells reestablished their ability to perform HR. From the results that CPT-induced fork slowing is HR dependent, we speculated that the effect of PARP-1 deletion on the replication fork could be reversed by concomitant deletion of Ku. Indeed, using PARP-1/Ku70−/− DT40 cells, we found that deletion of Ku reversed the effects of the PARP-1 mutant on the replication fork progression (Fig. 5 B). Thus, the entire distribution of fork rates during CldU labeling shifted to slower fork rates in the presence of CPT in the PARP/Ku double knockout cells (Fig. S3). To confirm the dependency of the PARP-1 phenotype on Ku, we investigated the rate of fork progression using PARP-1/Ku70−/− cells that stably expressed exogenous Ku70. As expected, the fork rates did not decrease after CPT addition, which is similar to the PARP-1 single mutant (Fig. 5 B and Fig. S3). Collectively, these results suggest that progression of the replication fork can be modulated by the HR pathway when replication forks collide with CPT-induced DNA damage. This activity clearly depends on PARP-1, which seems to function as a safeguard against the NHEJ pathway on stalled replication forks.
In this study, we found that PARP-1 can slow replication forks in response to DNA damage induced by CPT. We focused on the PARP-1–related repair pathways and found that the HR-dependent repair pathway may be involved in the modulation of fork progression on damaged templates in vertebrate cells (Fig. 5 C). Previous studies indicated that PARP-1 interacts with the Ku–DNA-PK complex that is required for the NHEJ pathway (Ariumi et al., 1999; Galande and Kohwi-Shigematsu, 1999; Li et al., 2004), and PARP-1 poly-ADP ribosylates Ku, decreasing the binding affinity of Ku to DSBs (Li et al., 2004). After these observations, Hochegger et al. (2006) suggested a biological role of the interaction of PARP-1 and NHEJ factors, proposing that PARP-1 minimized the suppressive effect of Ku and the NHEJ pathway on HR after replication fork collapse. Consequently, PARP-1 deficiency could result in reduced efficiency of HR because of the presence of Ku, resulting in enhanced progression of the replication fork. A previous study supports this idea, which found that replication fork arrest or slowing on damaged chromosomes was induced in an HR-dependent manner (Henry-Mowatt et al., 2003). Therefore, the reason why concomitant deletion of Ku70 in PARP-1−/− mutants resulted in fork slowing in response to CPT is likely to be reestablishment of HR-mediated DSB repair processes.
We think the reason why fork rates in PARP-1−/− cells were the same as those of wild-type cells in the presence of CPT because of the following points. A mutant of the checkpoint protein, Hus1, showed normal replication fork rates in the presence of CPT (Wang et al., 2004). This mutant is also deficient in the HR process (Wang et al., 2006). Intriguingly, PCNA dissociates from chromatin in response to CPT treatment in wild-type cells but not in the Hus1 mutant, indicating that restoration of chromatin binding of PCNA appears to be associated with a normal rate of fork progression in the presence of CPT. However, another study reported that Ku facilitated the association of PCNA to chromatin in response to DSBs (Park et al., 2004), so it is conceivable that restoration of chromatin binding of PCNA by Ku associates with normal fork rates on damaged DNA in PARP-1 or HR-defective cells. Indeed, we found that the chromatin-bound PCNA decreased in response to CPT but not in the presence of NU1025 (unpublished data). Heller and Marians (2006) showed that replication could be restarted and that leading-strand synthesis reinitiated downstream of an unrepaired block to leading-strand progression. If this occurs downstream of a DSB, de novo loading of PCNA by Ku might facilitate the restart. Further in-depth investigations are required to test this hypothesis.
We also suggest an alternative plausible model. Recently, the molecular details of a cytotoxic mechanism for CPT were suggested (Koster et al., 2007). They suggested that the accumulation of positive supercoils was induced by CPT ahead of the replication machinery and that DNA damages were induced within the supercoil. If PARP-1 or HR is deficient, Ku-mediated chromosomal translocation might occur within the supercoil. Presumably, the replication fork could then move on the translocated chromosome. However, we do not have any evidence to support this hypothesis.
In summary, we have shown that PARP-1 activity influences replication fork progression when replication forks collapse at CPT-induced single-strand gaps. The target of PARP-1 activity in this novel mechanism appears to be Ku, which interferes with HR-dependent DSB repair at the collapsed replication fork.
Materials and methods
Cell culture and drug treatment
HeLa cells, COS-7 cells, and m5S (Sasaki and Kodama, 1987) were grown in DME supplemented with 10% FBS at 37°C with 5% CO2. DT40 cells were grown in RPMI 1640 medium supplemented with 10% FBS, 1% chicken serum, and 10 μM β-mercaptoethanol at 39°C with 5% CO2.
Antibodies
Mouse anti-BrdU antibody was purchased from BD. Rat anti-BrdU antibodies were purchased from Oxford Biotechnology or Abcam. Mouse antiphosphohistone H2A.X (Ser139) antibody was purchased from Millipore. Mouse anti–PARP-1 and mouse anti–poly-ADP ribose antibodies were purchased from Trevigen. Goat anti–PARP-1 antibody was purchased from R&D Systems. Mouse anti–β-actin antibody was purchased from Sigma-Aldrich. Rhodamine-conjugated sheep antidigoxigenin antibody was purchased from Roche.
Colony formation assay
200 spontaneously growing HeLa cells were plated into each well of a 6-well dish. 24 h later, CPT (Topogen) was added at various concentrations to the medium in the presence or absence of 200 μM NU1025 (EMD). Cells were washed with PBS 22 h later and cultured in 10% FBS-containing medium for 6 or 7 d. Colonies consisting of >10 cells were counted.
Biochemical detection of poly-ADP ribose
Cells were grown on a coverslip (Matsunami Glass) and incubated for 1 h at 37°C in 56 mM Hepes, pH 7.5, 28 mM NaCl, and 0.01% digitonin in the presence or absence of 200 μM NU1025, and 25 μM FITC-NAD+ (Trevigen) was added with or without 10 μM CPT (Topogen). After 15 min, cells were fixed with ice-cold methanol/acetone (1:1) for 10 min. Fixed cells were counterstained with DAPI. For immunoblotting, 105 cells were lysed in 10 μl of 1× SDS gel loading buffer, and the lysate was loaded onto a 12% SDS-PAGE gel, transferred to nitrocellulose membrane (Biodyne), and detected with the mouse anti–FITC-HRP (1:2,500; Jackson ImmunoResearch Laboratories). Chemiluminescent signals were detected by the ECL system (GE Healthcare).
Plasmid constructs, transfection, and immunodetection of BrdU-labeled DNA
Expression constructs for PARP-1–EYFP and Topo I–DsRed were provided by M.S. Satoh (Laval University, Quebec City, Quebec, Canada), and expression construct PCNA-RFP was provided by M.C. Cardoso (Technische Universität Darmstadt, Darmstadt, Germany). Cells were plated in a 24-well plate containing 13-mm round coverslips (Thermo Fisher Scientific) or in a 6-well plate containing square coverslips (Matsunami Glass). After 24 h, 100 ng/ml expression constructs PARP-1–EYFP and 100 ng/ml Topo I–DsRed were transfected with Lipofectamine and Plus Reagent (Invitrogen) according to the manufacturer's manual. However, expression construct PCNA-RFP (20 ng/ml to COS-7 cells and 8 ng/ml to HeLa cells) was transfected instead of expression construct Topo I–DsRed. The cells were cultured for 24 h, pulse labeled with 25 μM BrdU (Sigma-Aldrich) for 5 min, and fixed in 4% PFA (Nacalai Tesque) in PBS at 25°C for 10 min. After washing in PBS, the cells were permeabilized in 0.5% Triton X-100 in PBS at 25°C for 10 min, washed three times in PBS, and incubated in blocking buffer (0.3% BSA and 0.02% Tween 20 in PBS) at 25°C for 10 min and in 50 μl of detection buffer (0.15% BSA, 0.01% Tween 20, 30 mM Tris-HCl, pH 8.0, 300 μM MgCl2, 1 mM 2-mercaptoethanol, and 0.5× PBS) containing the nucleases and the antibodies (20 μg/ml DNase I [Sigma-Aldrich], 700 U/ml Exonuclease III [Toyobo], and mouse monoclonal antibodies directed against BrdU; 1:100 [MBL International]) at 37°C for 30 min. After washing three times in PBST (0.1% Tween 20 in PBS) for 5 min, the cells were incubated in Alexa Fluor 647–conjugated goat anti–mouse IgG (1:250; Invitrogen) at 37°C for 30 min, washed three times in PBST, and counterstained with DAPI (20 ng/ml in PBS; Nacalai Tesque).
RNAi
10 nM siRNAs (Thermo Fisher Scientific) were transfected to 105 HeLa cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Cells were cultured for 48 or 72 h before preparation of total cellular proteins or replication labeling. The nucleotide sequences of the siRNA target sites in PARP-1 were as follows: siRNA against PARP-1 (a), 5′-AAGCCTCCGCTCCTGAACAAT-3′ (nt 2,401–2,421); siRNA against PARP-1 (b), 5′-AAGATAGAGCGTGAAGGCGAA-3′ (nt 2,671–2,691; Kameoka et al., 2004); and siRNA against XRCC1, 5′-AACUCGACUCACUGUGCAGAA-3′ (nt 151–171; Luo et al., 2004). Cell lysates were subjected on an 8 or 12% SDS-PAGE gel, transferred to nitrocellulose membrane, and detected with mouse anti–PARP-1 antibody (Trevigen) or β-actin (Sigma-Aldrich) as a control. After incubation in HRP-conjugated goat anti–mouse secondary antibody (MBL International), signals were enhanced by chemiluminescence with the ECL system (GE Healthcare). Cells grown on coverslips were fixed for immunofluorescence using the mouse anti–PARP-1 antibody and Alexa Fluor 488–conjugated goat anti–mouse IgG (Invitrogen).
Dynamic molecular combing and immunofluorescent detection
Genomic DNA was prepared and combed onto the silanated coverslips (Matsunami Glass) as described previously (Michalet et al., 1997) with modification (see following paragraph). 4 × 106 DT40 cells were pulse labeled for 20 min with 25 μM IdU (Sigma-Aldrich) and sequentially pulse labeled for 20 min with 250 μM CldU (Sigma-Aldrich) with or without 1 μM CPT. 2 × 106 m5S and HeLa cells were pulse labeled for 20 min with 100 μM IdU, washed with PBS twice, and pulse labeled for 20 min with 100 μM CldU with or without 1 μM CPT. NU1025 was pretreated for 2 h before IdU labeling. For preparation of genomic DNA, to remove the mitochondrial genome, the nuclei were extracted with buffer A (250 mM sucrose, 20 mM Hepes, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, pH 8.0, 1 mM EGTA, pH 6.8, 1 mM DTT, and 0.1 mM PMSF) before resuspension into low melting point agarose (Roche). Combed DNA molecules were heat denatured in 50% formamide (Roche) and 2× SSC at 72°C for 12 min. For immunodetection of IdU- and CldU-labeled DNA, denatured DNA molecules were incubated with mouse anti-BrdU monoclonal antibody (1:5; BD) and rat anti-BrdU monoclonal antibody (1:25; Oxford Biotechnology) for 1 h at 37°C. After washing with PBS and 0.05% Tween 20 for 5 min three times, they were incubated with Alexa Fluor 555–conjugated goat anti–mouse IgG (1:500; Invitrogen) and Alexa Fluor 488–conjugated rabbit anti–rat IgG (1:500; Invitrogen) for 30 min at 37°C. All antibodies were diluted in blocking solution (1% [wt/vol] blocking reagent in PBS and 0.05% Tween 20; Roche). After washing with PBS and 0.05% Tween 20 for 5 min three times, coverslips were mounted in VECTASHIELD (Vector Laboratories). To estimate the extension of DNA molecules, coverslips were prepared with λ-DNA, and the DNA molecules were stained with 6.7 mM YOYO-1 (Invitrogen) at 25°C for 1 h. YOYO-1–stained DNA molecules measured 21 ± 0.9 μm. As the virus genome is 48.5 kbp, the extension of DNA molecules is 2.32 ± 0.11 kbp/μm.
Microscopy
Nuclei were examined using a microscope (Axioplan 2 MOT; Carl Zeiss, Inc.) with a 63× Plan Apochromat NA 1.4 objective lens equipped with a charge-coupled device camera (MicroMAX; Princeton Instruments). For deconvolution analysis of the nucleus, z-stack images were captured at a step of 0.2 μm and processed using MetaMorph software (version 6.1; MDS Analytical Technologies).
Online supplemental material
Fig. S1 shows the effect of PARP inhibitor on replication fork progression in mouse m5S cells. Fig. S2 shows replication fork kinetics in HeLa cells treated with XRCC1 siRNA. Fig. S3 shows replication fork kinetics in BRCA2tr/− DT40 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806068/DC1.
Supplementary Material
Acknowledgments
We thank M.S. Satoh, M.C. Cardoso, and N. Aoki for materials, T. Saito, T. Kurokawa, K. Sakai, and A. Kohda for setup of molecular combing, H. Hochegger, J. Huberman, and K. Shirahige for valuable discussion and critical reading of the manuscript, and Y. Tsuji and K. Kagotani for encouragement.
This work was supported partly by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science (grant to K. Okumura and K. Sugimura), and the Sankyo Foundation of Life Science (grant to K. Okumura). S.-i. Takebayashi is supported by the Nakayama Foundation for Human Science.
Abbreviations used in this paper: CPT, camptothecin; DSB, double-strand break; EYFP, enhanced YFP; HR, homologous recombination; NHEJ, nonhomologous end joining; PARP, poly-ADP ribose polymerase; PCNA, proliferating cell nuclear antigen; SSB, single-strand break; SSBR, SSB repair; Topo I, topoisomerase I.
References
- Ariumi, Y., M. Masutani, T.D. Copeland, T. Mimori, T. Sugimura, K. Shimotohno, K. Ueda, M. Hatanaka, and M. Noda. 1999. Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene. 18:4616–4625. [DOI] [PubMed] [Google Scholar]
- Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235–1245. [DOI] [PubMed] [Google Scholar]
- Bowman, K.J., D.R. Newell, A.H. Calvert, and N.J. Curtin. 2001. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br. J. Cancer. 84:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott, K.W., S. Aoufouchi, P. Johnson, and S. Shall. 1996. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 24:4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S., M.F. Cheng, D. Trivedi, S.J. Petzold, and N.A. Berger. 1989. Camptothecin hypersensitivity in poly(adenosine diphosphate-ribose) polymerase-deficient cell lines. Cancer Commun. 1:389–394. [DOI] [PubMed] [Google Scholar]
- Dantzer, F., H.P. Nasheuer, J.L. Vonesch, G. de Murcia, and J. Menissier-de Murcia. 1998. Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res. 26:1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy, S.F., G.M. Saifi, M. Weinfeld, F. Johansson, T. Helleday, J.R. Lupski, and K.W. Caldecott. 2005. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 434:108–113. [DOI] [PubMed] [Google Scholar]
- Essers, J., H. van Steeg, J. de Wit, S.M. Swagemakers, M. Vermeij, J.H. Hoeijmakers, and R. Kanaar. 2000. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19:1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frouin, I., G. Maga, M. Denegri, F. Riva, M. Savio, S. Spadari, E. Prosperi, and A.I. Scovassi. 2003. Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1. A dynamic exchange of partners. J. Biol. Chem. 278:39265–39268. [DOI] [PubMed] [Google Scholar]
- Galande, S., and T. Kohwi-Shigematsu. 1999. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274:20521–20528. [DOI] [PubMed] [Google Scholar]
- Hatanaka, A., M. Yamazoe, J.E. Sale, M. Tanaka, K. Yamamoto, H. Kitano, E. Sonoda, K. Kikuchi, Y. Yonetani, and S. Takeda. 2005. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol. Cell. Biol. 25:1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller, R.C., and K.J. Marians. 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 439:557–562. [DOI] [PubMed] [Google Scholar]
- Henry-Mowatt, J., D. Jackson, J.Y. Masson, P.A. Johnson, P.M. Clements, F.E. Benson, L.H. Thompson, S. Takeda, S.C. West, and K.W. Caldecott. 2003. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell. 11:1109–1117. [DOI] [PubMed] [Google Scholar]
- Hochegger, H., D. Dejsuphong, T. Fukushima, C. Morrison, E. Sonoda, V. Schreiber, G.Y. Zhao, A. Saberi, M. Masutani, N. Adachi, et al. 2006. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25:1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka, M., S. Nukuzuma, A. Itaya, Y. Tanaka, K. Ota, K. Ikuta, and K. Yoshihara. 2004. RNA interference directed against Poly(ADP-ribose) polymerase 1 efficiently suppresses human immunodeficiency virus type 1 replication in human cells. J. Virol. 78:8931–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster, D.A., K. Palle, E.S. Bot, M.A. Bjornsti, and N.H. Dekker. 2007. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 448:213–217. [DOI] [PubMed] [Google Scholar]
- Leppard, J.B., Z. Dong, Z.B. Mackey, and A.E. Tomkinson. 2003. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell. Biol. 23:5919–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., S. Navarro, N. Kasahara, and L. Comai. 2004. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J. Biol. Chem. 279:13659–13667. [DOI] [PubMed] [Google Scholar]
- Luo, H., D.W. Chan, T. Yang, M. Rodriguez, B.P. Chen, M. Leng, J.J. Mu, D. Chen, Z. Songyang, Y. Wang, and J. Qin. 2004. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell. Biol. 24:8356–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet, X., R. Ekong, F. Fougerousse, S. Rousseaux, C. Schurra, N. Hornigold, M. van Slegtenhorst, J. Wolfe, S. Povey, J.S. Beckmann, and A. Bensimon. 1997. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 277:1518–1523. [DOI] [PubMed] [Google Scholar]
- Park, S.J., S.L. Ciccone, B. Freie, A. Kurimasa, D.J. Chen, G.C. Li, D.W. Clapp, and S.H. Lee. 2004. A positive role for the Ku complex in DNA replication following strand break damage in mammals. J. Biol. Chem. 279:6046–6055. [DOI] [PubMed] [Google Scholar]
- Plo, I., Z.Y. Liao, J.M. Barcelo, G. Kohlhagen, K.W. Caldecott, M. Weinfeld, and Y. Pommier. 2003. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst.). 2:1087–1100. [DOI] [PubMed] [Google Scholar]
- Pommier, Y., C. Redon, V.A. Rao, J.A. Seiler, O. Sordet, H. Takemura, S. Antony, L. Meng, Z. Liao, G. Kohlhagen, et al. 2003. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 532:173–203. [DOI] [PubMed] [Google Scholar]
- Sasaki, M.S., and S. Kodama. 1987. Establishment and some mutational characteristics of 3T3-like near-diploid mouse cell line. J. Cell. Physiol. 131:114–122. [DOI] [PubMed] [Google Scholar]
- Takata, M., M.S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., J. Guan, B. Hu, R.S. Weiss, G. Iliakis, and Y. Wang. 2004. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res. 32:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang, X., B. Hu, R.S. Weiss, and Y. Wang. 2006. The effect of Hus1 on ionizing radiation sensitivity is associated with homologous recombination repair but is independent of nonhomologous end-joining. Oncogene. 25:1980–1983. [DOI] [PubMed] [Google Scholar]
- Yung, T.M., S. Sato, and M.S. Satoh. 2004. Poly(ADP-ribosyl)ation as a DNA damage-induced post-translational modification regulating poly(ADP-ribose) polymerase-1-topoisomerase I interaction. J. Biol. Chem. 279:39686–39696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.