Abstract
The prevailing view that eukaryotic cells are restrained from intercellular exchange of genetic information has been challenged by recent reports on nanotubes, exosomes, apoptotic bodies, and nucleic acid–binding peptides that provide novel pathways for cell–cell communication, with implications in health and disease.
Introduction
Since the discovery of genetic transformation of pneumococci more than 60 years ago, the mechanisms of horizontal transfer of genetic material in bacteria have been firmly established (Chen and Dubnau, 2004). With the development of higher eukaryotes, the intercellular exchange of nucleic acid was restrained; however, throughout their evolution, plasma membrane carrier mechanisms have been conserved that allow viruses and other microbes to incorporate foreign genetic material and to cause deadly infectious and inflammatory diseases and cancer. Mechanistic studies of viral infection strategies have prepared the way for the development of viral as well as nonviral vectors for the transfer of therapeutic genetic material into human cells; such strategies are currently being tested in the clinical setting.
Recent reports from partly unrelated fields of cell biology research convey the intriguing idea that intercellular exchange of endogenous genetic material may occur through several distinct pathways in multicellular organisms. Here, we will discuss the potential role of nanotubes, exosomes, apoptotic bodies, and nucleic acid–binding peptides in the intercellular transfer of genetic information. In this context, the discoveries of systemic cosuppression in plants (Voinnet et al., 1998) and systemic RNA silencing (RNAi) in Caenorhabditis elegans (Fire et al., 1998), which hint toward a physiological role of nucleic acid transfer in eukaryotes, are of particular interest.
Systemic RNAi
A decade ago, it was reported that double-stranded RNA (dsRNA) injected into or fed to Caenorhabditis elegans could trigger systemic silencing of complementary transcripts throughout the recipient adult animal as well as in its progeny (Fire et al., 1998). In mutational screens, several genes involved in this process have been identified (Winston et al., 2002; Tijsterman et al., 2004). The most well-studied of these genes, sid-1 (systemic silencing deficient-1), encodes an 11-pass transmembrane protein (Winston et al., 2002) that acts as a membrane pore to mediate dsRNA transport over the plasma membrane (Feinberg and Hunter 2003). More recently, SID-2 was described to act as a single-pass transmembrane receptor for dsRNA endocytosis/transcytosis in intestinal epithelial cells of C. elegans (Winston et al., 2007). Sid-1 homologues have been found in humans and other mammals, though not in Drosophila melanogaster. An RNAi response can still be induced in cultured D. melanogaster S2 cells by supplying extracellular dsRNA (Clemens et al., 2000), which suggests the existence of several, potentially evolutionary distinct, pathways for systemic silencing signals. An RNAi screen to identify components necessary for exogenous dsRNA-triggered RNAi in S2 cells yielded several genes of functional importance, including components of the endocytic pathway (Saleh et al., 2006). Interestingly, internalization of cholesterol-conjugated siRNA into mouse liver cells was recently shown to involve the concerted effects of receptor-mediated endocytosis and the activity of one of the SID-1 mouse homologues (Wolfrum et al., 2007). Cholesterol-conjugated siRNA, when given intravenously, was incorporated into lipoprotein particles, which were subsequently endocytosed, making the siRNA available for SID-1–mediated import into the cytosol.
The exact molecular nature of the systemic silencing signal in both plants and C. elegans is ill-defined. Whether the hypothetical, systemically transmitted RNAs are long dsRNAs or short siRNAs, “naked” or bound to peptides or proteins, enclosed in membrane vesicles or something altogether different is a central question that remains to be answered. We will next discuss some studies that provide important clues as to how nucleic acids may be transferred between cells locally and at the systemic level.
Intercellular plasma membrane connections
The recently discovered membrane or tunneling nanotubes (TNTs) are exceedingly thin protrusions up to several micrometers long that can connect cells from several cell diameters apart. TNTs were originally described in cultured rat pheochromocytoma PC12 cells (Rustom et al., 2004) and immune cells (Onfelt et al., 2004), and were, in the former study, characterized as 50–200-nm wide actin-containing stretched tubes that provide membrane continuity between connected cells. Later, it was shown that cultured human macrophages exhibited two distinct types of TNTs (Onfelt et al., 2006): a thin, <0.7-μm actin-containing tube that supported unidirectional movement of the plasma membrane constituents, including surface attached pathogens; and a wider, >0.7-μm microtubule-containing tube that supported bidirectional transport of vesicles and organelles, e.g., endosomes and mitochondria. The actin-containing protrusions, termed cytonemes in the D. melanogaster wing imaginal disc, may represent a subclass of TNTs (Ramirez-Weber and Kornberg 1999). The first in vivo evidence of nanotubes in mammalian tissue was recently shown in the cornea (Chinnery et al., 2008).
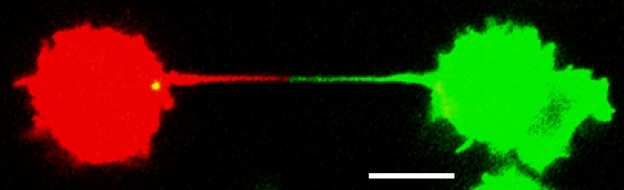
It was recently demonstrated that TNTs constitute a transmission route for HIV-1 particles between cultured Jurkat T cells (Sowinski et al., 2008). In this case, the TNTs were not continuous, as the connected cells were separated by a junction (Fig. 1). Regarding the possible natural cargos that use TNTs, much attention has been given to the transport of vesicles of endosomal origin (Rustom et al., 2004; Onfelt et al., 2006). This is of particular interest in the context of intercellular transfer of RNAs, given the tentative involvement of endosomal pathways in the transmission of RNAi in lower organisms. However, so far, there are no reports of the shuttling of an endogenous or experimentally overexpressed RNA between cells through this pathway. This has indeed been demonstrated to occur through so called plasmodesmata, which are involved in direct cell-to-cell communication in plants. Initial work showed that plant viruses encode movement proteins that mediate infectious spread of viral nucleic acids via plasmodesmata, which further led to the discovery that endogenous plant proteins, including transcription factors, use the same pathway to traffic between cells. Lucas et al. (1995) went on to show that the protein encoded by the maize knotted1 homeobox gene could selectively transfer its own mRNA to surrounding cells through plasmodesmata, providing early evidence of intercellular RNA transfer in higher eukaryotes (Lucas et al., 1995). From a developmental point of view, these findings are important, as they imply that cell fate is determined by position rather than by cell lineage. However, these studies were based on microinjection of fluorophore-labeled KNOTTED1 protein and mRNA into mesophyll donor cells; i.e., knotted1 mRNA may not be transferred by the same mechanism under physiological conditions. Nevertheless, RNA transfer was specific, as knotted1 antisense RNA as well as mRNA of an unrelated protein was poorly transferred to surrounding cells.
Figure 1.
TNT-connecting cells. Jurkat T cells differentially labeled with the membrane dyes DiO (green) and DiL (red) were cocultured. The connecting TNT contains a junction, as shown by the distinct separation of the membrane dyes. Other nanotubes have, however, been shown to mediate membrane continuity between cells. Reprinted by permission from Macmillan Publishers Ltd., Nature Cell Biology (copyright 2008; Sowinski et al., 2008), and D.M. Davis (Imperial College, London, England, UK). Bar, 10 μm.
Microvesicles and exosomes
Intriguingly, RNA-dependent RNA polymerases (RdRPs) found in C. elegans and plants are absent in the mammalian and insect genomes. RdRPs act as amplifiers of the RNAi effect, which allow a few short dsRNA stretches to trigger efficient knockdown of complementary single-stranded sequences. The potential dependence on RdRPs in systemic silencing in lower eukaryotes (Himber et al., 2003) would imply that a putative RNAi-based regulatory system in mammals is restricted to paracrine signaling events highly dependent on the local concentration of siRNA. However, an appealing yet hypothetical possibility of long-distance signaling using mRNA or siRNA mediators is the packaging of endogenous RNA species into exocytotic vesicles endowed with specific surface-targeting motifs; i.e., analogous to viral particles.
Over the past decades, the concept of cellular release of small membrane vesicles or exosomes (50–90 nm in diameter) and their role in intercellular communication have gained increasing attention. Comprehensive reviews of exosomes and mechanisms involved in exosome formation, secretion, and interaction with target cells is provided elsewhere (Lakkaraju and Rodriguez-Boulan 2008; Schorey and Bhatnagar 2008). For the purpose of this review, reports on exosome-mediated cell-to-cell transfer of nucleic acids are of particular interest. Ratajczak et al. (2006) showed that embryonic stem cells secrete membrane vesicles highly enriched in specific mRNAs, which can be transferred to and induce phenotypic changes in hematopoietic progenitor cells (Ratajczak et al., 2006). Recent work by another group suggested that mast cells secrete exosomes that contain a unique set of ∼1,300 different mRNAs (Valadi et al., 2007). Importantly, transferred mRNAs seemed biologically active, as their translation was demonstrated in recipient cells. Gene profile analysis showed significant differences in the level of transcripts isolated from exosomes and donor cells, which suggests that intricate selection mechanisms operate during the formation or intracellular sorting of exosomal RNA. Thus, the message transferred to neighboring cells via exosomes does not simply mirror the transcriptional status of the donor cell. Intriguingly, the exosome preparation also contained >100 different microRNAs (miRNAs). Considering the relative promiscuity of miRNA species for the binding of target mRNA, the impact of exosome-mediated transfer of miRNA on the translational machinery in receiving cells may be quite extensive. The microvesicle–exosomal pathway may thus constitute a well-designed mechanism for local and systemic intercellular transfer of information, with a complexity superior to that of direct cell-to-cell contacts or secreted soluble factors.
Intercellular DNA transfer
Although the discussions above deal with the possible transfer of RNA species (e.g., in the study by Valadi et al. [2007], no DNA could be detected in isolated exosomes), several reports indeed support the existence of horizontal DNA transfer between somatic cells. Bergsmedh et al. (2001) showed that DNA released as apoptotic bodies can be endocytosed/phagocytosed and integrated into the nuclei of recipient cells for expression. It was further suggested that activated oncogenes may be horizontally transferred by apoptotic bodies, and that transforming DNA was specifically propagated in host cells deficient in the major tumor suppressors p53 and p21. One interesting possibility is that horizontal DNA transfer through apoptotic bodies adds to more established mechanisms of genomic insults (e.g., oxygen radicals, alkylating agents, and ionizing radiation) that are involved in neoplastic transformation of somatic cells. Their results also give some support to the “genometastasis” hypothesis; i.e., that malignant dissemination is propagated by the escape of cancer cell–derived DNA rather than cancer cells per se from the primary tumor site to the circulation (García-Olmo et al., 2000). In this context, it is noteworthy that mammalian cells have been shown to secrete proteins with the ability to facilitate naked DNA uptake through endocytosis (Wittrup et al., 2007). Although it has been firmly established that tumor-derived nucleic acids circulate at significant levels and may have an important role in the diagnostics and prognostics of cancer disease (Diehl et al., 2008), the causal role of horizontal gene transfer in malignant progression remains a provocative speculation.
The possible pathophysiological role of horizontal DNA transfer has been, however, suggested by other studies investigating the pathogenesis of autoimmune disease. Previous studies by our group have shown that the antimicrobial peptide LL-37, which is widely expressed in epithelia, bone marrow, and the genitourinary tract of humans, forms stable complexes with DNA and translocates extracellular DNA to the nuclear compartment of recipient cells through a raft-associated endocytotic pathway (Sandgren et al., 2004). The ability of LL-37 to transfer DNA over the plasma membrane is a shared property within the growing family of so called cell-penetrating peptides (CPPs). These peptides, e.g., the Antennapedia homeobox peptide and the HIV-Tat transduction domain, are endowed with the unique ability to mediate efficient uptake of macromolecules into a wide variety of mammalian cells (Prochiantz 2008). A recent study of another group presented the novel concept of LL-37–mediated delivery of self DNA as an early pathogenic event in autoimmune disease (Lande et al., 2007). Normally, self DNA fails to trigger toll-like receptor 9 (TLR-9), an intracellular receptor that recognizes microbial DNA within endosomal compartments. However, the LL-37 peptide appears to break innate tolerance to self DNA by internalizing and retaining DNA in an early endocytotic compartment where TLR-9 resides, eventually resulting in T cell activation and autoimmune inflammation of the skin. Future studies will tell whether peptide-mediated intercellular transfer of self DNA is a general mechanism of autoimmune and inflammatory disease.
Conclusions and future directions
Our molecular understanding of endocytotic and intracellular sorting mechanisms, the dynamics of multivesicular bodies and exocytosis, and the composition of biological membranes have increased considerably over the past few years. As for the pathophysiological function of membrane transport pathways in mammalian cells, most studies have focused on the internalization and trafficking of foreign intruders, i.e., bacteria and viruses, and more lately on nonnatural, synthetic peptides and lipids for therapeutic nucleic acid delivery. The studies discussed here provide a framework for the existence of intrinsic membrane transport pathways for the intercellular exchange of nucleic acids locally and at the systemic level in multicellular organisms (Fig. 2). This raises the central question of whether the observed phenomena are traces of evolutionary remnants or if they indeed reflect significant biological functions in modern mammalian organisms. What could these functions be? Although it is easy to appreciate the potential versatility of a nucleic acid–based communication system in higher organisms, we can only speculate at this point on the specific signals that are transmitted and their physiological roles.
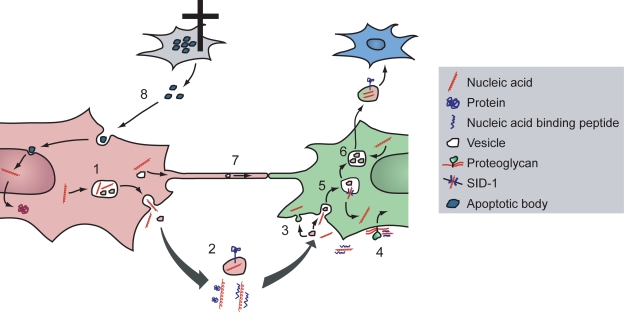
Figure 2.
Possible routes for nucleic acid exchange between cells. The classical mechanism of cellular communication via macromolecules is through secretion of signaling molecules. Secreted molecules are usually relayed through a secretory vesicular compartment (1) that subsequently fuses with the plasma membrane. Exosomes are released in a similar fashion, whereas microvesicles bud off directly from the plasma membrane. Such vesicles have been shown to contain nucleic acids (2) that hereby can be shuttled between cells. The mechanism of vesicular cargo uptake by recipient cells is largely unknown but may involve either direct membrane fusion or endocytosis (3). The internalization mechanisms of free peptide or protein bound nucleic acids have been better studied and may involve proteoglycan-dependent endocytosis (4). How endocytosed macromolecules escape the endosome and gain access to the cytoplasm remains ill-defined, but recent findings suggest a role for SID-1 in nucleic acid membrane transfer (5). Internalized vesicles might also intersect with the exosomal biogenesis machinery in late endosomes/multivesicular bodies (6), thus giving rise to compound vesicles that can deliver an integrated message to yet other cells (shown in blue). An alternative pathway for macromolecular shuttling between cells is through TNTs (7). Vesicles of endosomal origin are among the cargos demonstrated to be transported via this pathway. However, a multitude of other cargos, including nucleic acids and proteins, are potentially sorted for intercellular transport via TNTs. Finally, it has been demonstrated that apoptotic bodies released from tumor cells (8) can deliver oncogenic DNA and transform nonmalignant, surrounding cells.
Given the well-characterized involvement of the miRNA regulatory system in development and cancer, these processes are interesting candidates for using nucleic acid–based paracrine signaling. Development in particular demands a highly coordinated and specific signaling system where nucleic acids would be ideal. In this context, it is intriguing that recent studies suggest an involvement of RNA in epigenetic remodeling (Chandler 2007). Other examples may be tissue regeneration and fine tuning of the adaptive immune system. Taking cancer as an example of a multicellular, invasive organism, a communication route based on nucleic acid transfer would enable malignant cells to influence the surrounding nonmalignant cells and microenvironment in a highly specific and complex manner to assist the tumor in nutrient supply, invasion, and metastasis, as well as immune surveillance. In analogy to recent studies showing microvesicular transfer of constitutively active, mutated epidermal growth factor receptor (EGFR) between populations of malignant glioma cells (Al-Nedawi et al., 2008), it is tempting to speculate that separate clones within the same tumor similarly cooperate via vesicular exchange of RNA-based signals to promote proliferation, survival, and invasion. A general aspect of nucleic acid–based communication that appears particularly attractive, yet strictly hypothetical, is the potential of intracellular exchange of exosomal nucleic acids in, e.g., multivesicular bodies. Such mixing could in theory take place in several cells, one after the other, thus giving rise, in a single transporting vesicle, to a signal that integrates information from multiple cells.
Further characterization of TNTs, nucleic acid–transporting peptides, and exosomes should provide important clues to the emerging field of nonviral RNA and gene delivery vehicles. An exosome-like delivery vehicle appears promising, given that functional delivery of mRNAs and miRNAs between mammalian cells has been demonstrated (Valadi et al., 2007). A key question in the nucleic acid delivery field is how to improve passage of cargo molecules over the endolysosomal membrane into the cytosol. The study by Wolfrum et al. (2007) showing a function of SID-1 during endosomal escape of cholesterol-conjugated siRNA (Wolfrum et al., 2007) makes an interesting connection between systemic silencing mechanisms in C. elegans and pathways for nucleic acid delivery in mammals. We anticipate that future studies connecting various facets of cell biology research will revolutionize the way we perceive how cells communicate as well as the way we treat various diseases.
Acknowledgments
We apologize to all those authors whose important work could not be cited in this short overview due to space restrictions.
This work was funded by the Swedish Research Council, the Swedish Cancer Fund, the Lund University Hospital, and the Medical Faculty, Lund University (ALF).
Abbreviations used in this paper: dsRNA, double-stranded RNA; miRNA, microRNA; RdRP, RNA-dependent RNA polymerase; TNT, tunneling nanotube.
References
- Al-Nedawi, K., B. Meehan, J. Micallef, V. Lhotak, L. May, A. Guha, and J. Rak. 2008. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 10:619–624. [DOI] [PubMed] [Google Scholar]
- Bergsmedh, A., A. Szeles, M. Henriksson, A. Bratt, M.J. Folkman, A.L. Spetz, and L. Holmgren. 2001. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl. Acad. Sci. USA. 98:6407–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L. 2007. Paramutation: from maize to mice. Cell. 128:641–645. [DOI] [PubMed] [Google Scholar]
- Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241–249. [DOI] [PubMed] [Google Scholar]
- Chinnery, H.R., E. Pearlman, and P.G. McMenamin. 2008. Cutting edge: Membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J. Immunol. 180:5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, J.C., C.A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B.A. Hemmings, and J.E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, F., K. Schmidt, M.A. Choti, K. Romans, S. Goodman, M. Li, K. Thornton, N. Agrawal, L. Sokoll, S.A. Szabo, et al. 2008. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, E.H., and C.P. Hunter. 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 301:1545–1547. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver, and C.C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811. [DOI] [PubMed] [Google Scholar]
- García-Olmo, D., D.C. Garcia-Olmo, J. Ontanon, and E. Martinez. 2000. Horizontal transfer of DNA and the “genometastasis hypothesis”. Blood. 95:724–725. [PubMed] [Google Scholar]
- Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22:4523–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju, A., and E. Rodriguez-Boulan. 2008. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 18:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R., J. Gregorio, V. Facchinetti, B. Chatterjee, Y.H. Wang, B. Homey, W. Cao, Y.H. Wang, B. Su, F.O. Nestle, et al. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 449:564–569. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., S. Bouche-Pillon, D.P. Jackson, L. Nguyen, L. Baker, B. Ding, and S. Hake. 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 270:1980–1983. [DOI] [PubMed] [Google Scholar]
- Onfelt, B., S. Nedvetzki, K. Yanagi, and D.M. Davis. 2004. Cutting edge: Membrane nanotubes connect immune cells. J. Immunol. 173:1511–1513. [DOI] [PubMed] [Google Scholar]
- Onfelt, B., S. Nedvetzki, R.K. Benninger, M.A. Purbhoo, S. Sowinski, A.N. Hume, M.C. Seabra, M.A. Neil, P.M. French, and D.M. Davis. 2006. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J. Immunol. 177:8476–8483. [DOI] [PubMed] [Google Scholar]
- Prochiantz, A. 2008. Protein and peptide transduction, twenty years later a happy birthday. Adv. Drug Deliv. Rev. 60:448–451. [DOI] [PubMed] [Google Scholar]
- Ramirez-Weber, F.A., and T.B. Kornberg. 1999. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 97:599–607. [DOI] [PubMed] [Google Scholar]
- Ratajczak, J., K. Miekus, M. Kucia, J. Zhang, R. Reca, P. Dvorak, and M.Z. Ratajczak. 2006. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 20:847–856. [DOI] [PubMed] [Google Scholar]
- Rustom, A., R. Saffrich, I. Markovic, P. Walther, and H.H. Gerdes. 2004. Nanotubular highways for intercellular organelle transport. Science. 303:1007–1010. [DOI] [PubMed] [Google Scholar]
- Saleh, M.C., R.P. van Rij, A. Hekele, A. Gillis, E. Foley, P.H. O'Farrell, and R. Andino. 2006. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandgren, S., A. Wittrup, F. Cheng, M. Jonsson, E. Eklund, S. Busch, and M. Belting. 2004. The human antimicrobial peptide LL-37 transfers extracellular DNA plasmid to the nuclear compartment of mammalian cells via lipid rafts and proteoglycan-dependent endocytosis. J. Biol. Chem. 279:17951–17956. [DOI] [PubMed] [Google Scholar]
- Schorey, J.S., and S. Bhatnagar. 2008. Exosome function: from tumor immunology to pathogen biology. Traffic. 9:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski, S., C. Jolly, O. Berninghausen, M.A. Purbhoo, A. Chauveau, K. Kohler, S. Oddos, P. Eissmann, F.M. Brodsky, C. Hopkins, et al. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10:211–219. [DOI] [PubMed] [Google Scholar]
- Tijsterman, M., R.C. May, F. Simmer, K.L. Okihara, and R.H. Plasterk. 2004. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol. 14:111–116. [DOI] [PubMed] [Google Scholar]
- Valadi, H., K. Ekstrom, A. Bossios, M. Sjostrand, J.J. Lee, and J.O. Lotvall. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9:654–659. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., P. Vain, S. Angell, and D.C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 95:177–187. [DOI] [PubMed] [Google Scholar]
- Winston, W.M., C. Molodowitch, and C.P. Hunter. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 295:2456–2459. [DOI] [PubMed] [Google Scholar]
- Winston, W.M., M. Sutherlin, A.J. Wright, E.H. Feinberg, and C.P. Hunter. 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA. 104:10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittrup, A., S. Sandgren, J. Lilja, C. Bratt, N. Gustavsson, M. Morgelin, and M. Belting. 2007. Identification of proteins released by mammalian cells that mediate DNA internalization through proteoglycan-dependent macropinocytosis. J. Biol. Chem. 282:27897–27904. [DOI] [PubMed] [Google Scholar]
- Wolfrum, C., S. Shi, K.N. Jayaprakash, M. Jayaraman, G. Wang, R.K. Pandey, K.G. Rajeev, T. Nakayama, K. Charrise, E.M. Ndungo, et al. 2007. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 25:1149–1157. [DOI] [PubMed] [Google Scholar]