SUMMARY
Enzymatic utilization of nicotinamide adenine dinucleotide (NAD) has increasingly been shown to have fundamental roles in gene regulation, signal transduction, and protein modification. Many of the processes require the cleavage of the nicotinamide moiety from the substrate and the formation of a reactive intermediate. Using X-ray crystallography we show that human CD38, an NAD utilizing enzyme, is capable of catalyzing the cleavage reactions through both covalent and non-covalent intermediates, depending on the substrate used. The covalent intermediate is resistant to further attack by nucleophiles, resulting in mechanism-based enzyme inactivation. The non-covalent intermediate is stabilized mainly through H-bond interactions, but appears to remain reactive. Our structural results favor the proposal of a non-covalent intermediate during normal enzymatic utilization of NAD by human CD38 and provide structural insights into the design of covalent and non-covalent inhibitors targeting NAD utilization pathways.
INTRODUCTION
Nicotinamide adenine dinucleotide (NAD) is widely known to be a ubiquitous co-enzyme of oxidation-reduction reactions in cells. Accumulating evidence indicates, however, that it can function not only as a co-enzyme itself but also can serve as a substrate for multiple enzymes called NAD utilizing enzymes. The latter processes generally involve the enzymatic removal of its nicotinamide (Nic) moiety by specific NAD utilizing enzymes. The remaining adenine diphosphate (ADP)-ribosyl portion then forms a reactive intermediate with the catalyzing enzyme, which can be further used for multiple processes depending on the enzyme, such as protein ADP-ribosylation by some bacteria toxins (O'Neal et al., 2005) and mono-ADP-ribosyl transferases (Seman et al., 2004); histone deacetylation by sirtuin family proteins (Blander and Guarente, 2004; Sauve and Schramm, 2004); and the biosynthesis of the calcium mobilization messengers cyclic ADP-ribose (cADPR) and ADP-ribose (ADPR) by ADP-ribosyl cyclases (Guse, 2005; Howard et al., 1993; Lee, 2001). These processes are known to have important cellular and physiological functions in DNA repair (Lombard et al., 2005; Michan and Sinclair, 2007), transcriptional regulation (Blander and Guarente, 2004), cellular differentiation and proliferation, aging (Hassa et al., 2006), and calcium signaling (Lee, 2001; Lee et al., 1999).
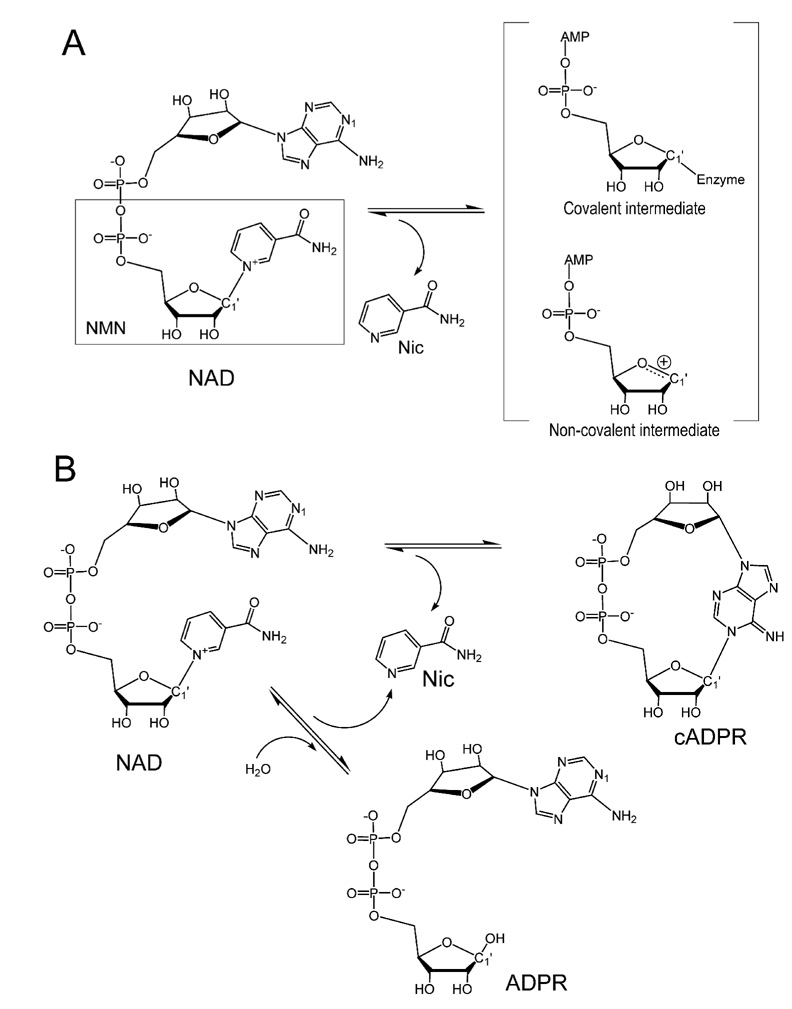
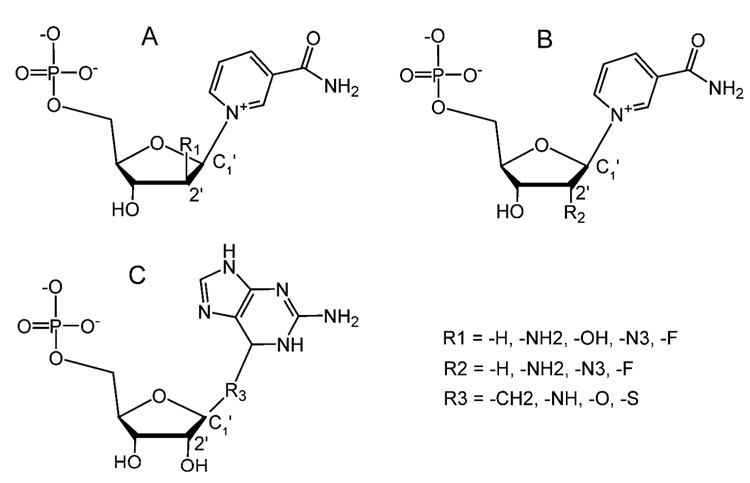
Although NAD is a substrate for multiple enzymes, the initial steps of the cleavage and release of the nicotinamide moiety are conserved. The nature of the subsequent intermediates formed, on the other hand, has been a widely debatable issue. Both covalent and non-covalent intermediates have been proposed (Figure 1A). In the former case, after the cleavage and the release of the nicotinamide, the remaining ribonucleotide forms a covalent bond with the catalytic residue (Sauve et al., 2000; Sauve and Schramm, 2002; Smith and Denu, 2006). In the non-covalent intermediate, it is proposed to be an oxocarbenium ion intermediate stabilized by non-covalent interactions (Berti et al., 1997; Handlon et al., 1994; Oppenheimer, 1994; Schuber and Lund, 2004; Tarnus et al., 1988; Tarnus and Schuber, 1987). As the characteristics of the intermediate determine the catalytic outcome of NAD utilization and are crucial for design of potent inhibitors for pharmacological purposes, it is important to characterize the chemical and structural nature of the intermediates.
Figure 1. Schematic diagram of the reactions of NAD catalysis.
A) Nicotinamide cleavage results in the formation of possible covalent and non-covalent intermediates. B) Reactions of forming cADPR or ADPR from NAD catalyzed by CD38.
In this study, we investigate the intermediates of CD38, a multifunctional molecule that is not only a lymphocyte antigen but also an NAD utilizing enzyme. As a member of NAD-utilizing enzymes of the ADP-ribosyl cyclase family (EC 3.2.2.5), human CD38 is a type II transmembrane ectoenzyme that catalyzes the conversions of NAD to cADPR and ADPR (Figure 1B) (Howard et al., 1993; Lee, 1994; Lee et al., 1989; Lee et al., 1993). Both products are calcium messenger molecules targeting different calcium channels and stores (reviewed in Lee, H.C. (Lee, 2001; Lee, 2004)). It has been proposed that after the release of the nicotinamide moiety, the intermediate shown in Figure 1A can either be attacked intra-molecularly (by the N1 atom of the adenine terminus) to form cADPR, or inter-molecularly (by a water molecule) to form ADPR, respectively (reviewed in Lee, H.C. (Lee, 2000; Lee, 2006)). In this study, we employed X-ray crystallography to investigate the nature of the intermediates formed during the catalysis of CD38. The results show that both covalent and non-covalent intermediates can be formed depending on the substrates. The structural results provide direct evidence for the pivotal role of the intermediate in determining subsequent reaction steps.
RESULTS
Covalent Intermediate
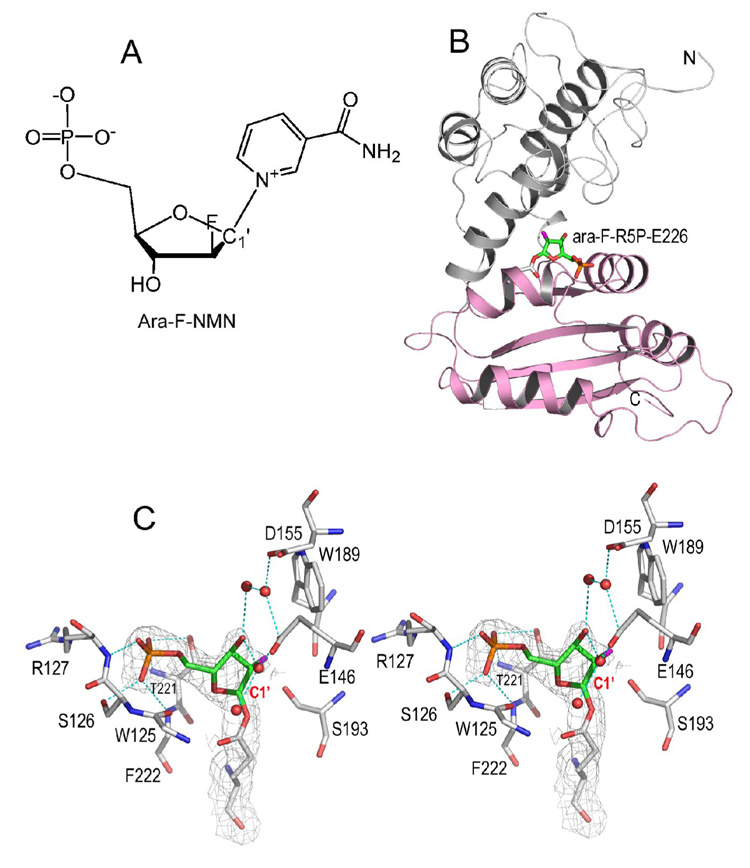
Nicotinamide mononucleotide (NMN) is a truncated version of the substrate NAD (Figure 1A) and can be hydrolyzed by CD38 to form ribose-5'-phosphate (R5P) (Sauve et al., 1998). We focused on this alternative reaction because the reaction intermediate should be structurally simpler without the adenine terminus of NAD. An NMN analog, ara-F-NMN (arabinosyl-2'-fluoro-2'-deoxynicotinamide mononucleotide), was used, which has a fluorine atom (2'-fluoro) substituting for the 2'-OH group in the trans orientation (Figure 2A). We show in Figure 2B that a covalent intermediate was formed after the release of the nicotinamide moiety. The covalent ara-F-R5P/wtCD38 complex was obtained by co-crystallization, and its structure was determined by X-ray crystallography at 2.0 Å resolution. The structure shows that the catalytic residue Glu226 forms a covalent acylal ester bond with the ara-F-R5P (Figure 2C). The covalent bond length is 1.6 Å, linking the carboxylate oxygen of Glu226 to the C1' carbon of the ara-F-R5P. Besides the covalent linkage, there are additional hydrogen bonding interactions between the enzyme and the phosphate group of ara-F-R5P, contributing to further stabilization of the covalent intermediate (Figure 2C). Another indication of the stability of the covalent intermediate is the observation that there are four water molecules trapped in the active site that are close to the intermediate (Figure 2C). These water molecules have H-bond interactions with protein residues Glu146, Asp155, as well as with the intermediate, ara-F-R5P. Nevertheless, these bound water molecules are not sufficient to promote the hydrolysis of the covalent intermediate to ara-F-ADP-ribose. The underlying reason is indicated by a close inspection of the conformation of the arabinosyl ring. The ring plane is seen to be almost parallel to the covalent bond, resulting in the covalent bond being shielded in a hydrophobic environment formed by residue Thr221. As a result, none of these four water molecules lies in a position suitable for nucleophilic attack of the covalent bond from the β-face of the ring. These structural features of the covalent intermediate indicate that it is highly stable and, once formed, prevents further catalysis by CD38, which is consistent with ara-F-NMN being a mechanism-based inhibitor with a Ki of 61 nM (Sauve et al., 2000).
Figure 2. Covalent intermediate after the cleavage of nicotinamide.
A) Chemical synthesized ara-F-NMN, a mechanism based analogue of substrate NMN. B) Overall structure of CD38 with its catalytic residue Glu226 covalently linked to ara-F-R5P. The two domains of CD38 are differently colored to show that the intermediate is right between the gulf of two domains. The covalent linkage is shown as sticks. C) A stereo presentation of active site structure showing the trapping of intermediate species ara-F-R5P. The 2Fo-Fc omit electron densities (with ara-F-R5P omitted during the calculation of the electron density map) for the trapped covalent intermediate is shown as gray isomesh at 1.0 σ. The active site residues are shown as sticks with their carbons atoms in gray. Ara-F-R5P, the remaining moiety after the release of leaving group nicotinamide from substrate, is also shown as sticks but with their carbon atoms in green. The covalent bond length between Glu226 and ara-F-R5P is 1.6 Å. Polar interactions between protein and ara-F-R5P are drawn as cyan dashed lines. Four water molecules observed in the active site are shown as red spheres.
Dynamics of the Covalent Intermediate
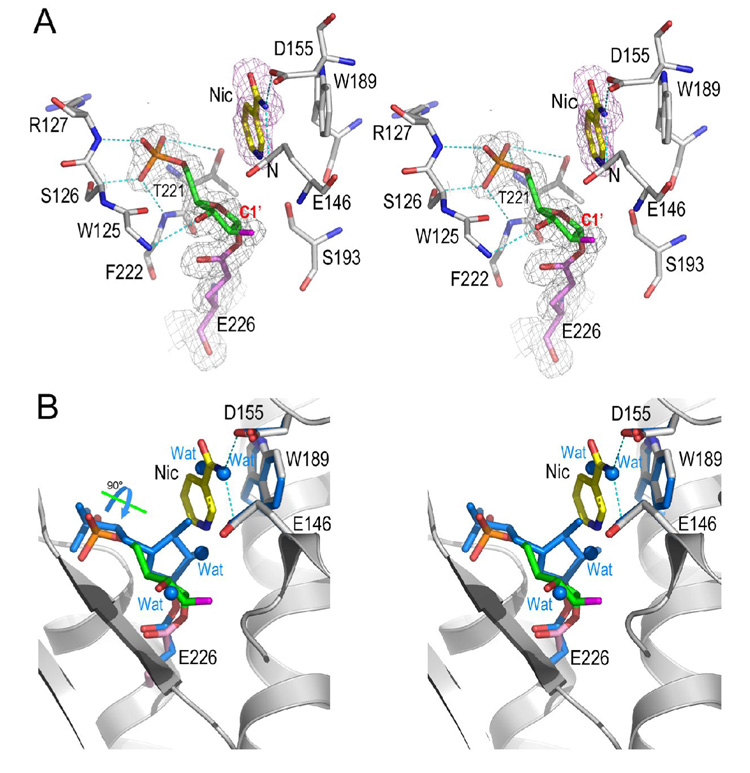
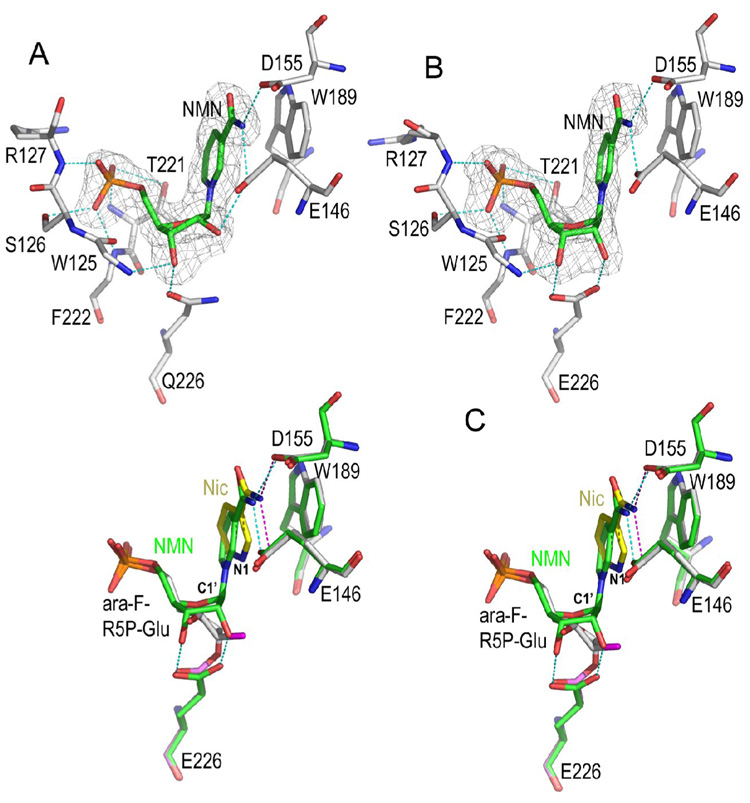
Despite its high potency, the inhibitory effect of ara-F-NMN, however, can be reversed in the presence of high concentrations of nicotinamide (Sauve et al., 2000). Based on the covalent intermediate structure described above, we reasoned that, to recover the enzyme’s activity, the arabinosyl ring of the intermediate should be able to adopt an alternative orientation so that its C1' atom is accessible to a nucleophile, such as nicotinamide. That is, the arabinosyl ring itself should be dynamic and be able to reposition itself upon the approach of a strong nucleophile. Although water is a good nucleophile in producing R5P from substrate NMN during its hydrolysis by CD38 (Sauve et al., 1998), it is not capable of doing so with ara-F-NMN as substrate (Figure 2C). Instead, the inhibition reversal by nicotinamide suggests that nicotinamide should be a better nucleophile to probe the possible dynamic behavior of the covalent intermediate. We introduced nicotinamide into the active site by soaking wild-type CD38 (wtCD38) crystals with 15 mM ara-F-NMN and 50 mM nicotinamide at 4 °C. The tertiary complex (ara-F-R5P/Nic/wtCD38) was obtained and the electron density defines clearly the entity of a nicotinamide together with the covalent intermediate in the active site (Figure 3A). The tertiary complex shows that nicotinamide is positioned and stabilized by both hydrophilic interactions with residues Glu146 and Asp155, as well as hydrophobic interactions with Trp189 through ring-ring stacking. These three residues have previously been identified to form a recognition site for the nicotinamide moiety of substrate NAD (or NGD), the adenine group of cADPR, and the guanine group of cGDPR (Liu et al., 2007; Liu et al., 2006). In the current complex, nicotinamide is on the β-face of the ara-F-R5P intermediate and is 3.7 Å away from the C1' carbon (Figure 3A), too far to elicit an efficient nucleophilic attack of the covalent intermediate. Nevertheless, the bound nicotinamide does interact strongly with the intermediate as shown in Figure 3B, where the covalent intermediates with and without nicotinamide are superimposed. It can be clearly seen that the binding of nicotinamide at the active site completely evacuates the water molecules seen in the nicotinamide-free covalent intermediate. Additionally, the recruitment of nicotinamide to its binding site appears to drive the rotation of the arabinosyl ring of the covalent intermediate from the nicotinamide-free position (marine carbon) to the nicotinamide bound position (green carbon). The large nicotinamide-induced rotational movement of the arabinosyl ring suggests that the covalent ara-F-R5P intermediate is structurally unstable, even though it is chemically resistant to hydrolysis. The strong interaction between nicotinamide and the covalent intermediate suggested by the structures are thus consistent with the biochemical evidence indicating high concentrations of nicotinamide can rescue CD38, although in a limited degree, after its inhibition by ara-F-NMN (Sauve et al., 2000).
Figure 3. Dynamics of covalent intermediate.
A) Structure of ara-F-R5P/Nic complex. With the pre-existence of covalent ara-F-R5P intermediate in the active site, nicotinamide, at high concentration, can bind to the active site through polar interactions to Glu146 and Asp155, and hydrophobic stacking interactions to Trp189. The 2Fo-Fc omit density covering ara-F-R5P and Glu226 is shown as gray isomesh contoured at 1.0 σ; the Fo-Fc electron density covering nicotinamide is show as purple isomesh contoured at 2.5 σ. Nicotinamide is 3.7 Å away to the reaction center atom C1′. B) Structural comparison of two covalent intermediates, with and without nicotinamide, to show the rearrangement of the covalent intermediate. Intermediate without nicotinamide (marine carbon atoms) was superimposed on intermediate (green carbon atoms) with the disturbance of high concentration of nicotinamide. Upon the binding of nicotinamide, the covalent ara-F-R5P intermediate dynamically rotates its arabinyl sugar group 90° around a free phosphate-sugar bond. Accordingly, four water molecules (marine spheres) are evacuated from the active site by this rotational movement.
Non-covalent Intermediate
Different from ara-F-NMN, NMN is not an inhibitor of CD38, but an efficient substrate that is hydrolyzed to nicotinamide and ribose-5'-phosphate (R5P) quickly with an Km of 149 µM and an Kcat of 512 s−1 (Sauve et al., 1998). These kinetic parameters are not in line with a stable covalent intermediate being formed from NMN, but suggest the possibility of a labile non-covalent intermediate instead. To trap this intermediate, we used a guanine containing molecule. We have previously shown that the guanine ring of NGD (nicotinamide guanine dinucleotide) can bind to the nicotinamide recognition site within the active site of CD38 and stabilize a non-covalent intermediate formed from the substrates NGD (Liu et al., 2006). The binding of the guanine ring at the nicotinamide binding site appears to block entry of water into the active site and thus prevent the hydrolysis of the non-covalent intermediate, extending its life-time for crystallographic study. Without intermediate-trapping, we found that NMN was readily hydrolyzed to R5P and nicotinamide. Neither R5P nor nicotinamide could be detected in the active site (data not shown).
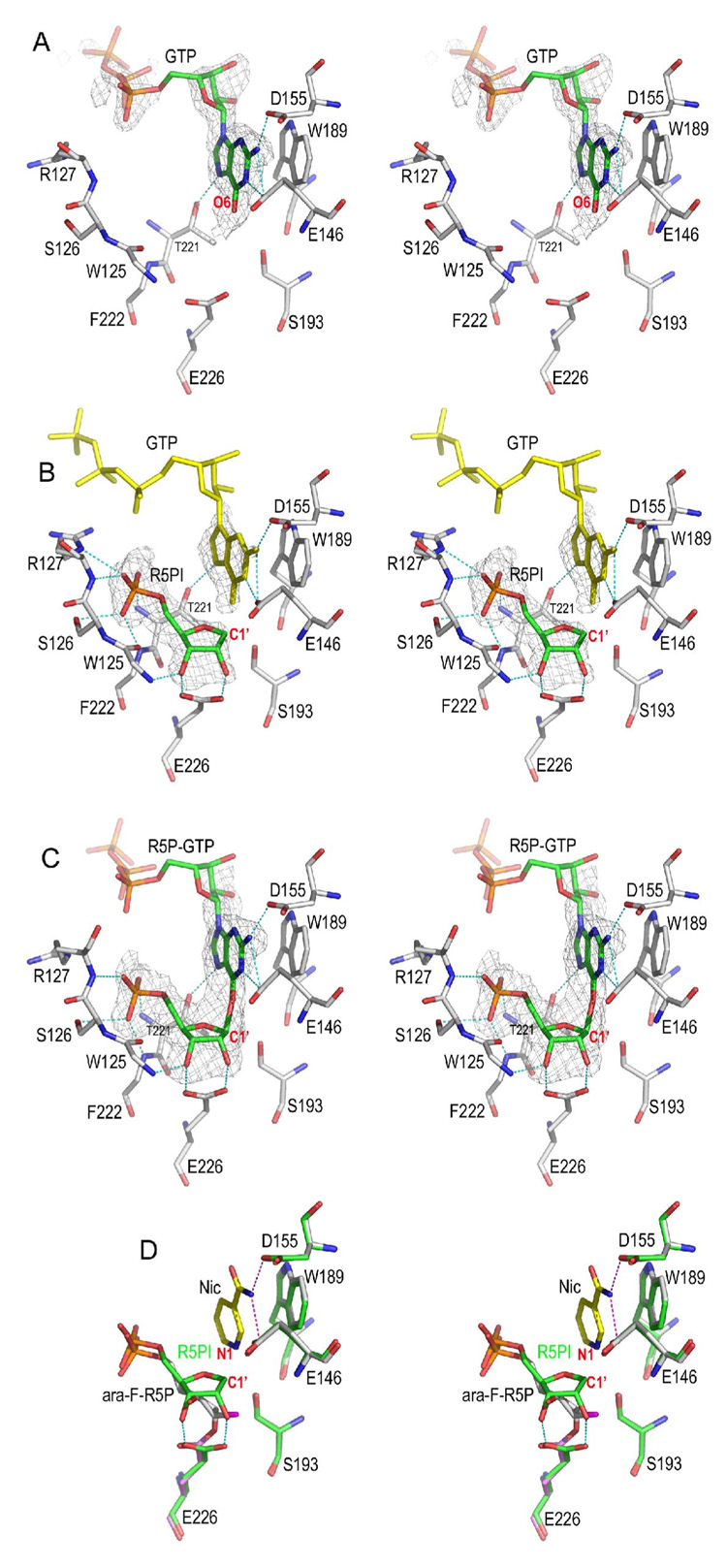
Here we used a heterogenic GTP molecule to trap the intermediate from NMN. We first determined the GTP/CD38 complex without NMN as shown in Figure 4A. The complex was formed by soaking CD38 crystals with GTP. As expected, the guanine ring of GTP occupies the nicotinamide recognition site defined by residues Glu146, Asp155, and Trp189, and forms extensive H bonds with Glu146, Asp155, and Thr221. The electron densities are unambiguous for the guanine ring and the adjacent ribose, but are poor for the remaining part of GTP, as it is out of the active site and is disordered.
Figure 4. Non-covalent intermediate trapped by GTP.
A) The GTP complex. GTP alone can inhibit CD38 by occupying the nicotinamide binding site defined by residues Glu146, Asp155, and Trp189. The Fo-Fc omit (with GTP omitted from the calculation of the electron density map) electron density is shown as gray isomesh contoured at 2.5 σ covering GTP.
B) The non-covalent R5PI intermediate trapped by GTP. The non-covalent R5P intermediate (R5PI) is show as sticks with its carbon green. R5PI forms two hydrogen bonds with the catalytic residues Glu226 and Trp125 main chain nitrogen. The Fo-Fc omit electron density is shown as gray isomesh contoured at 2.8 σ. The GTP molecule in the active site (yellow sticks) has almost the same conformation as in the GTP complex shown in A). The phosphate group of R5PI forms extensive H-bonds to residues Ser126, Arg127, Phe222, and Thr221.
C) A R5P-GTP adduct in which R5PI is attacked by GTP from the β-face to form a covalent bond. The bond distance between C1′ and GTP O6 is 1.9 Å. The Fo-Fc omit electron density (with R5P-GTP omitted from the calculation of the electron density map) is shown as gray isomesh and contoured at 2.8 σ. D) Structural comparison of covalent and non-covalent intermediates. The covalent intermediate (green carbon sticks) with nicotinamide trapped in the active site is used for the superimposition with R5PI intermediate (gray carbon sticks). Essential active site residues Glu146, Asp155, Trp189, and Ser193 align quite well whereas the catalytic residue Glu226 rotates its carboxylate group by 30° in order to form a covalent intermediate with ara-F-R5P.
To obtain the non-covalent intermediate, we soaked wtCD38 crystals in a solution of NMN and GTP at 4 °C for a short period (less than 3 min), followed by flash-freezing soaked crystals into liquid-nitrogen. This procedure allowed sufficient accumulation of the reaction intermediate species at the active sites. The corresponding crystal structure determined at 1.73 Å shows two CD38 molecules in the crystallographic asymmetric unit and the captured reaction species are shown in Figure 4B and 4C. In the active site of one molecule, we observed the electron densities corresponding to a GTP in the nicotinamide binding site (Figure 4B), as well as that corresponding to the R5P intermediate (R5PI) formed after the cleavage and release of nicotinamide. Different from the electron density observed for the covalent ara-F-R5P intermediate (Figure 2C or Figure 3A), there is no electron density between the R5PI and Glu226 (Figure 4B), indicating that the ribosyl C1' atom does not form covalent linkage to Glu226, but is instead at a distance of 3.5 Å from the catalytic residue Glu226. Moreover, the carboxylate group of Glu226 forms two H-bonds with the ribosyl 2'-, 3'- OH groups of the non-covalent intermediate (Figure 4B) that are absent in the covalent intermediate. It is clear that the intermediate formed with NMN is structurally distinct from the covalent intermediate formed with ara-F-NNM described above. This intermediate is, on the other hand, very similar to the non-covalent intermediate formed with NGD that we reported previously (Liu et al., 2006).
Non-covalent Intermediate Is Reactive
In the active site of the other CD38 molecule in the crystallographic asymmetric unit, we observed a reaction adduct R5P-GTP (Figure 4C) apparently formed by the nucleophilic attack of the R5PI (Figure 4B) by the guanine O6 of a bound GTP. The attack is from the β-face of the ribose, demonstrating a retaining feature of the catalysis by CD38. The bond distance between C1′-O6 is 1.9 Å, longer than the bond distance of a standard C-O bond (1.33Å), suggestive of a dissociative character of the reaction adduct. That the product R5P-GTP can be formed indicates that the non-covalent intermediate R5PI shown in Figure 4B is highly susceptible to further attack even in its crystalline state by any nucleophile nearby, be it the bound GTP (Figure 4C) or bound water (Figure. 2C). This is consistent with the kinetic data showing NMN hydrolysis catalyzed by CD38 is at a high rate with an Kcat of 512 s−1 (Sauve et al., 1998).
Figure 4D aligns and compares the structural features of the non-covalent (R5PI) and covalent intermediates (ara-F-R5P/Nic). In the covalent intermediate (ara-F-R5P/Nic), the arabinosyl ring of the ara-F-ribose apparently moved toward Glu226 to form the covalent linkage. In the non-covalent intermediate, the ribose forms two H-bonds with the carboxylate group of the catalytic residue Glu226. The superimposition of the structures of the two intermediates indicates that the N1 atom of the bound nicotinamide (in ara-F-R5P/Nic complex) is only 2.4 Å to the C1' carbon of the R5PI intermediate. The close proximity provides structural evidence that R5PI is likely to be susceptible to nucleophilic attack by nicotinamide. Indeed, when we soaked CD38 crystals in an NMN solution supplemented with a high concentration of nicotinamide (up to 200 mM), we could not observe any electron densities for either R5PI or nicotinamide in the active site. This is also consistent with nicotinamide being an efficient nucleophile in the base-exchange reaction catalyzed by CD38, with a Km of 0.92 mM and a kcat of 0.007 s−1 (Aarhus et al., 1995; Sauve and Schramm, 2002).
Based on the above structural analyses of the covalent and non-covalent intermedates, we can conclude that the reaction intermediate formed during the hydrolysis of regular substrates, such as NMN, NAD, or NGD, is more reasonably non-covalent. Covalent intermediate can be, and is, formed, but only with special substrate/inhibitor, such as ara-F-NMN.
The NMN/CD38 Complexes
To better understand the catalysis process, we also determined the Michaelis complex of the substrate NMN. Two different approaches were used to obtain NMN/CD38 complexes. The first one is to use a catalytically inactive mutant of CD38 (Munshi et al., 2000). We have shown that CD38 can be inactivated by mutating the catalytic residue Glu226 to Gln226, which should allow the binding of NMN to the active site without being cleaved, as we have previously done to obtain the Michaelis complexes with NAD and cADPR (Liu et al., 2007; Liu et al., 2006). Figure 5A shows that in the NMN/E226Q complex, the nicotinamide moiety of NMN binds to the site defined by residues Glu146, Asp155, and Trp189, the same site for the binding of nicotinamide in ara-F-R5P/Nic complex. The ribosyl 3'-OH group forms one H-bond with the carbonyl group of Gln226. The phosphate group has an almost identical interaction pattern as observed in the R5PI complex (Figure 5A & Figure 4B).
Figure 5. Structural determinants for nicotinamide-glycosidic bond cleavage.
A) NMN/E226Q complex. Substrate NMN is shown as sticks with its carbon atoms green. The Fo-Fc omit electron density (with NMN omitted from the calculation of the map) is shown as gray isomesh contoured at 2.5 σ. The nicotinamide moiety of NMN is recognized by its interactions to Glu-146, Asp-155, and Trp-189. Dashed lines colored in cyan show the specific polar interactions between NMN and the enzyme.
B) NMN/wtCD38 complex. The presentation scheme is the same as A).
C) Stereo representation of the structural comparison of the NMN/wtCD38 complex with the ara-F-R5P/nicotinamide complex. Both complexes are shown as sticks with green carbon for the NMN/wtCD38 complex and gray/magenta/yellow carbon for the ara-F-R5P/nicotinamide/wtCD38 tertiary complex. The alignment of both structures shows the structural determinants for the enzymatic cleavage of nicotinamide.
The second approach to obtain the NMN/CD38 complex is to use the trapping technique described above. We screened various guanine containing molecules, including GTP, GDP, guanosine, and guanine, for their ability to trap, by co-soaking each of them with NMN at low temperature for a short period. We found that the incubation of 5 mM guanine and 40 mM NMN with wtCD38 crystals followed by flash-freezing to liquid nitrogen temperature can accumulate sufficient amounts of the activated substrates in the active sites for the structural detection. The solved NMN/wtCD38 complex is shown in Figure 5B. In contrast to the NMN/E226Q complex (Figure 5A), the carboxylate group of Glu226 forms two H-bonds with the ribose. It is thus clear that the replacement of the carboxylate group in Glu226 with the corresponding amide results in the 2'-OH group of substrate NMN being at a distance of 4.4 Å away from the Nε2 atom of Gln226, too distant to form a hydrogen bond. These structural features are remarkably similar to those seen in the NAD/E226Q complex we reported previously (Liu et al., 2007; Liu et al., 2006) and further buttress the proposal we advanced previously that the distortion of ribose imposed by the two H-bonds between Glu226 and the ribose -OH groups is critically important for the catalytic activities of CD38.
Role of substrate distortion in N-glycosidic bond cleavage
The role of substrate distortion is further investigated in Figure 5C by aligning and comparing the ara-F-R5P/Nic and the NMN/wtCD38 complexes, both containing nicotinamide in its free and uncleaved forms (part of NMN), respectively. In the aligned structures, both the free nicotinamide and the uncleaved nicotinamide have polar interactions with residues Glu146 and Asp155 as well as non-polar interactions with residue Trp189. There is, however, one notable structural difference; namely, the free nicotinamide ring plane has a 16° rotation relative to the uncleaved nicotinamide ring plane of NMN (Figure 5C). The relative orientation of the two nicotinamide rings indicates that, in the Michaelis complex, the substrate NMN is distorted and its leaving group can be subjected to a force that preferentially drives the nicotinamide ring to swing away from the C1' carbon, displaying a dissociative character of the nicotinamide-glycosidic bond. The dissociative force includes hydrophobic interactions and aromatic stacking of the nicotinamide ring and the Trp189 indole ring, which has also been proposed to explain N-glycosidic bond cleavage in nucleosides (Versees et al., 2004).
Additionally, as described above, the H-bonds between the ribosyl 2'-OH and Glu226 flattens the ribose ring, absorbing the electron migration from the nicotinamide-glycosidic bond, and further destabilizing the substrate, effecting the cleavage of the nicotinamide-glycosidic bond. After cleavage, the R5P intermediate is stabilized by two H-bonds between the 2'-, 3'- OH groups and the catalytic residues (Figure 4B). The scenario described above is fundamentally different from the general belief that the glycosidic bond cleavage is via direct attack of the ribosyl C1’ by the catalytic residue. Substrate distortion and leaving group activation in NMN hydrolysis as the main driving forces for catalysis are also in accordance with the apparent absence of an appropriate general acid in the active site of CD38 that is capable of protonating the leaving group after the cleavage step, as would be required by a Schiff-acid catalysis mechanism.
DISCUSSION
The nature of the intermediate formed after nicotinamide-glycosidic bond cleavage is a highly debated topic. The two prevailing models of the intermediate are covalent and non-covalent, as shown in Figure 1A. We employ a direct approach of X-ray crystallography to resolve the issue. Using a model substrate NMN and its analogue, ara-F-NMN, we determined the structures of both types of intermediates, as well as the substrate Michaelis complexes at high resolution. The results advance our understanding of the catalytic mechanism of CD38 and provide substantial structural clues for the design of potential inhibitors for pharmacological purposes.
In the ara-F-R5P complex, the replacement of the 2'-OH of NMN with the 2'-fluro group apparently reduces the strength of the interaction between the carboxylate group of Glu226 and the arabinosyl ring, allowing relative movement of the arabinosyl ring toward Glu226 to form a covalent linkage. It may very well be that, after the dissociation of the nicotinamide from NMN, the intermediate is initially non-covalent in nature. However, without the stabilization from the 2'-OH via H-bonding, the carboxylate group of Glu226 can rotate and attack the labile C1' atom to form the covalently linked intermediate (Figure 5C). To do this, the side chain of Glu226 needs to rotate about 30° to make a linkage with the C1' of ara-F-ribose. The ara-F-ribose is seen to move deeper toward Glu226, facilitating the linkage (Figure 5C). Indeed, the arabinosyl ring of the covalent ara-F-R5P intermediate (nicotinamide free form) is rotated as much as 90° during its movement after the nicotinamide-glycosidic bond cleavage (Figure 3B). It is entirely possible that the substitution of the 2'-OH group by 2'-fluro from the β-face of the ara-F-NMN renders the molecule incapable of forming any H-bond with Glu226, disfavoring the stabilization of the initial non-covalent intermediate and promoting the formation of a more stable covalent intermediate instead. Clearly, once the covalent intermediate is formed, its stability disfavors further catalysis to occur and the enzyme is inhibited. Therefore, when NMN is used as substrate, the intermediate is non-covalent, and its hydrolysis rate is high with an Kcat of 512 s−1 (Sauve et al., 1998); while with ara-F-NMN, the intermediate is covalent and stable, and the enzyme is inhibited. Only in the presence of high concentrations of nicotinamide can the inhibition be reversed via a base exchange reaction, which has a measured Km value of 17 mM for nicotinamide (Sauve et al., 2000). Further evidence supporting our conclusion is from mutagenesis. The catalytically inactive E226Q mutant (Munshi et al., 2000), lacking the carboxylate group of Glu226, is not capable of forming a covalent intermediate. We indeed found that E226Q could not even bind ara-F-NMN, i.e. could not form an E226Q/ara-F-NMN complex when applying the same soaking protocol used for obtaining the E226Q/NMN complex (data not shown). We are thus proposing that, under normal conditions with the substrates NAD or NMN, the non-covalent intermediate shown in Figure 4B is both structurally and kinetically reasonable. This is also supported by biochemical data (Cakir-Kiefer et al., 2000). Our structural data strongly support the concept that the same enzyme can catalyze the nicotinamide-glycosidic bond cleavage through two entirely different reaction pathways depending on the substrate involved. The critical structural determinant on the substrate for selecting the catalytic pathways appears to be the 2'-OH, as that is the only difference between ara-F-NMN and NMN.
Structures presented in this study provide clues about why the 2’-OH is critical and point to the importance of H-bonding to both ribosyl OH groups during catalysis. In the R5PI complex, two ribose hydroxyl groups form two H-bonds with the catalytic residue Glu226. With this ribose configuration, the intermediate is non-covalent and is stabilized by the two H-bonds (Figure 4B). The extra H-bond with the 2'-OH group can sufficiently reduce the electronegativity of Oε2 of Glu226, rendering it incapable of forming a covalent bond with C1'. Consequently, the reaction proceeds through the non-covalent mechanism. The positioning of the non-covalent intermediate at the active site automatically allows efficient attack by a nucleophile from its β-face, as shown in Figure 4C. In the ara-F-R5P or ara-FR5P/Nic complex, there is no H-bond between Glu226 and 2'-position of the arabinosyl ring. Therefore, the Oε2 of the Glu226 is free to attack C1' carbon and form a covalent bond. This covalent linking process may be accelerated by the dynamic rotational movement of the arabinosyl ring as shown in Figure 2B. Likewise, Glu226 also rotates to facilitate the covalent bond formation (Figure 5C).
The chemistry of the intermediate for NAD hydrolysis has been favored to be an oxocarbenium ion based on independent results from kinetic isotope effects (Berti et al., 1997; Bull et al., 1978), catalysis of NAD pyridinium analogs (Tarnus and Schuber, 1987), 2'-substitutions effects of NAD ribose (Handlon et al., 1994), and preferential methanolysis over hydrolysis (MullerSteffner et al., 1996). In our study, structural evidence is provided for a noncovalent intermediate. We were able to apparently trap the intermediate in the active site by using a GTP molecule to block the entry of water. This noncovalent intermediate could be the proposed oxocarbenium ion, although our structural results do not definitively prove that. We clearly observed that the intermediate is likely to be formed via interactions between Glu226 and hydroxyl groups of the ribose. This is consistent with the proposed mechanism by Oppenheimer for the inductive stabilization of the oxocabenium ion intermediate via an interaction between the 2’-hydoxyl and an active site carboxylate (Oppenheimer, 1994). We would thus generalize and propose that for the ribosyl substrates, such as NMN, NAD, NGD, and nicotinamide ribose, while for arabinosyl substrates/analogs, such as ara-F-NMN, ara-F-NAD, ara-NAD, and 2'-deoxy-nicotinamide ribose, the enzyme would take the covalent reaction pathway.
The structural results in this study provide clues for the rational design of efficient covalent and noncovalent inhibitors of CD38 (Figure 6). Accordingly, the key to a potent covalent inhibitor is the efficient formation of a stable covalent intermediate. With ara-F-NMN as a model, modifications of the 2’-position of either the arabinosyl ring (Figure 6A) or the ribosyl ring (Figure 6B) would be the approach. The modifications should aim to prevent the formation of strong H-bond interactions between the inhibitors and the catalytic residue. Arabinosyl derivatives would be preferred as the modified group would be pointing away from Glu226. The cis-NH2 substitution of the ribosyl 2’-OH group, however, would not be a good choice as NH2 can still serve as a hydrogen donor to form H-bonds with the catalytic residue.
Figure 6. Rational designs of covalent and non-covalent inhibitors based on mechanism of catalysis.
A) Covalent inhibitors of 2'-substitutions of arabino configuration. B) Covalent inhibitors of 2'- substitutions of ribose configuration. Among these potential inhibitors, 2'-substitution by an -NH2 group might not be a good covalent intermediate as it can likely form an H-bond with the catalytic residue Glu226 to stabilize a non-covalent intermediate.
C) Non-covalent inhibitors: R5P-Guanine based inhibitors.
As for non-covalent inhibitors, we can model after the R5P-GTP adduct shown in Figure 4C. The electron density map of the adduct indicates that the R5P and the guanine portion are bound tightly and have well defined density, while the disordered triphosphate and ribose moieties are not recognized by the active site. This portion of the adduct can thus be deleted to make an inhibitor (Figure 6C) that can bind tightly to the active site and block normal substrates from entering. Candidates of 6-substititions of the guanine moiety could be a -CH2, -NH, -O, or -S group. One special feature of the adduct is that it possesses very similar stereochemistry to fludarabine phosphate, an effective drug for the treatment of chronic lymphocytic leukemia (Rai et al., 2000). Indeed, studies using knockout mice establish the importance of CD38 and its enzymatic activities in a wide range of physiological processes, including insulin secretion (Kato et al., 1999), α-adrenoceptor signaling in aorta (Mitsui-Saito et al., 2003), hormonal signaling in pancreatic acinar cells (Fukushi et al., 2001), migration of dendritic cell precursors (Partida-Sanchez et al., 2004), bone resorption (Sun et al., 2003), susceptibility to bacterial infection (Partida-Sanchez et al., 2001) as well as aberrant social behavior in mice due to a defect in oxyntocin secretion (Jin et al., 2007). The development of covalent and non-covalent inhibitors for CD38 should thus have physiological and pharmacological significance in biomedical research of CD38 related diseases in particular and NAD metabolism diseases in general.
SIGNIFICANCE
NAD is emerging as an important regulator for a wide variety of physiological functions, including signal transduction, gene regulation, and protein post-transcriptional modification. It can also be a substrate for the formation of several messenger molecules. The enzymatic reactions of NAD generally involve removal of the nicotinamide moiety and the formation of an intermediate with the catalyzing enzyme. Both covalent and non-covalent intermediates have been proposed. In this study, we resolved this issue by determining the structures of both the covalent and non-covalent intermediates as well as the Michaelis complexes of CD38. In the covalent intermediate, the reaction center, C1′, forms a 1.6 Å covalent bond with the catalytic residue Glu226. The covalent intermediate is dynamic and can undergo a large rotational movement induced by a nucleophile, nicotinamide. The covalent intermediate is highly stable, preventing further catalysis by CD38. In the non-covalent intermediate, the reaction center, C1′, is 3.5 Å from the catalytic residue. The non-covalent intermediate is stabilized through H-bonds between its ribosyl 2′-, 3′- OH groups and the catalytic residue, but is much more reactive than the covalent intermediate. The non-covalent intermediate can be readily attacked by a nearby nucleophile to form a covalent adduct from the β-face of the intermediate. By comparing the covalent and non-covalent intermediates, we propose that the determinant for selecting the two distinct intermediate pathways does not reside in the enzyme itself, but is, rather, the 2’-position of substrate’s pentose ring. We further propose that the non-covalent pathway is generally adopted during reactions involving natural substrates, such as NAD or NMN. The structural data presented provide clues for designing potent inhibitors for CD38, which should provide invaluable tools for scientific and diagnostic analysis of the wide range of physiological functions that are known to be regulated by CD38.
EXPERIMENTAL PROCEDURES
Synthesis of ara-F-NMN
Reagents were obtained from Aldrich or TCI and were used as supplied. 1H NMR spectrum was carried out at 600 MHz on a Varian Unity INOVA 600. Mass spectrum was obtained on a SHIMADZU Liquid Chromatography Mass Spectrometer LCMS-QP8000α. 2’-Fluoro-2’-deoxy-substituted nicotinamide arabino mononucleotide (ara-F-NMN) was synthesized according to published procedures (Sleath et al., 1991). 1H NMR (600 MHz, D2O) δ 9.46 (s, 1 H, N2), 9.31 (d, 1 H, N6), 9.01 (d, 1 H, N4), 8.31 (t, 1 H, N5), 6.76 (dd, 1 H, 1’), 5.57 (dt, 1 H, 2’), 4.64 (dt, 1 H, 3’), 4.47 (m, 1 H, 4’), 4.36 (m, 1 H, 5’), 4.24 (m, 1 H, 5’’). m/z 336.70 (M+, calculated 337.06).
Complexes Formation
Expression, purification and crystallization of wtCD38 and E226Q mutant proteins were performed with procedures as previously described (Liu et al., 2007; Liu et al., 2005; Munshi et al., 2000). The compound ara-F-NMN was synthesized as described (Sauve et al., 2000). GTP, NMN, and Nicotinamide were purchased from Sigma (Sigma-aldrich, St. Louis, USA). The Ara-F-R5P complex was obtained by co-crystallization of wtCD38 with ara-F-NMN. The Ara-F-R5P/Nic complex was formed by soaking wtCD38 crystals in soaking solution containing 15 mM ara-F-NMN, 50 mM nicotinamide, 100 mM MES, pH 6.0, 15% PEG4000, and 30% glycerol.
The GTP/CD38 complex was formed by soaking wtCD38 crystals with 5 mM GTP, 100 mM MES, pH 6.0, 15% PEG 4000, and 30% glycerol for 2 min at 4 °C. Trapping of the R5PI intermediate with GTP was done by directly soaking wtCD38 crystals in 2 µL soaking solution containing 40 mM NMN, 5 mM GTP, 100 mM MES, pH 6.0, 15% PEG 4000, and 30% glycerol, for 3 min at 4 °C.
The NMN/E226Q complex was formed by incubating E226Q mutant crystals in 2 µL soaking solution that contained 40 mM NMN, 100 mM MES, pH 6.0, 15% PEG 4000, 30% glycerol for 1 min at 0 °C. The trapping of the NMN/wtCD38 complex was done by incubating wtCD38 crystals with 2 µL soaking solution that contained 40 mM NMN, 5 mM guanine, 100 mM MES, pH 6.0, 15% PEG 4000, 30% glycerol for 7 min at 4 °C. The addition of 5 mM of guanine prevented the hydrolysis of NMN to R5P and was essential for the successful formation of NMN/wtCD38 complex.
X-ray Crystallography
X-ray diffraction data were collected at the Cornell High-Energy Synchrotron Source (CHESS) A1 station with crystals protected by a liquid nitrogen cryo-stream at 100 K. For each data set, a total of 360 images with an oscillation angle of 1° were collected from a single crystal using a Quantum Q-210 CCD detector. All data sets were processed by using the program HKL2000 suite (Otwinowski and Minor, 1997). The crystallographic statistics are listed in Table 1.
Table 1.
Crystallographic data and refinement statistics.
| Data collection | ara-F-R5P | ara-F-R5P/Nic | GTP | R5PI/GTP | NMN/E226Q | NMN/wtCD38 |
|---|---|---|---|---|---|---|
| Cell dimensions | 41.8 | 41.7 | 41.7 | 41.7 | 41.9 | 41.9 |
| a, b, c (Å)/ | 96.2 | 52.8 | 52.7 | 52.8 | 53.3 | 53.2 |
| α,β,γ(°) | 103.6/ | 65.4/ | 65.2 | 65.1/ | 65.7/ | 65.6/ |
| 79.5 | 106.1 | 106.1 | 106.1 | 106.1 | 106.2 | |
| 82.7 | 92.0 | 91.8 | 91.9 | 92.0 | 91.8 | |
| 86.9 | 95.1 | 95.0 | 95.3 | 95.1 | 95.1 | |
| Space group | P1 | P1 | P1 | P1 | P1 | P1 |
| Resolution (Å) | 30-2.00 | 30-1.64 | 30-1.60 | 30-1.73 | 30-1.90 | 30-1.80 |
| (2.07-2.00) | (1.70-1.64) | (1.66-1.60) | (1.79-1.73) | (1.98-1.90) | (1.86-1.80) | |
| Unique reflections | 90177 | 61988 | 65992 | 53519 | 41633 | 47229 |
| Multiplicity | 3.6 (3.2) | 3.5 (3.0) | 3.8 (2.9) | 3.6 (2.8) | 3.8 (2.9) | 3.4 (2.0) |
| I/sigma | 18.1 (2.5) | 17.5 (5.7) | 21.6 (2.5) | 25.8 (2.0) | 21.2 (2.9) | 12.6 (1.5) |
| Rmerge (%) | 6.4 (48.5) | 5.6 (18.9) | 5.3 (35.2) | 4.2 (45.8) | 9.0 (43.6) | 9.9 (44.3) |
| Completeness (%) | 86.7 (75.0) | 96.3 (86.9) | 95.5 (81.6) | 95.5 (82.2) | 97.5 (95.2) | 96.7 (87.2) |
| Refinement | ||||||
| Resolution (Å) | 20-2.00 | 20-1.65 | 20-1.60 | 20-1.73 | 20-1.90 | 20-1.80 |
| R factor (%) | 20.1 | 17.4 | 17.6 | 17.7 | 18.5 | 17.3 |
| Rfree factor (%) | 27.0 | 21.1 | 21.8 | 21.1 | 24.1 | 21.7 |
| Protein atoms | 12300 | 4100 | 4100 | 4100 | 4100 | 4100 |
| Water molecules | 770 | 494 | 409 | 399 | 384 | 378 |
| Ligands | 6 | 2+2 | 2 | 1+1+1 | 2 | 2 |
| Mean B (Å2) | 38.8 | 32.2 | 29.5 | 40.1 | 33.3 | 33.1 |
| R.M.S. deviations | ||||||
| Bond lengths (Å) | 0.022 | 0.013 | 0.029 | 0.016 | 0.016 | 0.016 |
| Bond angles (°) | 1.987 | 1.504 | 2.475 | 1.566 | 1.572 | 1.523 |
Values in parentheses are from the highest resolution shell.
Rmerge = Σ|I – <I>|/ΣI, where I is the integrated intensity of a given reflection.
R = Σ||Fobs| - |Fcalc||/Σ|Fobs|. Rfree was calculated using 5% of data excluded from refinement.
All complex structures were determined by difference Fourier calculation with the starting phases derived from the apo wtCD38 model (PDB ID: 1YH3). The models for all ligands were built manually in O (Jones et al., 1991) based on the σA weighted Fo-Fc difference electron density maps. For the ara-F-R5P complex, the restraints for covalent linkage between ligands and catalytic residue Glu226 were loosely restrained at 1.6 Å; for the ara-F-R5P/Nic complex, the catalytic residue Glu226 in both molecules was modeled as covalently linked to ara-F-R5P. Two nicotinamide molecules were modeled near ara-F-R5P ribose; for the GTP complex, the GTP molecule was modeled in both active sites from both molecules in the crystallographic asymmetric unit; for the R5PI/GTP complex, an R5P-GTP molecule was built in the active site of molecule, a trapped R5P intermediate and a GTP molecule were built in the active site of another molecule; for NMN complexed with wtCD38 and E226Q mutants, the NMN molecule was built separately for each case.
Structure refinements for these complexes were performed in program REFMAC (Murshudov et al., 1997) with manually modified stereochemical restraints generated from program PRODRG (Schuttelkopf and van Aalten, 2004). TLS group refinements were introduced to model data anisotropy. Solvents were added automatically by Arp/warp and manually inspected and modified in the program O. The refinement results and model statistics are listed in Table 1.
DATA DEPOSITION
Atomic coordinates and structure factors have been deposited at the Protein Data Bank (http://www.pdb.org) with the accession codes of 3DZF for ara-F-R5P complex, 3DZG for ara-F-R5P/ Nic complex, 3DZH for GTP complex, 3DZI for R5PI/GTP complex, 3DZJ for NMN/E226Q complex, and 3DZK for NMN/wtCD38 complex.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH to MacCHESS (RR01646) and H.C.L./Q.H. (GM061568). The crystallographic data were collected at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the NSF and NIH National Institute of General Medical Sciences under award DMR-0225180.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aarhus R, Graeff RM, Dickey DM, Walseth TF, Lee HC. ADP-ribosyl cyclase and CD38 catalyze the synthesis of a calcium-mobilizing metabolite from NADP. J Biol Chem. 1995;270:30327–30333. doi: 10.1074/jbc.270.51.30327. [DOI] [PubMed] [Google Scholar]
- Berti PJ, Blanke SR, Schramm VL. Transition state structure for the hydrolysis of NAD(+) catalyzed by diphtheria toxin. Journal of the American Chemical Society. 1997;119:12079–12088. doi: 10.1021/ja971317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bull HG, Ferraz JP, Cordes EH, Ribbi A, Apitzcastro R. Concerning Mechanism of Enzymatic and Non-Enzymatic Hydrolysis of Nicotinamide Nucleotide Coenzymes. Journal of Biological Chemistry. 1978;253:5186–5192. [PubMed] [Google Scholar]
- Cakir-Kiefer C, Muller-Steffner H, Schuber F. Unifying mechanism for Aplysia ADP-ribosyl cyclase and CD38/NAD(+) glycohydrolases. Biochem J. 2000;349:203–210. doi: 10.1042/0264-6021:3490203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi Y, Kato I, Takasawa S, Sasaki T, Ong BH, Sato M, Ohsaga A, Sato K, Shirato K, Okamoto H, Maruyama Y. Identification of cyclic ADP-ribose-dependent mechanisms in pancreatic muscarinic Ca(2+) signaling using CD38 knockout mice. J Biol Chem. 2001;276:649–655. doi: 10.1074/jbc.M004469200. [DOI] [PubMed] [Google Scholar]
- Guse AH. Second messenger function and the structure-activity relationship of cyclic adenosine diphosphoribose (cADPR) Febs J. 2005;272:4590–4597. doi: 10.1111/j.1742-4658.2005.04863.x. [DOI] [PubMed] [Google Scholar]
- Handlon AL, Xu C, Mullersteffner HM, Schuber F, Oppenheimer NJ. 2'-Ribose Substituent Effects on the Chemical and Enzymatic-Hydrolysis of Nad(+) Journal of the American Chemical Society. 1994;116:12087–12088. [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Grimaldi JC, Bazan JF, Lund FE, Santos-Argumedo L, Parkhouse RM, Walseth TF, Lee HC. Formation and hydrolysis of cyclic ADP-ribose catalyzed by lymphocyte antigen CD38. Science. 1993;262:1056–1059. doi: 10.1126/science.8235624. [DOI] [PubMed] [Google Scholar]
- Jin D, Liu HX, Hirai H, Torashima T, Nagai T, Lopatina O, Shnayder NA, Yamada K, Noda M, Seike T, et al. CD38 is critical for social behaviour by regulating oxytocin secretion. Nature. 2007 doi: 10.1038/nature05526. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kato I, Yamamoto Y, Fujimura M, Noguchi N, Takasawa S, Okamoto H. CD38 disruption impairs glucose-induced increases in cyclic ADP-ribose, [Ca2+]i, and insulin secretion. J Biol Chem. 1999;274:1869–1872. doi: 10.1074/jbc.274.4.1869. [DOI] [PubMed] [Google Scholar]
- Lee HC. Cyclic ADP-ribose: a calcium mobilizing metabolite of NAD+ Mol Cell Biochem. 1994;138:229–235. doi: 10.1007/BF00928466. [DOI] [PubMed] [Google Scholar]
- Lee HC. Enzymatic functions and structures of CD38 and homologs. Chem Immunol. 2000;75:39–59. doi: 10.1159/000058774. [DOI] [PubMed] [Google Scholar]
- Lee HC. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu Rev Pharmacol Toxicol. 2001;41:317–345. doi: 10.1146/annurev.pharmtox.41.1.317. [DOI] [PubMed] [Google Scholar]
- Lee HC. Multiplicity of Ca2+ messengers and Ca2+ stores: a perspective from cyclic ADP-ribose and NAADP. Curr Mol Med. 2004;4:227–237. doi: 10.2174/1566524043360753. [DOI] [PubMed] [Google Scholar]
- Lee HC. Structure and enzymatic functions of human CD38. Mol Med. 2006;12:317–323. doi: 10.2119/2006-00086.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Munshi C, Graeff R. Structures and activities of cyclic ADP-ribose, NAADP and their metabolic enzymes. Mol Cell Biochem. 1999;193:89–98. doi: 10.1007/978-1-4419-8740-2_13. [DOI] [PubMed] [Google Scholar]
- Lee HC, Walseth TF, Bratt GT, Hayes RN, Clapper DL. Structural Determination of a Cyclic Metabolite of Nad+ with Intracellular Ca-2+-Mobilizing Activity. Journal of Biological Chemistry. 1989;264:1608–1615. [PubMed] [Google Scholar]
- Lee HC, Zocchi E, Guida L, Franco L, Benatti U, De Flora A. Production and hydrolysis of cyclic ADP-ribose at the outer surface of human erythrocytes. Biochem Biophys Res Commun. 1993;191:639–645. doi: 10.1006/bbrc.1993.1265. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kriksunov IA, Graeff R, Lee HC, Hao Q. Structural basis for formation and hydrolysis of the calcium messenger cyclic ADP-ribose by human CD38. J Biol Chem. 2007;282:5853–5861. doi: 10.1074/jbc.M609093200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kriksunov IA, Graeff R, Munshi C, Lee HC, Hao Q. Crystal structure of human CD38 extracellular domain. Structure. 2005;13:1331–1339. doi: 10.1016/j.str.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kriksunov IA, Graeff R, Munshi C, Lee HC, Hao Q. Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis. J Biol Chem. 2006;281:32861–32869. doi: 10.1074/jbc.M606365200. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui-Saito M, Kato I, Takasawa S, Okamoto H, Yanagisawa T. CD38 gene disruption inhibits the contraction induced by alpha-adrenoceptor stimulation in mouse aorta. J Vet Med Sci. 2003;65:1325–1330. doi: 10.1292/jvms.65.1325. [DOI] [PubMed] [Google Scholar]
- MullerSteffner HM, Augustin A, Schuber F. Mechanism of cyclization of pyridine nucleotides by bovine spleen NAD(+) glycohydrolase. Journal of Biological Chemistry. 1996;271:23967–23972. doi: 10.1074/jbc.271.39.23967. [DOI] [PubMed] [Google Scholar]
- Munshi C, Aarhus R, Graeff R, Walseth TF, Levitt D, Lee HC. Identification of the enzymatic active site of CD38 by site-directed mutagenesis. J Biol Chem. 2000;275:21566–21571. doi: 10.1074/jbc.M909365199. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O'Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- Oppenheimer NJ. NAD hydrolysis: chemical and enzymatic mechanisms. Mol Cell Biochem. 1994;138:245–251. doi: 10.1007/BF00928468. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Partida-Sanchez S, Cockayne DA, Monard S, Jacobson EL, Oppenheimer N, Garvy B, Kusser K, Goodrich S, Howard M, Harmsen A, et al. Cyclic ADP-ribose production by CD38 regulates intracellular calcium release, extracellular calcium influx and chemotaxis in neutrophils and is required for bacterial clearance in vivo. Nat Med. 2001;7:1209–1216. doi: 10.1038/nm1101-1209. [DOI] [PubMed] [Google Scholar]
- Partida-Sanchez S, Goodrich S, Kusser K, Oppenheimer N, Randall TD, Lund FE. Regulation of dendritic cell trafficking by the ADP-ribosyl cyclase CD38: impact on the development of humoral immunity. Immunity. 2004;20:279–291. doi: 10.1016/s1074-7613(04)00048-2. [DOI] [PubMed] [Google Scholar]
- Rai KR, Peterson BL, Appelbaum FR, Kolitz J, Elias L, Shepherd L, Hines J, Threatte GA, Larson RA, Cheson BD, Schiffer CA. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N Engl J Med. 2000;343:1750–1757. doi: 10.1056/NEJM200012143432402. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Deng HT, Angeletti RH, Schramm VL. A covalent intermediate in CD38 is responsible for ADP-ribosylation and cyclization reactions. Journal of the American Chemical Society. 2000;122:7855–7859. [Google Scholar]
- Sauve AA, Munshi C, Lee HC, Schramm VL. The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry. 1998;37:13239–13249. doi: 10.1021/bi981248s. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Mechanism-based inhibitors of CD38: a mammalian cyclic ADP-ribose synthetase. Biochemistry. 2002;41:8455–8463. doi: 10.1021/bi0258795. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr Med Chem. 2004;11:807–826. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- Schuber F, Lund FE. Structure and enzymology of ADP-ribosyl cyclases: Conserved enzymes that produce multiple calcium mobilizing metabolites. Current Molecular Medicine. 2004;4:249–261. doi: 10.2174/1566524043360708. [DOI] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr Med Chem. 2004;11:857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- Sleath PR, Handlon AL, Oppenheimer NJ. Pyridine Coenzyme Analogs .3. Synthesis of 3 Nad+ Analogs Containing a 2'-Deoxy-2'-Substituted Nicotinamide Arabinofuranosyl Moiety. Journal of Organic Chemistry. 1991;56:3608–3613. [Google Scholar]
- Smith BC, Denu JM. Sir2 protein deacetylases: Evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Iqbal J, Dolgilevich S, Yuen T, Wu XB, Moonga BS, Adebanjo OA, Bevis PJ, Lund F, Huang CL, et al. Disordered osteoclast formation and function in a CD38 (ADP-ribosyl cyclase)-deficient mouse establishes an essential role for CD38 in bone resorption. Faseb J. 2003;17:369–375. doi: 10.1096/fj.02-0205com. [DOI] [PubMed] [Google Scholar]
- Tarnus C, Muller HM, Schuber F. Chemical Evidence in Favor of a Stabilized Oxocarbonium-Ion Intermediate in the Nad+ Glycohydrolase-Catalyzed Reactions. Bioorganic Chemistry. 1988;16:38–51. [Google Scholar]
- Tarnus C, Schuber F. Application of Linear Free-Energy Relationships to the Mechanistic Probing of Nonenzymatic and Nad+-Glycohydrolase-Catalyzed Hydrolysis of Pyridine Dinucleotides. Bioorganic Chemistry. 1987;15:31–42. [Google Scholar]
- Versees W, Loverix S, Vandemeulebroucke A, Geerlings P, Steyaert J. Leaving group activation by aromatic stacking: an alternative to general acid catalysis. J Mol Biol. 2004;338:1–6. doi: 10.1016/j.jmb.2004.02.049. [DOI] [PubMed] [Google Scholar]