Abstract
Biomolecular surface mapping methods offer an important alternative method for characterizing protein-protein and protein-ligand interactions in cases in which it is not possible to determine high resolution 3D structures of complexes. Hydroxyl radical footprinting offers a significant advance in footprint resolution compared to traditional chemical derivatization. Here we present results of footprinting performed with hydroxyl radicals generated on the nanosecond time scale by a laser-induced photodissociation of hydrogen peroxide. We applied this emerging method to a carbohydrate-binding protein, galectin-1. Since galectin-1 occurs as a homodimer, footprinting was employed to characterize the interface of the monomeric subunits. Efficient analysis of the MS data for the oxidized protein was achieved with the recently developed ByOnic software that was altered to handle the large number of modifications arising from side chain oxidation. Quantification of the level of oxidation has been achieved by employing spectral intensities for all of the observed oxidation states on a per-residue basis. The level of accuracy achievable from spectral intensities was determined by examination of mixtures of synthetic peptides, related to those present after oxidation and tryptic digestion of galectin-1. A direct relationship between side chain solvent accessibility and level of oxidation emerged that enabled the prediction of the level of oxidation given the 3D structure of the protein. The precision of this relationship was enhanced through the use of average solvent accessibilities computed from 10 ns molecular dynamics simulations of the protein.
Keywords: AMBER, ByOnic, galectin-1, MD simulations, oxidative surface mapping, solvent accessible surface area, SASA
Introduction
There has been increasing interest in the development of high-throughput methods for characterizing protein-ligand contact surfaces (footprinting). One approach employs hydrogen-deuterium exchange mass spectrometry (H/D-Ex MS) method [1-5], in which amide protons in the backbone of a protein are allowed to undergo exchange in deuterated water. To detect and quantify the level of deuterium incorporation by mass spectrometry typically necessitates proteolytic digestion under conditions that attenuate hydrogen back-exchange, such as low pH and low temperature. However, under these conditions, complete digestion of the protein is difficult to achieve [6]. Additionally, while deuterium exchange is altered in the presence or absence of a ligand, it is a complex phenomenon that depends heavily on protein folding kinetics and local amino acid sequence. It is also worth noting that the backbone amide protons are not necessarily differentially shielded by the presence of a ligand, that is, they may be shielded by side chains in both the free and bound form of a protein-ligand complex. Chemical-derivatization methods, such as biotinylation [7,8] or acetylation [9,10], which focus on the side chains, have been employed to probe protein-protein, protein-ligand or protein-nucleic acid interactions. The advantage of these methods over deuterium-exchange is that the modification is irreversible, facilitating analysis, whereas perhaps the most notable limitation is that the only a few residues or types of residues will be labeled in any protein. Thus, the resolution of the contact surface is extremely low. An alternative chemical derivatization method that employs hydroxyl radicals to oxidize the side chains appears to offer several advantages. Firstly, hydroxyl radicals are highly reactive and result in covalent modifications that facilitate a high throughput proteomic type analysis of the product. Further, in contrast to H/D exchange, the extent of hydroxyl radical oxidation, for any given amino acid, appears to depend directly on solvent accessibility [11-13]. Thus, this type of labeling should be useful in identifying ligand contact regions. Lastly, in contrast to other chemical derivatization methods, depending on the manner in which they are generated and on exposure times, hydroxyl radicals can react with all amino acid side chains. Methods to generate hydroxyl radicals in biological environments, include: direct synchrotron radiolysis of water [14-17], photodissociation of dilute hydrogen peroxide induced by high energy laser [18-20], UV [13,21] or γ-irradiation [22]. Alternative methods include, Fenton chemistry [23-25], whereby hydroxyl radicals are generated by contact between hydrogen peroxide and metal-bound ethylenediaminetetraacetic acid, or by high voltage discharge within an electrospray ion source [26,27]. In several methods, a major drawback is the availability or complexity of the instrumentation. While Fenton chemistry is an affordable method that has been utilized in the structural analysis of nucleic acids [28-30], and has been also applied to study proteins [31,32] it also requires long incubation times and chemical additives. For a weak interaction, as is typical for many carbohydrate-protein complexes, a long incubation, or exposure time would allow the ligand to undergo multiple exchanges with the protein surface, during which the contact region would be exposed to reactive radicals. Additionally, rapid reaction is necessary to ensure that the oxidation of the amino acids take place before any unfolding or other conformational changes of the protein occurs. To minimize these issues, we believe pulsed laser-induced dissociation of hydrogen peroxide is advantageous due to the relatively rapid speed of hydroxyl radical generation.
Previous studies by Hambly [19,20] and Aye [18] have demonstrated the utility of generating hydroxyl radicals by a single laser pulse of dilute hydrogen peroxide. The radical concentration achieved from a single pulse of a dilute peroxide solution (1% or less) is adequate to achieve significant levels of protein surface oxidation [18,19]. This is an important observation since the resulting oxidation leads to protein denaturation [18] and, thus, although subsequent irradiation would ensure high oxidation levels, it would not be appropriate for 3D structural studies. In the limit, multiple laser pulses can also lead to backbone cleavage [33].
High resolution footprinting necessitates not only the identification of the sites of oxidation (on a per amino acid residue basis), but also the quantitation of the level or rate of oxidation at each site. Mass spectrometry provides a powerful tool for this characterization, although since the majority of side chains can exhibit multiple oxidation states data analysis is challenging. In a typical protein sequencing analysis, the protein is digested by a specific protease, such as trypsin, and the resulting fragments sequenced by tandem MS methods. This approach relies heavily on a knowledge of the masses of each amino acid and any post-translation modifications. Commonly used proteomics software, such as SEQUEST [34] or Mascot [35], are not optimized to handle the extensive number of permutations generated by the range of possible side chain oxidation states. To address this issue, we employed a program (ByOnic [36]), which has been specifically modified to handle the large number of amino acid modifications associated with hydroxyl radical oxidation. ByOnic combines de novo sequencing and database searching to match the peptide fragments to a sequence provided by the user.
Although relative peptide abundances in a protein digest may be estimated by spectral counting, the quantitation of proteomics data benefits from more sophisticated treatments [37]. Here we employ a label-free approach to quantify the subtle differences in pre-residue oxidation levels arising from reaction with hydroxyl radical. Proteolysis of an oxidized protein leads to multiple instances of peptides with the same sequence, which differ only in the extent of oxidation. To quantify the level of oxidation at any given site requires the identification of all peptides with oxidation at that site, using tandem MS. We compute the percentage of oxidation at each amino acid (%AAox) from the LC-MS signal intensities of each peptide containing a specific oxidized amino acid (Iox), relative to the total of all intensities associated with that peptide sequence (Iox + Inon-ox) using the following straightforward relationship:
| (1) |
To ascertain the suitability of our use of spectral intensities for quantitation of oxidation levels, as well as to indicate the extent to which a quantitative relationship exists between oxidation and side chain solvent accessibility, we have applied hydroxyl radical footprinting to a small globular protein for which extensive x-ray structural data exists. Galectin-1 is a homodimer member of the galectin family found in many animals including humans, which has the ability to bind oligosaccharide ligands containing galactosyl residues [38-42]. Galectins are the target of many studies because of their involvement in various biological processes such as cell adhesion, differentiation, regulation of cell growth, regulation of apoptosis, cell activation, mRNA splicing, inflammation, and induction of Ca2+ dependent signal transduction pathways [43-46]. Due to its small size, and the availability of x-ray structures from various species, with various ligands, galectin-1 is a suitable protein for mass spectrometric analysis and surface mapping method development.
The interpretation of hydroxyl radical surface mapping data, in terms of 3D structural properties, is based on the assumption that those residues that are exposed to solvent will react more readily with hydroxyl radicals than buried or otherwise shielded residues. To assist in the analysis of the MS data, we have employed molecular dynamics (MD) simulations to predict the time-averaged solvent accessible surface area (<SASA>) of each side chain [5]. In contrast to x-ray structural data, MD simulations enable the generation of a realistic ensemble of side chain orientations in solution at experimental temperature, which can be particularly important when employing homology models [47]. Additionally, MD refined side chain ensembles have been shown to lead to better correlation with experimentally-determined levels of oxidation [5], and so provide a robust basis for estimating <SASA> values. This computational approach also allows us to separate the galectin-1 homodimer into its two subunits and predict the solvent accessibility of the residues in the interface region. In addition, molecular modeling techniques [48-50] can be particularly useful in generating the 3D fold of proteins of interest when no experimentally-derived structure is available, and in this study we examine the extent to which a homology-based model of galectin-1 can be used to interpret oxidative footprinting data. In principle, oxidative surface mapping could provide a valuable alternative to NMR or x-ray analysis for validating predictions from such popular computational methods as homology modeling and automated ligand docking. To employ footprinting effectively in this regard, a quantitative understanding of the relationship between solvent accessibility and extent of oxidation must be developed. We are approaching this problem by quantifying the experimental data and by employing a robust method to estimate side chain <SASA>.
Experimental
Sample preparation
Galectin-1 expressed by Chinese hamster ovary (CHO) cells with the sequence characterized by Cho & Cummings [39], was purified as described previously [39], and stored under reducing conditions in presence of dithiothreitol (DTT) [51], Bio-Rad Laboratories (Hercules, CA). Prior to laser exposure, DTT was removed by dialysis using Spectra/Por RC Float-A-Lyzer, MW 3,500, Spectrum Laboratories, Inc. (Rancho Dominguez, CA). Galectin-1 concentration was adjusted to 0.25 mM by adding a ten times diluted phosphate buffer saline (0.1 × PBS), Mediatech, Inc. (Herndon, VA). Immediately prior to laser exposure, the protein solution was mixed with hydrogen peroxide (10%, by dilution of 30% solution w/w), Sigma-Aldrich (St. Louis, MO) to a final concentration of 1%, to a final volume of 8 μL (final protein concentration approximately 0.22 mM).
Synthetic peptides, in greater than 98% purity were purchased from Biomatic Corporation (Wilmington, DE) and stored as a lyophilized powder. Prior to laser exposure, each peptide was re-suspended in 25 mM ammonium bicarbonate buffer to a concentration of 0.75 mM. An aliquot (4.2 μL) of the peptide solution was added to a solution of hydrogen peroxide (10%) and 25 mM ammonium bicarbonate (both Sigma-Aldrich) and adjusted to a final concentration of 1% H2O2, and the total volume of 8 μL (final peptide concentration approximately 30 mM). Purified water (18.2 MΩ) for all re-suspensions and buffer preparations was obtained from an in-house NANOpure Diamond system.
Laser photolysis
Hydroxyl radicals were generated by exposure to radiation from a Compex 110 KrF excimer laser operating at 248 nm, Lambda Physik, Coherent Inc. (Santa Clara, CA). A microcentrifuge tube (1.5 mL) containing 8 μL of the protein, or peptide, hydrogen peroxide solution was aligned along the beam axis to ensure the maximal contact of the laser beam with the protein solution, at a distance of 12 cm from the laser source. Each sample was exposed to a single 30 ns laser pulse with the adjusted laser power of 110 mJ/pulse. In the case of multiple pulses, a pulse rate of 5 Hz was employed. Immediately after irradiation, the sample was flash-frozen in liquid nitrogen and lyophilized for 1.5 hr to remove any remaining H2O2 and water.
Gel electrophoresis
Prior to gel electrophoresis, the lyophilized samples were re-suspended in 0.1 × PBS buffer. A premixed running electrophoresis Laemmli sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate (SDS), 25% glycerol, 0.01% Bromophenol Blue) with addition of 5% reducing agent β-mercaptoethanol (both Bio-Rad Laboratories) was added to the PBS-sample solution in a 2:1 v/v ratio. A 4-10% Tris-Tricine SDS gel (Bio-Rad Laboratories) was run under two buffer systems, anode (0.2 M Tris-HCl, pH 8.9) and cathode (0.1 M Tris, 1 M Tricine, 1% SDS, pH 8.25) [52] to identify any possible protein fragments. Electrophoresis voltage and current conditions were 150 V and 100 A, respectively, and the length of the run was 1 hr 15 min. After completion of the run, the gel was washed in nanopure water and stained via a silver staining procedure, using SilverSNAP® Stain Kit II, Pierce (Rockford, IL) following the manufacturer’s protocol.
Proteolytic digestion
In order to prepare the sample for mass spectrometric analysis, the lyophilized protein (or peptide) was re-suspended in 15 μL (50 mM) ammonium bicarbonate (Sigma-Aldrich) digest buffer. Dithiothreitol (Bio-Rad Laboratories) was added to a final DTT concentration of 10 mM, and the solution heated in a water bath for 1 hr at 60° C. Iodoacetamide (IAA, Sigma-Aldrich) was added to the sample to give a final IAA concentration of 50 mM, and the sample incubated for 1 hr at room temperature. Finally, sequencing grade modified trypsin, Promega (Madison, WI) was added in a 1:30 (protease:protein) ratio. The pH of the mixture was determined by pHydrion Papers, Micro Essential Laboratory (Brooklyn, NY) to be between 6 and 8. The sample was then incubated for 18 hrs at 37° C. Digestion was quenched by removing the trypsin and salts from the solution by passing through a C-18 ZipTip, Millipore Corporation (Bedford, MA). The galectin-1 digest was eluted from the ZipTip with 10 μL of mixed solvent acetonitrile:water:formic acid (50:49.9:0.1 (v/v)), dried on a speedvac, and resuspended in 10 μL (0.1%, v/v) aqueous solution of formic acid (Sigma-Aldrich) and stored at 6° C before analyzed.
ESI-FT AND MALDI-TOF
Single mass spectra were obtained by high resolution MALDI-TOF, 4700 Proteomics Analyzer, Applied Biosystems (Lincoln, CA). In a typical experiment, 0.5 μL of each sample was mixed with 0.6 μL of 10 mg/mL α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) dissolved in ethanol, spotted on a MALDI plate, Applied Biosystems (Foster City, CA) and analyzed in a reflector positive mode with 4,200-fixed laser intensity.
Tandem mass spectrometry was performed on a Finnigan LTQ-FT hybrid linear ion trap/Fourier transform ion cyclotron resonance (FTICR) mass spectrometer (7-Tesla) with an electrospray ionization source (ESI), Thermo Electron (San Jose, CA), connected to an Agilent 1100 capillary liquid chromatograph (Palo Alto, CA) through a C18 column, 15 cm × 150 μm, Micro-Tech Scientific (Vista, CA). To elute peptides from the column, mobile phases A, 99.9% H2O/0.1% formic acid (v/v), and B, 99.9% acetonitrile/0.1% formic acid (v/v) were used. In a 130 min run, a gradient from 5-80% (v/v) B was employed at a flow rate of 1 μL/min. MS/MS spectra were acquired on the nine most abundant ion precursors from each MS scan. Raw data sets were converted into mzXML format, followed by conversion to pkl format, using RedAW and mzXML2Other [53].
Peak assignment and quantitation of oxidation
Established software for identification of MS/MS peaks from proteolytic digests, such as Mascot [35] and SEQUEST [34] are not well suited for the analysis of data sets that include a large number of mass modifications, as arise from hydroxyl radical oxidation. Emerging programs, such as MyriMatch [54] and ByOnic [36], have more flexibility in this regard. For peak assignment we employed ByOnic to search against a database containing only the CHO galectin-1 sequence [39], augmented by common mass modifications, as associated with carbamidomethylation of cysteine and oxidation of methionine, as well as 14 potential mass shifts arising from side chain oxidation. This number of mass modifications leads to over 45 potential oxidation products (Table 1). Most of the expected oxidation products have been previously characterized by Chance’s laboratory [11,55,56] and some of the less common modifications have been described by Berlet et al. [57] and Kubota et al. [58]. ByOnic includes three features not found in most database-search programs that make it especially suitable for identifying multiply modified peptides. First, it recalibrates m/z measurements based on well-identified peaks from an initial pass through the data. We defined a well-identified peak to be a singly charged b- or y-ion from a peptide identification with p-value smaller than 0.001. Unlike SEQUEST or Mascot, ByOnic’s scoring algorithm counts a match with 0.1 Da error more heavily than one with 0.2 Da error; hence recalibration offers improved sensitivity and specificity. Second, to improve efficiency, each type of modification has a user-defined limit on the number of occurrences per peptide. For example, up to five modifications per peptide were allowed, with at most four oxidations on reactive residues (cysteine, methionine, histidine, tryptophan, leucine, isoleucine, valine, arginine, phenylalanine, and tyrosine), and at most one on less reactive residues. This is in contrast to other methods that allow a peptide to carry any number of each type of enabled modification. Third, results are double-checked with a “wild-card” modification: this type of search allows any integer change, within user-settable mass limits, to any one residue in each peptide. A wild card can be enabled along with any other set of modifications, and often finds unanticipated chemical artifacts such as sodiation, over-alkylation [59], and carbamidomethylated N-termini. Therefore, ByOnic was chosen as the main database search tool for this study. In general searches were performed with the following search parameters: specificity for tryptic fragments, fragment ion tolerance of 0.35 Da (after recalibration), peptide tolerance varied with parent charges (1.5 Da per charge), and including all of the mass modifications listed in Table 1. All oxidation positions were confirmed by tandem MS.
Table 1.
Observeda mass changes from side chain oxidation
| Residue | Potential mass changes | Associated modifications [55,57] |
|---|---|---|
| C | -16; +32, +48 | hydroxy, sulfonic acid |
| M | +16; -32; +32 | sulfoxide, aldehyde, sulfone |
| W | +16, +32, +48; +20 | hydroxy, ring opening, hydroxykynurenin |
| Y | +16, +32, +48 | hydroxy |
| F | +16, +32, +48 | hydroxy |
| H | +16; +5, -10, -23, -22 | oxo, ring opening |
| L | +16; +14 | hydroxy, carbonyl |
| I b | +16; +14 | hydroxy, carbonyl |
| V | +16; +14 | hydroxy, carbonyl |
| P | +16; +14; -30 | hydroxy, carbonyl, pyrrolidone |
| R | -43; +16; +14 | deguanidination, hydroxy, carbonyl |
| Kb | +16; +14 | hydroxy, carbonyl |
| E | -30; +16; +14 | decarboxylation, hydroxy, carbonyl |
| Q | +16; +14 | hydroxy, carbonyl |
| D | -30; +16 | decarboxylation, hydroxy |
| Nb | +16 | hydroxy |
| S | +16; -2 | hydroxy; - H2O |
| Tb | -2 | - H2O |
| Ab | +16 | hydroxy |
Observed mass changes shown in boldface
Low level of oxidation observed (< 2%)
Quantitation of the level of oxidation at each amino acid position was performed using an in-house program that first matched the peptide masses from the LC-MS spectra to the sequences previously identified by ByOnic. The level of oxidation for any given amino acid was then computed by summing the spectral intensities for all peptides containing that amino acid, using the LC-MS spectral intensities for the peptides. The percentage of oxidation on a per-residue basis was then readily computed from Equation 1 and a representative example of this procedure is presented in Figure S1. For reasons of sensitivity, levels of oxidation were computed from LC-MS data rather than from MALDI-TOF data. Although this required a more complex data analysis, it generated a more complete list of oxidation states and therefore led to a more complete understanding of the relationship between solvent accessibility and level of oxidation.
Molecular modeling
Since there are no crystal structures of CHO galectin-1, a homology model was generated that was subsequently employed in MD simulations. A template structure (pdb id: 1W6P [60]) for the homology model that has 90% sequence identity with CHO galectin-1 was identified by searching the National Center for Biotechnology Information (NCBI) BLAST database (www.ncbi.nlm.nih.gov) and retrieved from the Research Collaboratory for Structural Biology (RCSB) data base [61]. The model was built using the Molecular Operating Environment (MOE), Chemical Computing Group Inc. (www.chemcomp.com) program. In order to calculate the <SASA> of each side chain in galectin-1, a 10-ns MD simulation of the complete protein (homodimer), as well as of one subunit of the protein (monomer) was performed using the all-atom AMBER 9 [62] force field with the PARM99 protein parameters [63]. All histidine residues were considered as neutral and were protonated at the Nε position. Prior to the MD simulations, the protein was solvated by 10,600 TIP3P [64] water molecules for the homodimer, and 5,637 waters for monomer. Counter ions were added to neutralize charge (four Na+ per monomer) using the tLEAP module of AMBER. Each system was subjected to energy minimization (1000 steps of steepest descent and 1000 of conjugate gradient) and heated from 0 K to the simulation temperature of 300 K over 50 ps. The simulation was performed using the NPT ensemble, at 1 atm using a 2-fs integration time step. The atomic co-ordinates were stored every 10 ps for analysis, for a total of 1000 snapshots. SASA values were calculated for individual snapshots employing the NACCESS program [65], with <SASA> and standard deviations computed over the 10 ns simulation.
Results and Discussion
Effect of number of laser exposures, and hydrogen peroxide concentration on protein integrity
To ensure a high resolution footprint, it is necessary to minimize the duration of exposure to hydroxyl radical relative to ligand binding kinetics; yet, to observe oxidation an adequate level of hydroxyl radicals is essential. Laser induced radical formation has the potential to satisfy these requirements [18]. Additionally, the extent of side chain oxidation may be enhanced by increasing the number of laser exposures, or by increasing the concentration of peroxide [18]; however, extreme oxidation conditions can lead to protein disintegration via back bone cleavage [33]. To examine the effect of these variables on protein integrity, the experiment was performed with 1, 5, 15, and 25 laser exposures, and under a range of hydrogen peroxide concentrations and analyzed by SDS-PAGE.
In the case of galectin-1, backbone cleavages were observed (Figure S2) after multiple laser pulses, especially in cases of 15 and 25 shots, which on the SDS-PAGE gel can be seen as smaller fragments that migrated between 14 and 5 kDa. In addition, there is an obvious decrease in the intensity of galectin-1 at 14.5 kDa after 25 laser pulses which can be attributed to extensive protein degradation. In contrast, the SDS-PAGE analysis indicates that increasing the concentration of hydrogen peroxide does not affect protein integrity with a single laser pulse. However, to avoid the potential for protein denaturation [18], we elected to employ a single laser exposure with a 1% peroxide concentration.
Oxidation experiments applied to a synthetic peptide
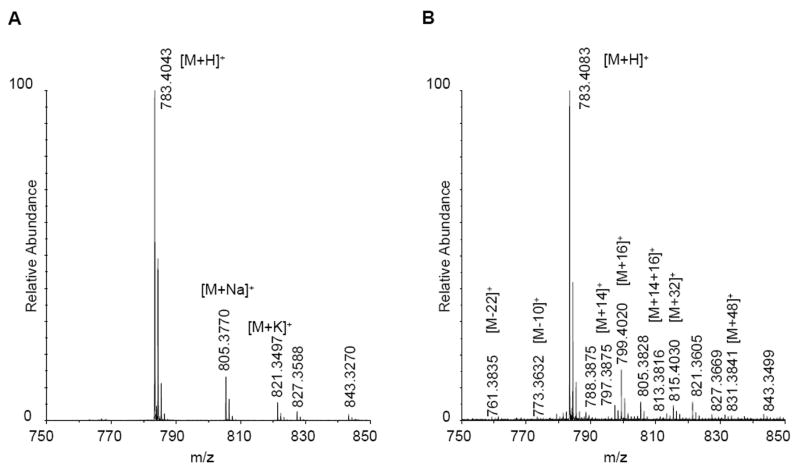
To confirm that our experimental conditions were able to generate the expected oxidation states without leading to any backbone fragmentation (as suggested by the SDS-PAGE data) a synthetic fragment of galectin-1 (LHFNPR, peptide-1) was subjected to hydroxyl radical oxidation. Peptide-1 corresponds to positions 44-49 in galectin-1 and with m/z 783.4 (molecular weight of 782.91 Da). An examination of the MALDI-TOF data for the oxidized peptide confirmed that there was no backbone cleavage. As expected, the most abundant product was the peptide with a single oxidation (mass change of +16 Da, m/z 799.4), while the second most abundant product contained two oxidations (+32 Da, m/z 815.4) (Figure 1). Subsequent MS/MS analysis confirmed that most of the expected oxidation states were present in the sample. The difference between the MALDI-TOF and LC-MS experiments was in the ability of the FT instrument to detect products of oxidation that were present in very low amounts. By tandem mass spectrometry, histidine oxidation states with mass changes of +16, -10 and -22 Da were identified, as well as leucine oxidation products with +16 and +14 mass changes, and proline with addition of 14 Da. Phenylalanine was found in three oxidation states with addition of 16, 32, and 48 Da. Some of these higher oxidation products were also identified in the MALDI-TOF spectrum, but in such low quantities that in other circumstances these would be most likely considered as noise.
Figure 1.
Peptide-1 before (A) and after oxidation (B), indicating oxidation of phenylalanine (m/z 799) and histidine (m/z 761 and 773).
Not all of the possible oxidation states were identified for the residues in peptide-1, for example neither arginine nor asparagine were oxidized. Nonetheless, the extent of oxidation was deemed sufficient to warrant application of this method to intact galectin-1. Prior to that, however, we continued with the examination of synthetic peptides in order to determine the extent to which the level of oxidation could be quantified from spectral intensities.
Quantitation of level of oxidation
In order to confirm that spectral intensities may be employed to quantify the extent of oxidation, as defined in Equation 1, samples of two additional synthetic peptides, which represent oxidized fragments of galectin-1, were prepared in known concentrations. In peptide-2 (LHYNPR), the phenylalanine has been replaced by a tyrosine in position three. This substitution results in a net change of 16 Da and is equivalent to an oxidation product of the phenylalanine. Similarly, peptide-3 (LDFNPR), is also an oxidized form of peptide-1. In peptide-3, the histidine was replaced by an aspartic acid, which is a net mass change of -22 Da. At an advanced stage of histidine oxidation, the ring of the histidine side chain opens, and two amino groups and a carbonyl group are replaced by a carboxyl group, which results in a loss of 22 Da [11,15]. A summary of the peptide molecular weights, amino acid replacements and the net molecular weight changes is presented in Table 2.
Table 2.
Synthetic peptides used in quantification studies
| Peptide | Sequence | m/z | Mass Change | Oxidation Equivalent |
|---|---|---|---|---|
| 1 | LHFNPR | 782.91 | Non-oxidized | |
| 2 | LHYNPR | 798.91 | +16 Da | F3Y |
| 3 | LDFNPR | 760.85 | -22 Da | H2D |
Mixtures of each of the peptides containing representative oxidations (peptide-2 and peptide-3) with the non-oxidized material (peptide-1) were prepared in the following ratios peptide-1:peptide-2 (60:40) and peptide-1:peptide3 (33:66). Additionally, a sample containing all three peptides was prepared in a ratio of peptide-1:peptide-2:peptide3 (50:25:25). From the peak intensities of the peptides in each sample, the levels of oxidation for phenylalanine and histidine were determined (Equation 1, Table 3). The intensities for all peptides in each sample were determined using both MALDI-TOF and LC-MS experiments. To be consistent with the proposed analysis of galectin-1, all MS data for the synthetic samples were processed using the ByOnic software, followed by intensity summation on a per-residue basis using an in-house program. In general, both mass spectrometric methods resulted in close agreement with each other and with the known concentrations. Quantitation using the LC-MS data was only slightly less accurate than the high resolution MALDI data. In the case of the phenylalanine oxidation (peptide-1:peptide-2) a maximum error of 2% with respect to the known concentrations was observed, while for the histidine oxidation (peptide-1:peptide-3) a slightly higher error of 6% was noted. In the third test case, the ratio of all three peptides was determined. Again, there was generally good agreement between the known ratios and the intensity-determined values for each MS method. As seen in the simple mixtures, the measured level of phenylalanine oxidation (24% by MALDI-TOF, 26% by LC-MS) was in slightly better agreement with the known concentration (25%) than for the histidine oxidation product (22% by MALDI-TOF, 28% by LC-MS). The fact that the ESI and Maldi data were in good agreement with each other and with the expected values suggests that each of the peptides displayed similar ionization efficiencies, in spite of the fact that the “oxidation” of histidine to aspartate (-22) might be expected to impact the net charge. Generally, the dominant charge state for these peptides under the ionization conditions used was +1. Previous studies of a peptide containing both a histidine and an arginine side chain indicated that the first protonation occurred on exclusively on the arginine [66]. Eliminating the histidine would leave the arginine as the dominant basic site, and replacing it with an aspartate should not significantly alter the surface activity in the droplet for the ESI experiments [67,68]. Therefore, under electrospray ionization conditions where the +1 charge state dominates, it is not surprising to find that the replacement of a histidine with an aspartate does not significantly alter the ionization efficiency of the arginine-containing peptide.
Table 3.
Oxidation level equivalents for mixtures of synthetic peptides
| Method | Pep-1 and Pep-2 F oxid – 40% | Pep-1 and Pep-3 H oxid – 66% | Pep-1, pep-2 and pep-3 F and H oxid – 25 % each | |
|---|---|---|---|---|
| m/z 799 oxidation (%) | m/z 761 oxidation (%) | m/z 799 oxidation (%) | m/z 761 oxidation (%) | |
| MALDI-TOF | 41.9 (1.2)a | 64.1 (2.8) | 24.5 (0.8) | 26.5 (0.7) |
| LC-MS | 42.8 (2.1) | 66.4 (6.6) | 23 (1.2) | 27.4 (2.7) |
Standard deviation in parentheses
Extent of oxidation of galectin-1 from single laser exposure
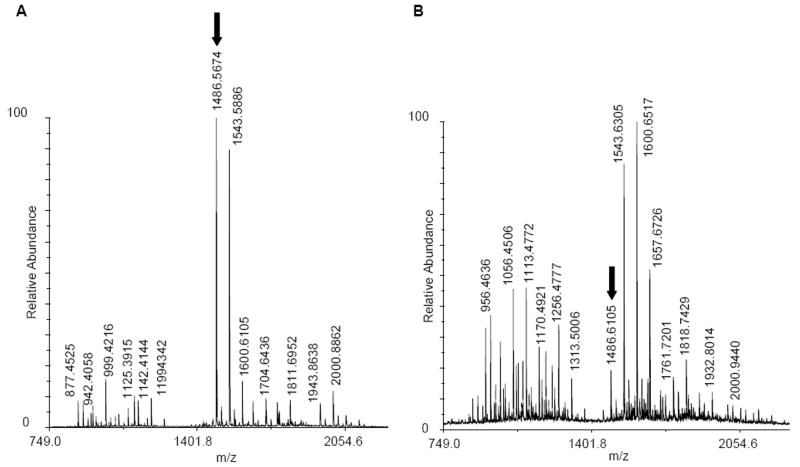
An examination of the high resolution MALDI-TOF spectra of the tryptic digest of oxidized galectin-1 clearly indicated that a single laser pulse in the presence of 1% hydrogen peroxide induced significant oxidation of the protein (Figure 2). As a result of the oxidation, intensities of non-oxidized peptides decreased, for example in the peptides with m/z 1486, 1142, 1041 and 942, while numerous new peaks resulting from side chain oxidation appeared. To fully characterize the sites of oxidation, tandem mass spectrometry with further fragmentation of peptides was performed.
Figure 2.
MS spectrum of tryptic digestion of galectin-1 before oxidation (A), and after oxidation (B). Oxidation leads to more facile proteolysis, formation of peaks from oxidation products, and a decrease in the abundance of non-oxidized peptides (see peak indicated by arrows).
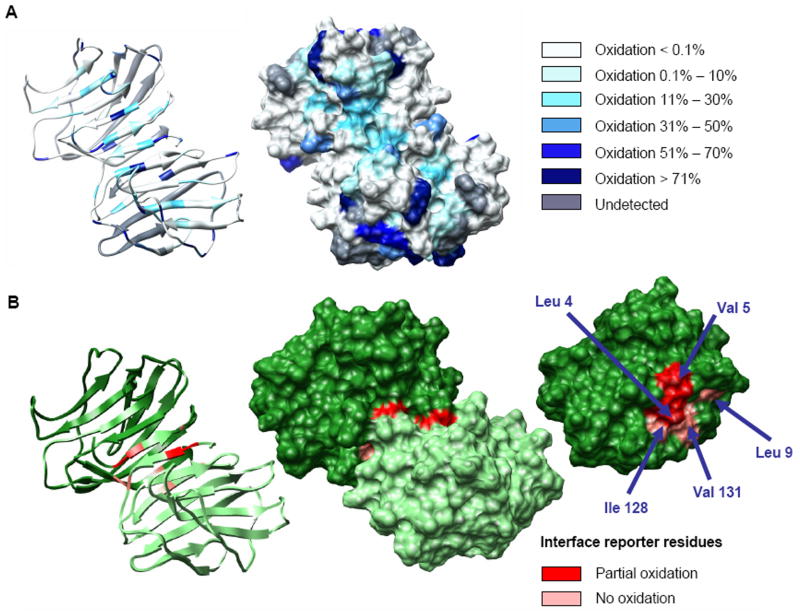
Employing ByOnic to search against a database comprised of the sequence of galectin-1, allowing for 14 known mass changes from oxidation, led to the observation of 43 unique side chain oxidation products (Table 1). Oxidation of the intact protein led to the identification of additional oxidation products, relative to the data obtained for synthetic peptide-1, for example, for arginine and asparagine. An example of the appearance of an oxidized peptide in the tandem MS spectrum of a tryptic peptide from galectin-1 (FNAHGDANTIVCNSK) before and after laser exposure is presented in Figure 3. The MS/MS data clearly identified the oxidation of the histidine residue (-22 Da). Sequence coverage of galectin-1 as determined by MS/MS sequencing of tryptic fragments was satisfactory over triplicates of the experiments (approximately 85% on average). Of these identified residues, approximately 42% displayed oxidation after a single laser exposure (Figure 4a). A quantified list of the total oxidation level observed for each residue is presented in Table 4.
Figure 3.

Tandem mass spectra of a tryptic peptide from galectin-1 in non-oxidized (A) and oxidized (B) forms indicating peaks associated with a 22 Da mass loss, typical of histidine oxidation.
Figure 4.
Per-residue oxidation levels plotted on the solvent accessible surface of dimeric galectin-1 (A). Galectin-1 dimer interface in detail. Reporter residues are indicated in shades of red according to the measured level of side chain oxidation. From left: ribbon structure of the homodimer, solvent accessible surface structure of the homodimer, and monomeric subunit illustrating interfacial residues (B).
Table 4.
Per-residue oxidation levels and side chain <SASA> values for galectin-1
| Residue | Oxidation [%] | <SASA> [Å2] | Residue | Oxidation [%] | <SASA> [Å2] | ||
|---|---|---|---|---|---|---|---|
| dimer | dimer | monomer | dimer | dimer | monomer | ||
| A1 | 0.0 (0.0)a | 57 (14) | 82 (2) | K28 | 0.0 (0.0) | 112 (11) | 114 (8) |
| A6 | 0.1 (0.1) | 8 (4) | 34 (4) | K36 | 0.1 (0.0) | 99 (13) | 115 (8) |
| A24 | 0.0 (0.0) | 38 (8) | 17 6) | K63 | 0.0 (0.0) | 33 (7) | 31 (4) |
| A27 | 0.0 (0.0) | 10 (4) | 0 (0) | K99 | n.d. | 87 (12) | 86 (7) |
| A51 | 0.0 (0.0) | 18 (6) | 21 (4) | K107 | 0.0 (0.0) | 97 (8) | 101 (12) |
| A55 | 0.0 (0.0) | 40 (6) | 46 (4) | K127 | 0.0 (0.0) | 119 (15) | 134 (17) |
| A67 | 0.0 (0.0) | 62 (4) | 64 (2) | K129 | 0.0 (0.0) | 75 (18) | 113 (20) |
| A76 | 0.0 (0.1) | 44 (12) | 41 (5) | L4 | 56.1 (12.4) | 16 (8) | 67 (8) |
| A94 | n.db | 57 (5) | 57 (3) | L9 | 0.0 (0.0) | 3 (2) | 14 (4) |
| A116 | 0.0 (0.0) | 35 (11) | 27 (6) | L11 | 13.3 (8.6) | 8 (6) | 3 (2) |
| A121 | 1.9 (3.1) | 11 (3) | 11 (2) | L17 | 0.0 (0.0) | 3 (3) | 1 (1) |
| A122 | 0.0 (0.0) | 6 (5) | 4 (1) | L32 | 0.0 (0.0) | 11 (6) | 5 (2) |
| A132 | 1.5 (2.6) | 15 (6) | 50 (4) | L34 | 0.0 (0.1) | 4 (3) | 1 (1) |
| C2 | 41.9 (9.7) | 85 (9) | 71 (4) | L41 | 12.5 (7.2) | 5 (3) | 3 (2) |
| C16 | 18.7 (4.0) | 29 (10) | 34 (9) | L43 | 20.7 (6.6) | 3 (2) | 1 (1) |
| C42 | 54.9 (7.8) | 1 (2) | 0 (1) | L96 | n.d. | 1 (2) | 0 (0) |
| C60 | 5.8 (1.4) | 2 (2) | 0 (0) | L100 | 8.9 (7.8) | 10 (8) | 0 (0) |
| C88 | n.d. | 23 (8) | 15 (5) | L112 | n.d. | 25 (10) | 12 (3) |
| C130 | 0.0 (0.0) | 25 (8) | 68 (7) | M114 | 94.0 (2.5) | 21 (9) | 27 (7) |
| D26 | 0.0 (0.0) | 100 (6) | 88 (12) | M120 | 83.8 (10.2) | 6 (5) | 3 (2) |
| D37 | 2.6 (0.5) | 55 (8) | 50 (5) | N8 | 0.0 (0.0) | 49 (10) | 109 (4) |
| D54 | 0.0 (0.0) | 23 (4) | 16 (2) | N10 | 0.0 (0.0) | 86 (13) | 86 (6) |
| D64 | 0.0 (0.0) | 65 (9) | 76 (5) | N33 | 0.0 (0.1) | 20 (4) | 24 (2) |
| D92 | n.d. | 67 (6) | 70 (5) | N39 | 0.9 (1.3) | 61 (8) | 67 (4) |
| D95 | n.d. | 45 (8) | 38 (5) | N40 | 0.0 (0.0) | 29 (8) | 33 (6) |
| D102 | 1.5 (1.3) | 98 (16) | 72 (4) | N46 | 0.6 (1.1) | 10 (4) | 8 (2) |
| D123 | 1.0 (0.9) | 38 (10) | 65 (5) | N50 | 0.0 (0.0) | 90 (6) | 96 (6) |
| D125 | 0.0 (0.0) | 69 (9) | 29 (6) | N56 | 0.0 (0.0) | 87 (9) | 82 (5) |
| E15 | 0.0 (0.0) | 68 (30) | 37 (11) | N61 | 0.0 (0.1) | 11 (4) | 10 (2) |
| E22 | 0.0 (0.0) | 86 (13) | 67 (10) | N65 | 0.0 (0.0) | 99 (7) | 109 (3) |
| E71 | 0.1 (0.2) | 66 (8) | 49 (4) | N110 | 0.0 (0.0) | 1 (1) | 5 (1) |
| E74 | 10.4 (11.1) | 37 (10) | 30 (5) | N113 | 0.0 (0.1) | 89 (9) | 118 (5) |
| E86 | n.d. | 50 (14) | 35 (8) | N118 | 0.0 (0.0) | 47 (8) | 58 (5) |
| E105 | 0.0 (0.0) | 75 (10) | 77 (9) | P13 | 0.0 (0.0) | 36 (9) | 37 (5) |
| E115 | 0.0 (0.0) | 104 (15) | 119 (9) | P25 | 55.1 (9.0) | 96 (4) | 91 (5) |
| E134 | n.d. | 95 (14) | 77 (18) | P47 | 0.1 (0.1) | 1 (1) | 0 (0) |
| F30 | 0.1 (0.1) | 7 (3) | 2 (2) | P75 | 91.6 (7.5) | 103 (4) | 60 (4) |
| F45 | 18.5 (12) | 7 (5) | 2 (2) | P78 | n.d. | 36 (12) | 26 (3) |
| F49 | 3.5 (2.8) | 8 (3) | 10 (2) | P81 | n.d. | 38 (7) | 54 (6) |
| F77 | 94.8 (7.6) | 14 (7) | 15 (3) | P101 | 7.4 (4.1) | 56 (17) | 33 (2) |
| F79 | n.d. | 10 (6) | 3 (1) | P109 | 35.9 (7.4) | 68 (7) | 57 (3) |
| F91 | n.d. | 4 (2) | 1 (1) | Q80 | n.d. | 114 (12) | 122 (6) |
| F106 | n.d. | 18 (6) | 14 (2) | Q93 | n.d. | 87 (20) | 94 (19) |
| F108 | n.d. | 5 (3) | 0 (0) | R20 | 98.9 (0.4) | 118 (14) | 109 (10) |
| F126 | 0.0 (0.0) | 6 (5) | 6 (3) | R48 | 6.1 (1.7) | 31 (8) | 7 (3) |
| F133 | n.d. | 21 (9) | 70 (13) | R73 | n.d. | 78 (6) | 83 (8) |
| G3 | 0.0 (0.0) | 8 (3) | 23 (2) | R111 | 54.4 (9.1) | 65 (8) | 66 (4) |
| G14 | 0.0 (0.0) | 36 (4) | 31 (2) | S7 | 5.1 (1.4) | 37 (6) | 54 (5) |
| G21 | 0.0 (0.0) | 1 (1) | 0 (0) | S29 | 0.0 (0.0) | 14 (6) | 21 (3) |
| G35 | 0.0 (0.0) | 0 (0) | 0 (0) | S38 | 35.9 (2.0) | 55 (6) | 53 (3) |
| G53 | 0.0 (0.0) | 38 (4) | 41 (1) | S62 | 4.2 (0.4) | 4 (2) | 5 (1) |
| G66 | 0.0 (0.0) | 19 (5) | 10 (2) | S83 | n.d. | 28 (7) | 35 (4) |
| G69 | 0.0 (0.0) | 20 (3) | 18 (1) | T57 | 0.0 (0.0) | 24 (5) | 29 (3) |
| G82 | n.d. | 14 (4) | 18 (3) | T70 | 0.0 (0.0) | 102 (6) | 104 (3) |
| G103 | 0.0 (0.0) | 31 (4) | 23 (4) | T84 | n.d. | 60 (10) | 76 (13) |
| G124 | 0.0 (0.0) | 11 (6) | 24 (2) | T90 | n.d. | 41 (11) | 47 (5) |
| H44 | 4.5 (1.1) | 33 (8) | 27 (4) | T97 | n.d. | 19 (6) | 23 (7) |
| H52 | n.d. | 116 (17) | 113 (8) | V5 | 18.6 (8.6) | 38 (10) | 78 (8) |
| H72 | 92.1 (14.8) | 74 (11) | 50 (5) | V19 | 0.0 (0.0) | 4 (3) | 1 (1) |
| H104 | 5.0 (5.7) | 62 (19) | 87 (11) | V23 | 0.0 (0.0) | 2 (2) | 0 (0) |
| I58 | 0.0 (0.0) | 2 (2) | 0 (0) | V31 | 15.8 (1.3) | 32 (7) | 28 (2) |
| I89 | n.d. | 8 (5) | 3 (2) | V59 | 0.0 (0.0) | 4 (2) | 2 (1) |
| I98 | n.d. | 3 (3) | 0 (1) | V85 | n.d. | 10 (6) | 6 (1) |
| I117 | 0.0 (0.0) | 1 (1) | 1 (1) | V87 | n.d. | 7 (4) | 2 (1) |
| I128 | 0.0 (0.0) | 14 (10) | 13 (3) | V131 | 0.0 (0.0) | 12 (6) | 28 (8) |
| K12 | 0.0 (0.0) | 107 (15) | 97 (10) | W68 | 96.9 (3.3) | 105 (9) | 108 (8) |
| K18 | 0.0 (0.0) | 64 (10) | 109 (9) | Y119 | 17.3 (22.1) | 32 (6) | 31 (4) |
Standard deviation in parentheses
Oxidation level not determined due to excessive uncertainty in measurement or peak assignment
Because some residues will be on the protein surface, and therefore exposed to hydroxyl radicals, while others will be occluded in the core of the protein, it is important to determine the percentage of residues that might be expected to react, based on their exposure on the surface of the protein, and on their chemical susceptibility to oxidation. In order to relate the oxidation data to the 3D structure of the protein, it is necessary to determine the accessibility of each amino acid side chain to hydroxyl radical. Since hydroxyl radical is essentially the same size as water, it is reasonable to assume that, for a given side chain, the extent of oxidation will be related to SASA of the side chain. This assumption has been qualitatively demonstrated by previous hydroxyl radical oxidation studies [19,21]. While SASA values may be determined directly from any 3D structure, to account for side chain motion in solution at room temperature, we believe a more robust approach is to determine average <SASA> values by MD simulation of the protein, rather than from a single static structure, such as a homology model or x-ray structure. As indicated from the standard deviations in the <SASA> values computed from the MD data (Table 4), side chain motion can lead to significant variations, which are not taken into account when only a single structure is employed in the analysis.
Relationship between side chain oxidation levels and solvent accessibility
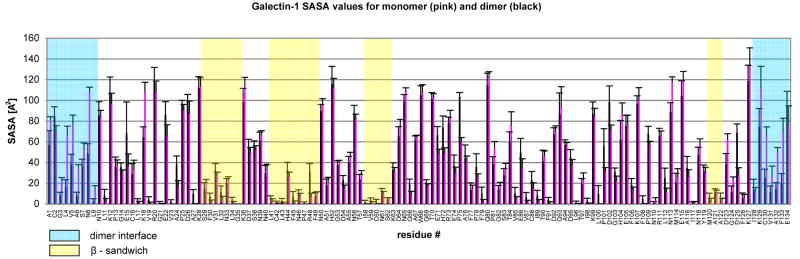
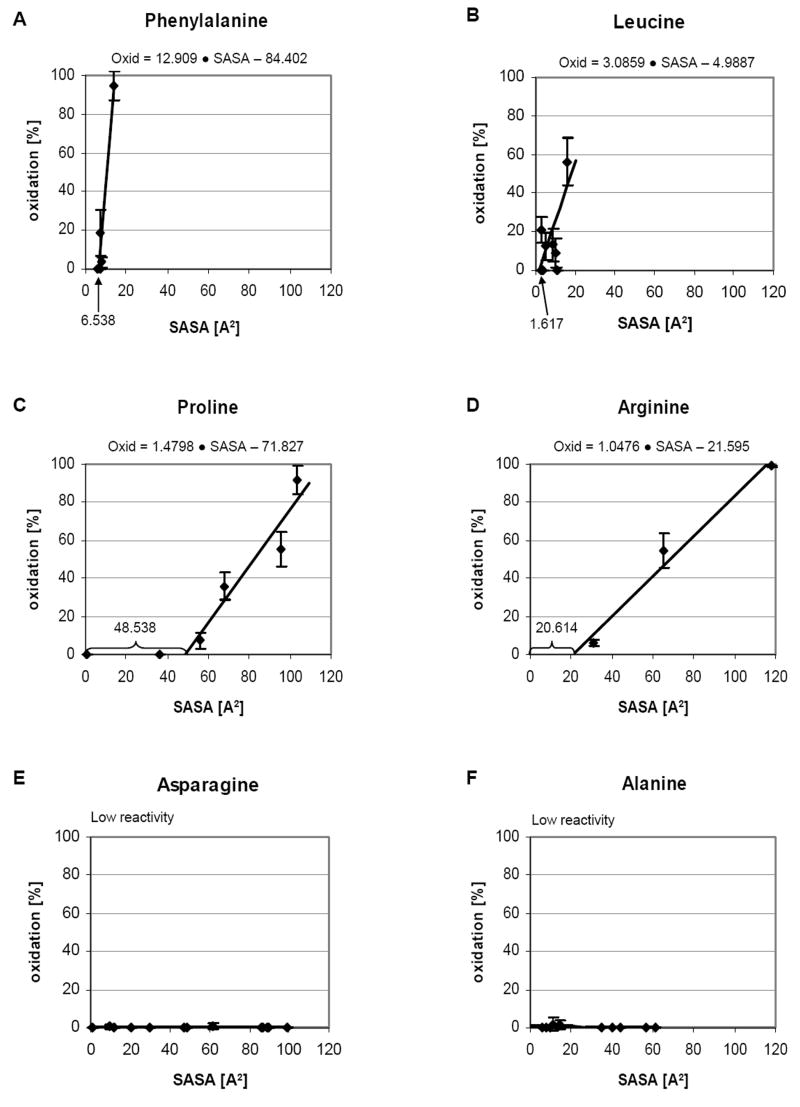
From 1000 snapshots extracted from a 10 ns MD simulation of the galectin-1 homodimer, the average <SASA> of each amino acid side chain for all 134 residues was calculated (Figure 5 and Table 4). To quantify the correlation between the observed oxidation levels and the calculated <SASA> values, the experimental and theoretical data were compiled for each amino acid side chain. By plotting oxidation level versus <SASA> value, for each side chain of the same type, a linear relationship between <SASA> and extent of oxidation was observed (Figure 6). Additionally, for most side chains, a minimum solvent exposure appeared to be required to enable any oxidation to occur. The level of exposure required varied according to the side chain, from a low of 2 Å2 for leucine to a high of approximately 48 Å2 for proline. The relationship naturally also depends on the chemical susceptibility of the side chain to oxidation, and to a far lesser extent on the local primary sequence [69]. Additionally, under the conditions employed here, some side chains remained essentially unoxidized regardless of their exposure. From the slopes of the plots of <SASA> versus percent oxidation, the side chains could be divided into categories according to their reactivity (high, medium or low). For example, highly reactive residues such as phenylalanine could be characterized by 12.9 % oxidation/Å2, whereas a medium reactivity residue, such as arginine or proline would result in only 1-2 % oxidation/Å2. This simple analysis neglects the precise details of the atomic site of reactivity, but nevertheless demonstrated the direct dependence of oxidation level on side chain exposure. From each reactivity group, two plots of the oxidation level dependence on the <SASA> are shown in Figure 6. The extent to which these relationships are transferable to other proteins is currently under investigation. However, the analysis presented here agrees well with earlier characterization of side chain reactivity to hydroxyl radical [55,56]. Generally highly reactive amino acids include cysteine, methionine, the aromatics and leucine. The medium reactivity category includes arginine and proline, while the remaining amino acids such as asparagine, aspartic acid, serine, lysine, glutamine, glutamic acid, threonine and alanine have low reactivities. Having derived a quantitative relationship between SASA and percent oxidation, it is possible in principle to predict the extent of oxidation that should be observed, given the 3D structure of a protein. To test the accuracy of such predictions, as well as the sensitivity of the experimental method to 3D structure, these techniques were applied to the characterization of the interface region between monomeric domains in galectin-1.
Figure 5.
Solvent accessible surface area (<SASA>) for each of the 134 side chains in galectin-1. Values for monomer (pink) and dimer (black) domains are reported. Shaded regions indicate the dimer interface (blue). Other regions of low exposure include the sheets associated with the β-sandwich structure (yellow).
Figure 6.
Percentage of oxidation versus side chain <SASA> for selected amino acids. Highly reactive side chains (A, B) are characterized by steep slopes (%Ox/Å2) and low minimal exposure values (Å2). Moderately reactive side chains (C, D) require significant exposure before oxidization may occur. Side chains that are inert under these experimental conditions (E, F) show no sensitivity to level of exposure.
Characterization of the galectin-1 dimer interface
Most amino acid side chains were found to be reactive to some extent under the current experimental conditions (Table 1); however, the following residues were consistently observed to be sensitive to oxidation: cysteine, methionine, tryptophane, tyrosine, phenylalanine, histidine, leucine, valine, proline, arginine. Although an experiment, in which differences in oxidation levels between the monomer and dimer were observed, would be well suited to characterize the dimer interface, galectin-1 cannot be obtained in monomeric form. In contrast, computationally, both species are amenable to examination. Thus, by examining the difference in the computed <SASA> values from independent 10 ns MD simulations of the dimer and monomer, the interface region was readily identified (residues 1-9 and 128-143), in agreement with those residues identified in the x-ray structure of the dimer [60]. A subset of reporter residues in the interface (L4, V5, L9, I128, V131 and F133) were identified on the basis that they were predicted to be at least partially shielded from solvent in the interface, and that were likely to be reactive. The observed oxidation levels for these residues are presented in Table 5 and illustrated in Figure 4b. With the exception of F133, which was not identified in the MS/MS data, the oxidation levels were markedly lower for these interfacial residues than would be expected for exposed residues. Notably, the predicted oxidation levels for the dimer, based on the relationships derived above, were in good agreement with the observed values.
Table 5.
Experimentally-measured and <SASA>-derived oxidation levels for reporter residues in the galectin-1 dimer interface
| Residue | Oxidation [%] | <SASA> [Å2] | ||
|---|---|---|---|---|
| measured | predicted | dimer | monomer | |
| L4 | 56 | 44 | 16 | 67 |
| L9 | 0 | 4 | 3 | 14 |
| V5 | 19 | 18 | 38 | 78 |
| V131 | 0 | 4 | 12 | 28 |
| I128 | 0 | 0 | 14 | 13 |
The observation that L4 and V5 are significantly oxidized (56% and 19%, respectively) despite being associated with the interface [60], suggests that easily oxidized side chains must be highly excluded from solvent in order to prevent their oxidation, under these reaction conditions. Both L4 and V5 have reduced solvent accessibilities relative to their <SASA> in the galectin-1 monomeric domain (approximately 75% and 50%, respectively); however, this is not sufficient to prevent their partial oxidation. By employing the linear relationship between percentage oxidation and SASA it is possible to predict the level of oxidation for each of the residues in the interface (Table 5). Notably the predicted oxidation levels for L4 (44%) and V5 (18%) are in good agreement with the observed values. These relationships need to be confirmed by analysis of further proteins; however, they appear to offer a promising approach to relating observed oxidation to 3D structure.
As expected, residues in the interface with very low <SASA> values were not found to be oxidized, for example L9, I128 and V131. Other residues that are associated with the dimer interface, but which have very low reactivity levels did not get oxidized regardless their solvent exposure (A1, A6, S7, N8, K129, E134), which illustrates the benefit of identifying a sub-set of reporter residues prior to attempting to interpret the oxidation data purely in terms of 3D structure effects.
Recent studies have employed oxidative footprinting data to guide computational molecular docking, however it was concluded that footprinting data alone was insufficient to provide a definitive choice of model [70]. It is anticipated that the quantitative relationships derived here to relate oxidation levels to 3D structure will be valuable in such approaches. Additionally, we have demonstrated that given an accurate structure for the complex, it is possible to use <SASA> values to predict oxidation levels that are in good agreement with experimental data. Thus, data from oxidative footprinting are likely to be particularly useful in validating independently generated molecular complexes, by for example automated docking procedures.
Conclusion
A combined experimental and theoretical approach directly applicable to protein surface mapping has been presented. A linear relationship between the level of side chain oxidation and average <SASA>, derived from MD simulation data, has been derived. Notably, each side chain has been found to require a minimum level of exposure before it will be oxidized under the mild conditions employed here. By considering this minimal exposure requirement, with the known side chain reactivities, potential reporter groups may be identified. Knowledge of the expected reporter groups can be used to provide an estimate of the expected surface oxidation levels and is therefore of significance to the study of protein-protein and protein-ligand interactions. Although not all amino acid side chains react under the conditions presented here, the level of surface coverage, in terms of those residues that were exposed to solvent and that were not inert to oxidation was approximately 70%. This level of coverage is far greater than would be achieved using traditional chemical derivatization methods, such as biotinylation or acetylation, wherein only a few residues can act as reporter groups. Equally significant to good coverage is the ability to quantify the level of oxidation and relate that directly to per-residue <SASA> values. This ability significantly elevates the level of footprint resolution, which is key to the practical application of this method in characterizing protein complexes. Such quantification facilitates the identification of occluded surfaces and should provide a powerful tool for determining the 3D structures of complexes that are not amenable to analysis by traditional experimental structural methods.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (RR05357 and GM55230) for financial support. We would also like to thank to M. A. Duncan (UGA) for providing access to the excimer laser and T. Cheng (UGA) for helping with its set up. R.J.W wishes to thank M. R. Chance (Case Western) for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Englander JJ, Del Mar C, Li W, Englander SW, Kim JS, Stranz DD, Hamuro Y, Woods VL., J Protein Structure Change Studied by Hydrogen-Deuterium Exchange, Functional Labelling, and Mass Spectrometry. Proc Natl Acad Sci USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King D, Bergmann C, Orlando R, Benen JA, Kester HC, Visser J. Use of Amide Exchange Mass Spectrometry to Study Conformational Changes Within the Endopolygalacturonase II-Homogalacturonan-Polygalacturonase Inhibiting Protein System. Biochemistry. 2002;41:10225–10233. doi: 10.1021/bi020119f. [DOI] [PubMed] [Google Scholar]
- 3.King D, Lumpkin M, Bergmann C, Orlando R. Studying Protein-Carbohydrate Interactions by Amide Hydrogen/Deuterium Exchange Mass Spectrometry. Rapid Commun Mass Spectrom. 2002;16:1569–1574. doi: 10.1002/rcm.746. [DOI] [PubMed] [Google Scholar]
- 4.Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem. 2001;Suppl 37:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 5.Zheng X, Wintrode PL, Chance MR. Complementary structural mass spectrometry techniques reveal local dynamics in functionally important regions of a metastable serpin. Structure. 2008;16(1):38–51. doi: 10.1016/j.str.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Seyfried NT, Atwood JA, III, Almond A, Day AJ, Orlando R, Woods RJ. Fourier transform mass spectrometry to monitor hyaluronan–protein interactions: use of hydrogen/deuterium amide exchange. Rapid Commun Mass Spectrom. 2007;21:121–131. doi: 10.1002/rcm.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker DL, Seyfried NT, Li H, Orlando R, Terns RM, Terns MP. Determination of Protein-RNA Interaction Sites in the Cbf5-H/ACA Guide RNA Complex by Mass Spectrometric Protein Footprinting. Biochemistry. 2008;47(6):1500–1510. doi: 10.1021/bi701606m. [DOI] [PubMed] [Google Scholar]
- 8.Shell SM, Hess S, Kvaratskhelia M, Zou Y. Mass spectrometric identification of lysines involved in the interaction of human replication protein a with single-stranded DNA. Biochemistry. 2005;44(3):971–8. doi: 10.1021/bi048208a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Schuster MC, Sfyroera G, Geisbrecht BV, Lambris JD. Solution Insights into the Structure of the Efb/C3 Complement Inhibitory Complex as Revealed by Lysine Acetylation and Mass Spectrometry. J Am Soc Mass Spectrom. 2008;19(1):55–65. doi: 10.1016/j.jasms.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shkriabai N, Patil SS, Hess S, Budihas SR, Craigie R, Burke TR, Le Grice SFJ, Kvaratskhelia M. Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry. Proc Nat Acad Sci USA. 2004;101(18):6894–6899. doi: 10.1073/pnas.0400873101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takamoto K, Chance MR. Radiolytic Protein Footprinting with Mass Spectrometry to Probe the Structure of Macromolecular Complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–76. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 12.Maleknia SD, Wong JW, Downard KM. Photochemical and electrophysical production of radicals on millisecond timescales to probe the structure, dynamics and interactions of proteins. Photochem Photobiol Sci. 2004;3(8):741–8. doi: 10.1039/b315904c. [DOI] [PubMed] [Google Scholar]
- 13.Sharp JS, Guo J-T, Uchiki T, Xu Y, Dealwis C, Hettich RL. Photochemical Surface Mapping of C14S-Sm11p for Constrained Computational Modeling of Protein Structure. Anal Biochem. 2005;340:201–212. doi: 10.1016/j.ab.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Xu G, Chance MR. Radiolytic Modification of Sulfur- Containing Amino Acid Residues in Model Peptides: Fundamental Studies for Protein Footprinting. Anal Chem. 2005;77:2437–2449. doi: 10.1021/ac0484629. [DOI] [PubMed] [Google Scholar]
- 15.Xu G, Takamoto K, Chance MR. Radiolytic Modification of Basic Amino Acid Residues in Peptides: Probes for Examing Protein- Protein Interactions. Anal Chem. 2003;75:6995–7007. doi: 10.1021/ac035104h. [DOI] [PubMed] [Google Scholar]
- 16.Guan J-Q, Vorobiev S, Almo SC, Chance MR. Mapping the G-Actin Binding Surface of Cofilin Using Synchrotron Protein Footprinting. Biochemistry. 2002;41:5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 17.Maleknia SD, Brenowitz M, Chance MR. Millisecond Radiolytic Modification of Peptides by Synchrotron X-rays Identified by Mass Spectrometry. Anal Chem. 1999;71:3965–3973. doi: 10.1021/ac990500e. [DOI] [PubMed] [Google Scholar]
- 18.Aye TT, Low TY, Sze SK. Nanosecond Laser-Induced Photochemical Oxidation Method for Protein Surface Mapping with Mass Spectrometry. 2005 doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- 19.Hambly DM, Gross ML. Laser Flash Photolysis of Hydrogen Peroxide to Oxidize Protein Solvent-Accessible Residues on the Microsecond Timescale. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Hambly D, Gross M. Laser Flash Photochemical Oxidation to Locate Heme Binding and Conformational Changes in Myoglobin. Int J Mass Spectrom. 2007;259(13):124–129. [Google Scholar]
- 21.Sharp JS, Becker JM, Hettich RL. Analysis of Protein Solvent Accessible Surfaces by Photochemical Oxidation and Mass Spectrometry. Anal Biochem. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 22.Sharp JS, Tomer KB. Analysis of the oxidative damage-induced conformational changes of apo- and holocalmodulin by dose-dependent protein oxidative surface mapping. Biophys J. 2007;92(5):1682–92. doi: 10.1529/biophysj.106.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platis IE, Ermacora MR, Fox RO. Oxidative polypeptide cleavage mediated by EDTA-Fe covalently linked to cysteine residues. Biochemistry. 1993;32(47):12761–7. doi: 10.1021/bi00210a027. [DOI] [PubMed] [Google Scholar]
- 24.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical ‘footprinting’. Curr Opin Struct Biol. 2002;12(5):648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 25.Greiner DP, Hughes KA, Meares CF. Radiolytic protein surface mapping. Biochem Biophys Res Commun. 1996;225(3):1006–8. doi: 10.1006/bbrc.1996.1285. [DOI] [PubMed] [Google Scholar]
- 26.Maleknia SD, Chance MR, Downard KM. Electrospray-assisted modification of proteins: a radical probe of protein structure. Rapid Commun Mass Spectrom. 1999;13(23):2352–8. doi: 10.1002/(SICI)1097-0231(19991215)13:23<2352::AID-RCM798>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 27.Wong JW, Maleknia SD, Downard KM. Hydroxyl radical probe of the calmodulin-melittin complex interface by electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2005;16(2):225–33. doi: 10.1016/j.jasms.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Berens C, Streicher B, Schroeder R, Hillen W. Visualizing metal-ion-binding sites in group I introns by iron(II)-mediated Fenton reactions. Chem Biol. 1998;5(3):163–175. doi: 10.1016/s1074-5521(98)90061-8. [DOI] [PubMed] [Google Scholar]
- 29.Hampel KJ, Burke JM. Time-Resolved Hydroxyl-Radical Footprinting of RNA Using Fe(II)-EDTA. Methods. 2001;23(3):233–239. doi: 10.1006/meth.2000.1134. [DOI] [PubMed] [Google Scholar]
- 30.Tullius TD, Dombroski BA. Hydroxyl radical “footprinting”: high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Nat Acad Sci USA. 1986;83(15):5469–73. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heyduk E, Heyduk T. Mapping protein domains involved in macromolecular interactions: a novel protein footprinting approach. Biochemistry. 1994;33(32):9643–50. doi: 10.1021/bi00198a033. [DOI] [PubMed] [Google Scholar]
- 32.Sharp JS, Becker JM, Hettich RL. Protein Surface Mapping by Chemical Oxidation: Structural Analysis by Mass Spectrometry. Analyt Biochem. 2003;313:216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 33.Garrison WM. Reaction Mechanisms in the Radiolysis of Peptides, and Proteins. Chem Rev. 1987;87:381–398. [Google Scholar]
- 34.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 35.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Bern M, Cai Y, Goldberg D. Lookup Peaks: A Hybrid of de Novo Sequencing and Database Search for Protein Identification by Tandem Mass Spectrometry. Anal Chem. 2007;79(4):1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 37.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An Assessment of Software Solutions for the Analysis of Mass Spectrometry Based Quantitative Proteomics Data. J Proteome Res. 2008;7(1):51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 38.Barondes SH, Cooper DNW, Gitt MA, Leffler H. Galectins. Structure and Function of a Large Family of Animal Lectins. J Biol Chem. 1994;269:20907–20810. [PubMed] [Google Scholar]
- 39.Cho M, Cummings RD. Galectin-1, a beta-galactoside-binding lectin in Chinese hamster ovary cells. I. Physical and chemical characterization. J Biol Chem. 1995;270(10):5198–206. doi: 10.1074/jbc.270.10.5198. [DOI] [PubMed] [Google Scholar]
- 40.Hirabayashi J, Kasai K. Effect of amino acid substitution by sited-directed mutagenesis on the carbohydrate recognition and stability of human 14-kDa beta-galactoside-binding lectin. J Biol Chem. 1991;266(35):23648–53. [PubMed] [Google Scholar]
- 41.Kasai K, Hirabayashi J. Galectins: a family of animal lectins that decipher glycocodes. J Biochem. 1996;119(1):1–8. doi: 10.1093/oxfordjournals.jbchem.a021192. [DOI] [PubMed] [Google Scholar]
- 42.Stillman BN, Hsu DK, Brewer CF, Johnson P, Liu FT, Baum LG. Galectin-3 and Galectin-1 Bind Distinct Cell Surface Glycoprotein Receptors to Induce T Cell Death. J Immunol. 2006;176:778–789. doi: 10.4049/jimmunol.176.2.778. [DOI] [PubMed] [Google Scholar]
- 43.Fitzner B, Walzel H, Sparmann G, Emmrich J, Liebe S, Jaster R. Galectin-1 is an inductor of pancreatic stellate cell activation. Cell Signal. 2005;17(10):1240–7. doi: 10.1016/j.cellsig.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Liu FT. Galectins: a new family of regulators of inflammation. Clin Immunol. 2000;97(2):79–88. doi: 10.1006/clim.2000.4912. [DOI] [PubMed] [Google Scholar]
- 45.Walzel H, Blach M, Hirabayashi J, Arata Y, Kasai K-i, Brock J. Galectin-induced activation of the transcription factors NFAT and AP-1 in human Jurkat T-lymphocytes. Cell Signal. 2002;14(10):861–868. doi: 10.1016/s0898-6568(02)00035-9. [DOI] [PubMed] [Google Scholar]
- 46.Wang JL, Gray RM, Haudek KC, Patterson RJ. Nucleocytoplasmic lectins. Biochem Biophys Acta. 2004;1673(12):75–93. doi: 10.1016/j.bbagen.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Dyekjaer JD, Woods RJ. In: NMR Spectroscopy and Computer Modeling of Carbohydrates. Recent Advances. Vliegenthart JFG, Woods RJ, editors. Vol. 930. American Chemical Society; Washington: 2006. p. 18. [Google Scholar]
- 48.Cozzetto D, Tramontano A. Relationship between multiple sequence alignments and quality of protein comparative models. Proteins. 2005;58(1):151–7. doi: 10.1002/prot.20284. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Fuentes N, Rai BK, Madrid-Aliste CJ, Fajardo JE, Fiser A. Comparative protein structure modeling by combining multiple templates and optimizing sequence-to-structure alignments. Bioinformatics. 2007;23(19):2558–65. doi: 10.1093/bioinformatics/btm377. [DOI] [PubMed] [Google Scholar]
- 50.Ginalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol. 2006;16(2):172–7. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Fouillit M, Levi-Strauss M, Giudicelli V, Lutomski D, Bladier D, Caron M, Joubert-Caron R. Affinity purification and characterization of recombinant human galectin-1. J Chromatogr B. 1998;706(1):167–171. doi: 10.1016/s0378-4347(97)00336-8. [DOI] [PubMed] [Google Scholar]
- 52.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 53.Pedrioli PGA, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu WM, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nature Biotech. 2004;22(11):1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 54.Tabb DL, Fernando CG, Chambers MC. MyriMatch: Highly Accurate Tandem Mass Spectral Peptide Identification by Multivariate Hypergeometric Analysis. J Proteome Res. 2007;6(2):654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan J-Q, Chance MR. Structural Proteomics of Macromolecular Assemblies Using Oxidative Footprinting and Mass Spectrometry. TRENDS Biochem Sci. 2005;30:583–592. doi: 10.1016/j.tibs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Xu G, Chance MR. Radiolytic Modification and Reactivity of Amino Acid Residues Serving as Structural Probes for Protein Footprinting. Anal Chem. 2005;77:4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 57.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272(33):20313–6. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 58.Kubota K, Yoneyama-Takazawa T, Ichikawa K. Determination of sites citrullinated by peptidylarginine deiminase using 18O stable isotope labeling and mass spectrometry. Rapid Commun Mass Spectrom. 2005;19(5):683–8. doi: 10.1002/rcm.1842. [DOI] [PubMed] [Google Scholar]
- 59.Boja ES, Fales HM. Overalkylation of a protein digest with iodoacetamide. Anal Chem. 2001;73(15):3576–82. doi: 10.1021/ac0103423. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Lucendo MF, Solis D, Andre S, Hirabayashi J, Kasai K, Kaltner H, Gabius HJ, Romero A. Growth-regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-site Mutations and their Impact on the Thermodynamics of Ligan Binding. J Mol Biol. 2004;343:957–970. doi: 10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 61.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr D. 2002;58(Pt 6 No 1):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 62.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco: 2006. [Google Scholar]
- 63.Wang J, Cieplak P, Kollman PA. How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J Comput Chem. 2000;21(12):1049–1074. [Google Scholar]
- 64.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Phys Chem. 1983;79:926–935. [Google Scholar]
- 65.Hubbard SJ, Thornton JM. NACCESS, 2.1.1. University College London; London: 1993. [Google Scholar]
- 66.Sullards MC, Reiter JA. Primary and Secondary Locations of Charge Sites in Angiotensin II (M+2H)(2+) Ions Formed by Electrospray Ionization. J Am Soc Mass Spectrom. 2000;11(1):40–53. doi: 10.1016/S1044-0305(99)00115-4. [DOI] [PubMed] [Google Scholar]
- 67.Bull HB, Breese K. Surface-Tension of Amino-Acid Solutions -Hydrophobicity Scale of Amino-Acid Residues. Arch Biochem Biophys. 1974;161(2):665–670. doi: 10.1016/0003-9861(74)90352-x. [DOI] [PubMed] [Google Scholar]
- 68.Zhou SL, Cook KD. A Mechanistic Study of Electrospray Mass Spectrometry: Charge Gradients Within Electrospray Droplets and Their Influence on Ion Response. J Am Soc Mass Spectrom. 2001;12(2):206–214. doi: 10.1016/S1044-0305(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 69.Sharp JS, Tomer KB. Effects of anion proximity in peptide primary sequence on the rate and mechanism of leucine oxidation. Anal Chem. 2006;78(14):4885–93. doi: 10.1021/ac060329o. [DOI] [PubMed] [Google Scholar]
- 70.Kamal JK, Chance MR. Modeling of Protein Binary Complexes Using Structural Mass Spectrometry Data. Protein Sci. 2007;17:1–16. doi: 10.1110/ps.073071808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.