Abstract
The nuclear lamins function in the regulation of replication, transcription, and epigenetic modifications of chromatin. However, the mechanisms responsible for these lamin functions are poorly understood. We demonstrate that A- and B-type lamins form separate, but interacting, stable meshworks in the lamina and have different mobilities in the nucleoplasm as determined by fluorescence correlation spectroscopy (FCS). Silencing lamin B1 (LB1) expression dramatically increases the lamina meshwork size and the mobility of nucleoplasmic lamin A (LA). The changes in lamina mesh size are coupled to the formation of LA/C-rich nuclear envelope blebs deficient in LB2. Comparative genomic hybridization (CGH) analyses of microdissected blebs, fluorescence in situ hybridization (FISH), and immunofluorescence localization of modified histones demonstrate that gene-rich euchromatin associates with the LA/C blebs. Enrichment of hyperphosphorylated RNA polymerase II (Pol II) and histone marks for active transcription suggest that blebs are transcriptionally active. However, in vivo labeling of RNA indicates that transcription is decreased, suggesting that the LA/C-rich microenvironment induces promoter proximal stalling of Pol II. We propose that different lamins are organized into separate, but interacting, microdomains and that LB1 is essential for their organization. Our evidence suggests that the organization and regulation of chromatin are influenced by interconnections between these lamin microdomains.
Keywords: Lamins, chromatin, RNA polymerase II transcription, chromosome organization
The nuclear lamina lines the inner membrane of the nuclear envelope (NE) and contributes to the size, mechanical stability, and shape of the nucleus (Lammerding et al. 2006). The major structural proteins of the lamina are the nuclear lamins, consisting of the A-type lamins lamin A (LA) and lamin C (LC) and the B-type lamins lamin B1 (LB1) and lamin B2 (LB2) (Krohne et al. 2005). Although the majority of lamin proteins are concentrated in the lamina, a small fraction is also found throughout the nucleoplasm (Moir et al. 2000). Little information is available on the structure and/or the assembly state of the lamins within nuclei, although there are reports that they can form filamentous structures in some types of cells (Aebi et al. 1986; Schermelleh et al. 2008). The lamina localization of LA and LB is disrupted in nuclei following the introduction of an N-terminal deletion mutant of LA (Goldman et al. 1992), suggesting that they are part of the same structure or separate interdependent structures. Fluorescence resonance energy transfer (FRET) experiments show that LA and LB1 can interact with each other both homotypically and heterotypically in live cells (Delbarre et al. 2006). These studies suggest that A- and B-type lamins polymerize into homopolymers that interact with homopolymers of the opposite type within the lamina and the nucleoplasm. In vitro, lamins can form both homotypic and heterotypic interactions of varying stability (Schirmer and Gerace 2004). Furthermore, it has been suggested that the variation in expression of lamin subtypes in different cells could be related to the tissue-specific properties of the lamin polymers (Schirmer and Gerace 2004).
Lamins and lamina structure are important for the organization of chromatin, DNA replication, and transcription (Dechat et al. 2008). Lmna−/− mice show a loss of chromatin integrity associated with deformations or blebbing of the NE and lamina (Sullivan et al. 1999). Furthermore, mutations in human LMNA that cause progeria, a premature aging disease, induce an abnormal thickening of the lamina, nuclear blebbing, and a loss of peripheral heterochromatin correlated with histone methylation changes and the mislocalization of chromosomes (Goldman et al. 2004; Shumaker et al. 2006; Meaburn et al. 2007). Gene expression and chromosome positioning are abnormal in lmnb1Δ/Δ mice and in LB1 silenced cells (Malhas et al. 2007; Tang et al. 2008), while disruption of lamina organization or depletion of B-type lamins inhibits DNA replication and RNA polymerase II (Pol II) transcription (Dechat et al. 2008). The effects of lamins on chromatin structure and function may be direct through the interaction of lamins with histones or with other chromatin-binding proteins (Dechat et al. 2008).
The nuclear lamina is also thought to play important roles in gene regulation at the nuclear periphery (Schneider and Grosschedl 2007). The prevalence of heterochromatin in the lamina region in some cells and observations that several silenced genes are localized to this area have led to the suggestion that the nuclear periphery is a repressive environment for transcription. Recent studies in which chromosomes are artificially tethered to the NE using LB1, lamina-associated polypeptide (LAP) 2β, or emerin fused to the lacI repressor protein found that some genes are suppressed when closely associated with the NE, but that others are not (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008). The variable regulation of gene expression may be due to differences in the experimental methods or the regulation of gene expression at the nuclear periphery could be gene-specific (Finlan et al. 2008).
Using a combination of high-resolution confocal microscopy and fluorescence correlation spectroscopy (FCS), we show that A- and B-type lamins form interconnected networks within the lamina and the nucleoplasm. To investigate the structure and function of these lamin networks, we silenced the expression of either LA/C, LB1, or LB2. Our results demonstrate that the individual lamins form distinct nuclear compartments in mammalian cells and that there are specific roles for each lamin in chromatin organization and gene expression. The results also provide new insights into the mechanisms responsible for the numerous LMNA mutations causing a wide range of human diseases.
Results
A- and B-type lamins form separate structures in the lamina and in the nucleoplasm
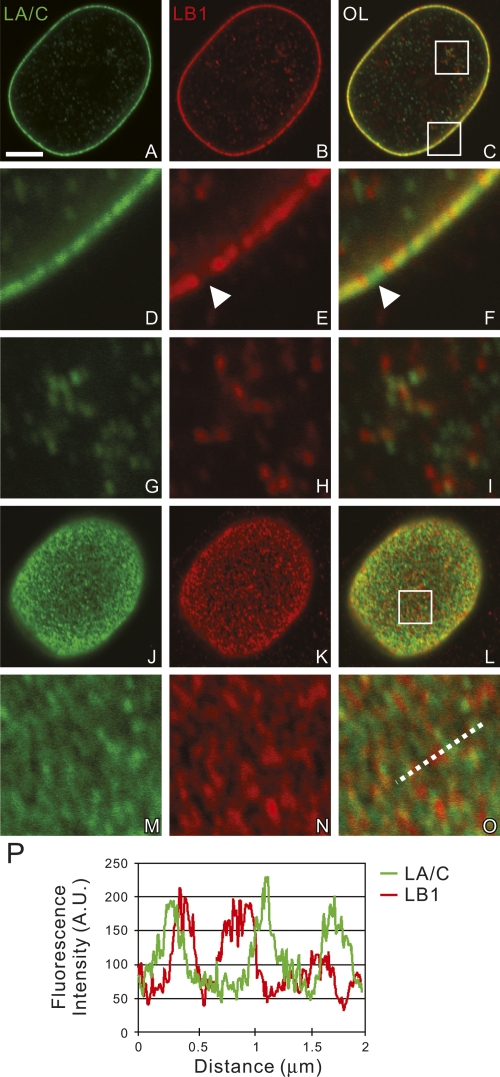
In mid-plane sections of HeLa nuclei, immunolocalized LA/C, LB1, and LB2 were distributed in discontinuous patterns in the lamina (Fig. 1A–F; Supplemental Fig. S1A–F). Gaps in either LB1 or LB2 staining (Fig. 1D–F, arrowheads) were frequently filled with LA/C and vice versa. Occasional points of colocalization were also apparent (Fig. 1D–F). The nucleoplasm had a reduced fluorescence signal relative to the lamina with LA/C and LB1/2 forming foci and short fibrous structures that were mostly separate with occasional points of colocalization (Fig. 1G–I; Supplemental Fig. S1G–I). At the upper and lower surfaces of nuclei, each of the lamins appeared as a complex meshwork of densely packed interconnected fibers and foci (Fig. 1J–P; Supplemental Fig. S1J–P). The A- and B-type lamin meshworks were similar in appearance, but were mainly separate from each other with some points of colocalization (Fig. 1P). Similar separation of the A- and B-type lamin meshworks was also observed in human foreskin fibroblasts (Supplemental Fig. S6).
Figure 1.
The distributions of LA/C and LB1 were determined by double-label confocal immunofluorescence. LA/C (green) and LB1 (red), from a single nucleus are seen in an equatorial section (A–I) and the nuclear surface (J–O). The areas indicated by white squares in C and L are enlarged fivefold in D–I and M–O, respectively. (P) A line scan showing the fluorescence intensity profile along the dotted line in O. These images demonstrate that lamins form mainly separate networks with some overlapping regions. Bar, 5 μm.
Silencing LB1 alters LA/C and LB2 meshworks and induces nuclear blebs in the lamina
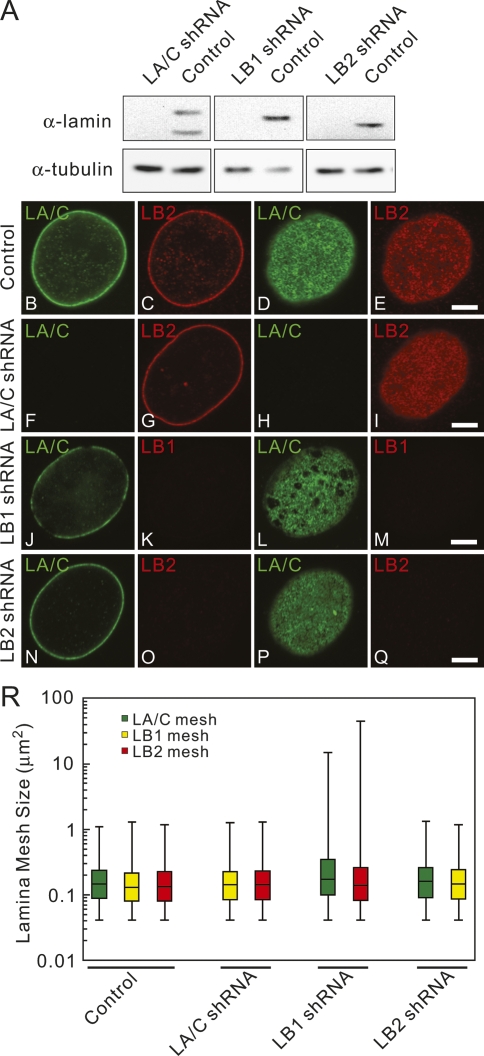
The points of partial colocalization between the A- and B-type lamin meshworks suggest that they may be structurally linked in the lamina. Previous FRET experiments also suggested a possible close interaction between A- and B-type lamins in the lamina (Delbarre et al. 2006). The physical interaction between A- and B-type lamins in the nucleus was tested experimentally by silencing the expression of LA/C, LB1, or LB2 using shRNAs in HeLa cells. At 4–5 d following transfection, the amount of targeted lamin was decreased by ∼75%–90% without an apparent effect on the expression of the remaining lamins as determined by immunoblotting (Fig. 2A). Lamins in individual cells were silenced to nearly undetectable levels as determined by immunostaining (Supplemental Fig. S2A–L; data not shown). Immunofluorescence analysis of the lamina showed that silencing of either LA/C or LB2 did not change the appearance of the remaining lamin meshworks compared with controls transfected with scrambled sequences or nonsilenced cells (Fig. 2B–I, N–Q). However, ∼75% of the nuclei in LB1 silenced cells showed a dramatic enlargement of the mesh sizes in some regions of the lamina for both LA/C and LB2 (Fig. 2J–M,R), while the remaining ∼25% showed no detectable changes. An enlarged mesh is defined as a lamina containing mesh sizes larger than 1.3 μm2, which was the largest mesh size observed in control cells (Fig. 2R). The LA/C and LB2 meshworks were always enlarged together in the same nucleus following LB1 silencing; enlargement of only one meshwork was never observed. The mesh sizes were increased up to 55-fold for LA/C and 125-fold for LB2 (Fig. 2R). The reason for this wide range of increase in mesh sizes is unknown. In contrast, no obvious changes in the nucleoplasmic lamin structures were observed following the silencing of each of the lamins (data not shown).
Figure 2.
The effects of silencing LA/C, LB1, or LB2 in HeLa cells were determined by confocal immunofluorescence. (A) Immunoblots of equivalent protein loadings show lamin expression levels in whole cell lysates of control and silenced cells. LA/C and LB1 or LB2 were localized in silenced cells and controls. (B–Q) Silencing vector is shown on left side. LA/C is shown in green, while LB1 and LB2 are shown in red. Controls are cells transfected with a scrambled sequence shRNA. (B,C) Equatorial region. (D,E) Surface of the same nucleus. Bar, 5 μm. (R) Mesh size variations within the lamina in control and silenced cells. In controls, LA/C, and LB2 silenced cells, the mesh size ranged from the limit of resolution (0.04 μm2, assuming that the shape of the minimal mesh is square) to 1.3 μm2 (median: 0.13–0.15 μm2 for all lamins). The data are displayed using box plots. The bars indicate the maximum and minimum mesh sizes measured. The line separating the rectangular region is the median of all measurements. The top region of the rectangle shows the distribution of 50%–75% of the mesh size measurements; whereas the bottom part of the rectangle shows the distribution of 25%–50% of the mesh size measurements. Images were taken with a PlanApochromat 100×, 1.40 NA oil immersion lens. In LB1 silenced cells, the mesh size ranged from 0.04 μm2 to 15 μm2 for LA/C and 45 μm2 for LB2 (n = 2000–3000 in 10 nuclei, median: 0.6–0.7 μm2).
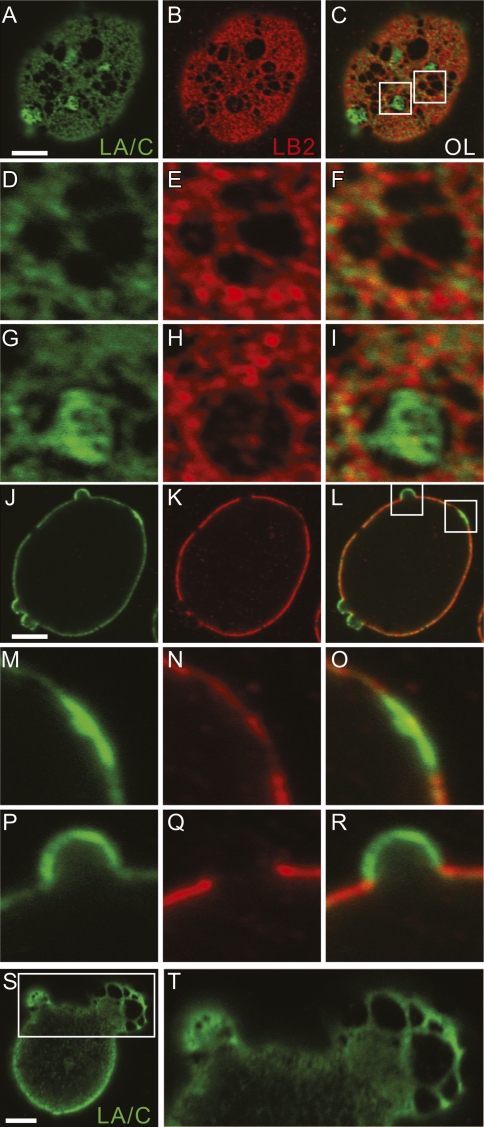
The structural alterations in the LA/C and LB2 meshworks after LB1 silencing were examined in greater detail. Within enlarged regions, the LA/C and LB2 meshworks frequently appeared to be collinear with a few sites of overlap (Fig. 3A–I). In many cells, some of the larger open areas contained LA/C-rich islands of small meshworks connected to the surrounding LA/C meshwork (Fig. 3A–C, G–I). LB2-rich islands within the open areas of the meshwork were never observed. Through-focus series revealed that these LA/C-rich structures had two forms: flat areas or plaques on the nuclear surface and outpocketings or blebs of the NE (Figs. 3J–R; Supplemental Fig. S2M). In all cases, blebs were enriched in LA/C, with little or no staining with LB2 antibody. Blebs devoid of all lamins were not observed. The LA/C lamina in the blebs frequently contained meshworks enlarged to the same degree as seen in nonblebbed regions of LB1 silenced cells (Fig. 3S,T). Blebs varied in appearance from smaller semicircular profiles (Fig. 3P–R) to larger structures with long axes of up to 13 μm (Fig. 3S,T). Furthermore, the larger blebs contained larger LA/C meshes, and the smaller blebs contained smaller meshes in their lamina regions (Fig. 3S,T). The enrichment of LA/C in both plaques and blebs and the increased mesh size of blebs suggested that blebs may have emerged from the plaques (Fig. 3M–R). Of the ∼75% of LB1 silenced cells with enlarged LA/C and LB2 meshworks, 31% formed blebs (Supplemental Fig. S2G–I). The ∼25% of silenced cells that did not contain enlarged lamina meshworks were never observed to contain blebs. In addition, blebs were not found in controls, or in LA/C or LB2 silenced cells (Supplemental Fig. S2A–F,J–L). The number of these structures seen in the nucleus of an LB1 silenced cell varied from one to three, while the largest appeared in nuclei with a single bleb. In addition to HeLa cells, we also observed similar increases in meshwork size and bleb formation in the nuclei of other human cell types including U-2 OS and MCF-7 (data not shown). Interestingly, nuclear pore complexes were not seen along the surface of blebs, but were located primarily along the fibers comprising the LB2 meshwork (Supplemental Fig. S3A–I). These results suggest that LB1 is required to maintain nuclear shape and organize the structures of the LA/C and LB2 meshworks and that B-type lamins are involved in positioning nuclear pore complexes within the lamina.
Figure 3.
LA/C and LB2 were colocalized in LB1 silenced cells. LA/C and LB2 are shown in green and red, respectively. A single nucleus with an enlarged meshwork in the lamina and several small blebs are seen in different focal planes in A–R. Z-stacks of fluorescence images were taken, and the nuclear surface and the equatorial planes are shown in A–I and J–R, respectively. Indicated regions of the lamina mesh in C are enlarged and shown in D–I. Other regions within the lamina shown in a midplane section in L are seen at higher magnification in M–O depicting an LA/C-rich plaque and a small LA/C-rich bleb (shown in P–R). Another nucleus of an LB1 silenced cell containing two blebs is seen in S and T. The larger of these blebs has a significantly enlarged LA/C meshwork; see higher-magnification view of the rectangular area in S, which is enlarged ∼2.4-fold in T. Bars, 5 μm.
FCS reveals that A- and B-type lamins form separate, but interacting, nucleoplasmic structures
The enlarged LA/C and LB2 meshwork observed after LB1 silencing could be related to changes in their dynamic properties. Previous fluorescence recovery after photobleaching (FRAP) analyses demonstrated that the lamins incorporated into the lamina have little or no mobility, while a fraction of nucleoplasmic LC is mobile (Broers et al. 1999). However, FRAP analyses require significant overexpression of GFP-tagged lamins, potentially inducing artifacts. To investigate the dynamic properties of the lamins under more physiological conditions, we carried out FCS measurements. This technique permits the spatial and temporal analysis of diffusion coefficients of extremely low levels of GFP-tagged lamins in nuclei (<100 kilocounts per second, kcps), equivalent to concentrations of <20 molecules (<0.1 μM) per detection volume (Pack et al. 2006).
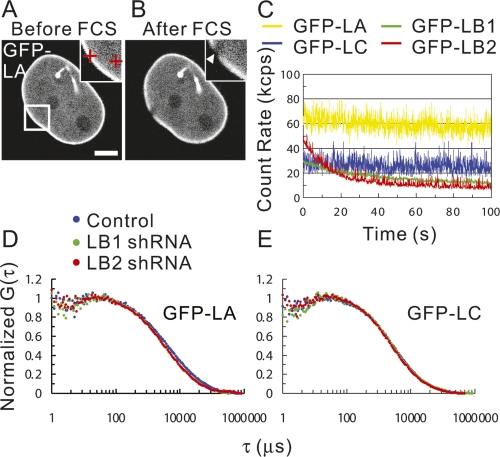
In order to determine the effects of silencing, the diffusion coefficients of each GFP-tagged lamin were first determined by FCS in nonsilenced cells expressing extremely low levels of protein. FCS measurements were not possible for any of the GFP-lamins within the lamina due to photobleaching (Fig. 4A,B; Hof et al. 2005). For the nucleoplasmic lamins, GFP-LB1 and LB2 were photobleached, but GFP-LA and LC diffused rapidly (Fig. 4C). Quantitative analysis of the FCS results (Fig. 4 C; Hess et al. 2002) revealed two mobile fractions of nucleoplasmic GFP-LA: a fast fraction of ∼53% of the protein moved with a diffusion coefficient of 5.0 ± 0.3 μm2/sec, and a slow fraction of ∼47% of the protein moved with a diffusion coefficient of 0.38 ± 0.04 μm2/sec. GFP-LC was also present in two mobile fractions: a fast (5.6 ± 0.3 μm2/sec) and a slower fraction (0.65 ± 0.06 μm2/sec), each consisting of ∼50% of the mobile protein. These mobile fractions comprised ∼90% of GFP-LA and GFP-LC in the nucleoplasm, while the remaining fractions (∼10%) were relatively immobile as they photobleached during FCS measurements. Based on the diffusion coefficients of the mobile fractions of nucleoplasmic GFP-LA and GFP-LC, estimates of nucleoplasmic viscosity, and the assumption that these fractions are globular in shape, we estimated their apparent molecular weights (Pack et al. 2006). For the fastest GFP-LA and GFP-LC fractions, the estimated molecular weight is ∼1.3 MDa. Similar estimates of the slower-moving fractions of GFP-LA and GFP-LC correspond to 2.9 GDa and 830 MDa, respectively. However, these latter estimates of molecular weights of the largest components in the slower-moving fractions cannot be attributable to free diffusion as it has been demonstrated that the cutoff for diffusion in nucleoplasm is 2–2.5 MDa (Gorisch et al. 2005).
Figure 4.
The mobilities of GFP-LA, LB1, LB2, and LC in live HeLa cells were measured by FCS. Fluorescence images of GFP-LA before and after FCS measurements are shown in A and B, respectively. Cross-hairs in the inset in A represent points measured by FCS. An arrowhead in the inset in B indicates the zone within the lamina bleached during FCS measurement. Bar, 5 μm. (C) Representative fluorescence fluctuations of GFP-LA, LB1, LB2, and LC in the nucleoplasm plotted as a function of time (yellow line for GFP-LA, blue line for GFP-LC, green line for GFP-LB1, and red line for GFP-LB2). (D,E) Note that LB1 and LB2 are rapidly photobleached. Mobilities of GFP-LA and LC in LB1 and LB2 silenced cells were measured by FCS. Averaged normal fluorescence autocorrelation curves of GFP-LA and GFP-LC in the nucleoplasm of control, LB1, and LB2 silenced cells were calculated from the fluorescence fluctuations and plotted as a function of correlation time τ (in microseconds) (blue dots for control cells, green dots for LB1 silenced cells, and red dots for LB2 silenced cells). G(τ) is the fluorescence autocorrelation function.
We next determined the effects of silencing individual lamins on the mobilities of the remaining lamins. Both GFP-LB1 and GFP-LB2 remained immobile in the lamina and the nucleoplasm when LA/C expression was silenced (data not shown). Likewise, the silencing of either LB1 or LB2 did not lead to changes in the mobilities of GFP-LA, GFP-LC, or the remaining B-type lamins in the lamina at least over the 15-sec time frame used for FCS measurements. However, when the mobilities of lamins are measured over longer time frames (∼300 min) by FRAP analyses, the recovery halftime of GFP-LA/C following LB1 silencing increases (Tang et al. 2008). Within the nucleoplasm, FCS analyses following LB1 or LB2 silencing showed an approximate twofold increase in the mobility of the slow fraction of GFP-LA (0.7–0.8 ± 0.1 μm2/sec, P < 0.05) (Fig. 4D), while the mobilities of the fast-moving fractions of GFP-LA (Fig. 4D) and of both fast and slow fractions of GFP-LC were unaltered (Fig. 4E). It should be noted that the FCS results for LB1 silencing were the same in cells with or without nuclear blebs. The significantly different mobilities of the A- and B-type lamins in the nucleoplasm provide further support that they form separate structures.
LB1 silencing alters chromatin organization: euchromatin is enriched in blebs
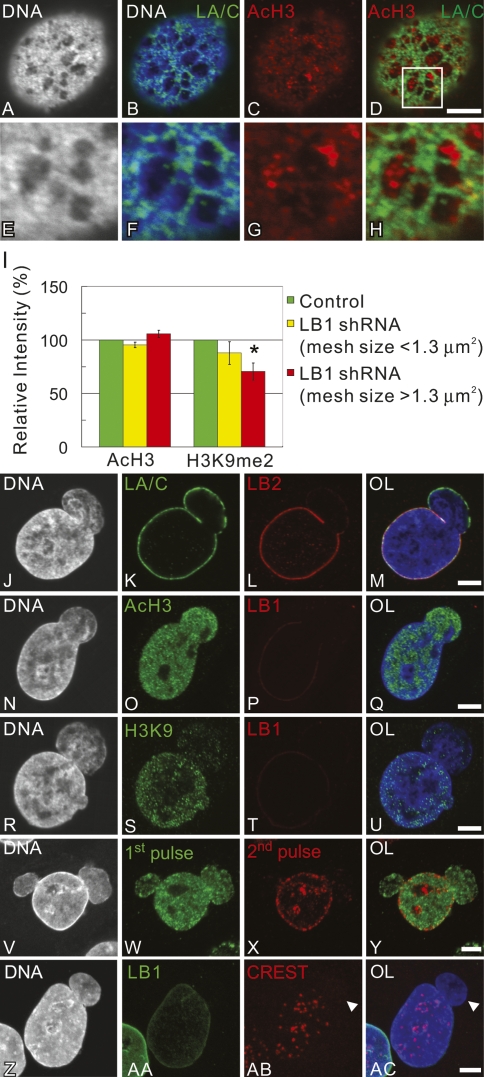
During our characterization of changes in the lamina after LB1 silencing, we observed that the DNA immediately beneath the large open areas of the meshworks and within the blebs stained weakly with Hoechst compared with areas directly under the lamin meshworks (Fig. 5A,B,E,F, J–M). Hoechst preferentially binds AT-rich DNA, which is typically enriched in heterochromatic regions (Hilwig and Gropp 1972), suggesting that DNA in the lightly stained areas is packaged as euchromatin. Immunofluorescence with anti-acetylated histone H3 (AcH3), a mark for transcriptionally active euchromatin (Trojer and Reinberg 2007), showed an enrichment of AcH3 in ∼87% of the enlarged lamina mesh regions (Fig. 5C,D,G,H) and in 100% of the blebs (Fig. 5N–Q). In contrast, the facultative heterochromatic mark histone H3 dimethylated at Lys 9 (H3K9me2) (Trojer and Reinberg 2007) was absent from ∼93% of the blebs, but was present in the main nuclear body (Fig. 5R–U). We were also able to correlate the overall increase in mesh size induced by LB1 silencing with more global changes in histone acetylation and H3K9me2. When the lamina mesh size increased above 1.3 μm2 (see Fig. 2R), there was an ∼30% reduction in fluorescence intensity with anti-H3K9me2 (P < 0.05) and an increase of ∼5% with anti-AcH3 (Fig. 5I) over the entire nucleus. There were no detectable changes in either H3K9me2 or AcH3 in the LB1 silenced cells without enlarged LA/C meshworks or blebs (Fig. 5I). These data show that the alterations in the structure of the lamina induced by LB1 silencing affect the normal organization of heterochromatin and euchromatin in interphase nuclei.
Figure 5.
(A–H) The state of chromatin modifications associated with the enlarged meshwork of the LA/C lamina in LB1 silenced cells assessed by confocal immunofluorescence. Silenced cells were processed for double-label immunofluorescence using anti-LA/C (green) and anti-AcH3 (red), and counterstained with Hoechst 33258 (white/blue). A region of mesh within the lamina in D is enlarged and shown in E–H. (J–Y) A series of equatorial confocal immunofluorescence images of LB1 silenced nuclei with blebs. In J–M, the bleb at the top right of the nucleus contains LA/C, but is devoid of LB2. In N–Q, AcH3 is present in the bleb, which is weakly stained with Hoechst 33258 and is devoid of LB1. R–U depict a nucleus with a bleb that is devoid of H3K9me2 (H3K9) and LB1 and stains very weakly with Hoechst 33258. V–Y show early (green) and mid to late replication patterns (red) visualized by double replication labeling. DNA is counterstained with Hoechst 33258 (blue). (Z–AC) Z-projected images of centromeres in a LB1 silenced cell with a nuclear bleb are visualized by immunostaining with CREST antiserum (red). DNA is counterstained with Hoechst 33258 (blue). Bar, 5 μm. Arrowhead indicates the position of the bleb. (I) Fluorescence intensities of AcH3 and H3K9me2 in nuclei of control and LB1 silenced cells. In each case, the fluorescence intensity in 40–150 nuclei was determined in each of two experiments. The relative fluorescence intensities in control cells and in LB1 silenced cells with normal meshes (<1.3 μm2) and large meshes (>1.3 μm2) are plotted in the histogram as green, yellow, and red bars, respectively. The error bar represents ±SD. The bar with an asterisk shows significant changes (P < 0.05) in the relative fluorescence intensity of AcH3 and H3K9me2 in LB1 silenced cells compared with those of control.
Since euchromatin replicates in early S phase, we also determined the effects of LB1 silencing on the replication states of chromatin by double replication labeling to visualize any alterations in early and mid-to-late replicating sites (Aten et al. 1992). When nuclei with blebs were analyzed, ∼84% of the blebs contained only early replicating chromatin (Fig. 5V–Y). The remaining ∼16% contained a few foci of late-replicating chromatin restricted to regions proximal to the main nuclear body (data not shown). Importantly, the main bodies of blebbed nuclei contained apparently normal early/mid-late replication patterns (Fig. 5V–Y). In order to characterize the chromatin content of the blebs in more detail, LB1 silenced cells were stained with CREST antiserum to localize centromeres (Shumaker et al. 2006). We hypothesized that the blebs, which contained only early replicating chromatin (Fig. 5V–Y), might be devoid of centromeres since pericentric heterochromatin is late replicating (O’Keefe et al. 1992). The results showed that >85% of the blebs were completely devoid of centromeres (Fig. 5Z–AC, arrowheads). In the remaining ∼15%, only one or two centromeres were located just inside the most proximal region of the bleb, while the remaining centromeric regions were localized to the main nuclear body away from the lamina (Fig. 5Z–AC).
LB1 silencing alters Pol II activity
The presence of AcH3 and the almost complete absence of H3K9me2 in the blebs induced by LB1 silencing suggest that these regions may be more transcriptionally active relative to other areas of the nucleus. Pol II was present in the blebs but was not enriched relative to the nuclear body (Fig. 6B–E). In contrast, there was a dramatic enrichment of activated hyperphosphorylated forms of Pol II (Pol IIo S2 and S5) labeling in ∼80% of the blebs (Fig. 6F–I; data not shown). In comparison with control cells, there was an ∼59% increase in fluorescence staining intensity for Pol IIo S2 (P < 0.05) and ∼30% for Pol IIo S5 (P < 0.05) in LB1 silenced nuclei with enlarged meshworks, but devoid of blebs (Fig. 6A). As was the case for H3K9me2 and AcH3 (see above), there was a positive correlation between LA/C lamina mesh size and Pol IIo fluorescence intensity (Fig. 6A). Nuclei in LB1 silenced cells with normal mesh size did not display a significant change in Pol IIo staining compared with nonsilenced cells (Fig. 6A). Immunoblotting of extracts from LB1 silenced cells showed a 1.7-fold increase in Pol IIo compared with controls confirming the overall increase in Pol IIo fluorescence intensity (Supplemental Fig. S4A).
Figure 6.
Transcriptional activity in LB1 silenced cells. (A) Total fluorescence intensities of Pol II, Pol IIo S2, and Pol IIo S5 in nuclei of control and LB1 silenced cells were measured using wide field microscopy. In each case, the fluorescence intensity in 40–150 nuclei was determined in each of two experiments. The relative fluorescence intensities in control cells and LB1 silenced cells with normal meshes (<1.3 μm2) and large meshes (>1.3 μm2) are plotted in the histogram as green, yellow, and red bars, respectively. Error bars represent ±SD. The bars with an asterisk mark experiments with significant changes (P < 0.05) in relative fluorescence intensity of Pol II and Pol IIo in LB1 silenced cells compared with those of controls. (B–E) Total Pol II in a nucleus with one bleb. Pol II is present both in the bleb and throughout the main nuclear body. (F–I) Pol II hyperphosphorylated on Ser 2 in the CTD (Pol IIo S2), LB1, and DNA showing the enrichment of Pol IIo in the bleb. (J–Q) RNA synthesis was visualized by BrU incorporation. Note the virtual absence of BrU incorporation into the bleb region (J–M), and the incorporation of BrU into a nucleolus in one of the few nuclei in which a nucleolus has entered the bleb (N–Q). (R–U) Localization of H3K4, LB1, and DNA demonstrating that this histone mark is present both in blebs and the nuclear body. (V–Y) Localization of H3K36, LB1, and DNA showing the absence of this histone mark within blebs. Bars, 5 μm.
In order to confirm the apparent increase in Pol II activity in LB1 silenced cells, we labeled nascent RNA by BrU incorporation in vivo (Pellizzoni et al. 2001). There were no obvious differences in BrU incorporation between control nuclei and LB1 silenced cells with normal meshworks or in bleb-free nuclei with enlarged lamina meshworks (data not shown). Surprisingly, ∼94% of the blebs showed almost no BrU incorporation, even though incorporation into the nuclear body appeared normal (Fig. 6J–M). Nucleoli were present in a small number of blebs, as demonstrated by the absence of Hoechst staining or antibody staining for Ki67, a nucleolar marker (Verheijen et al. 1989; data not shown). These nucleoli had levels of BrU incorporation similar to those in the nuclear body, even though there was little or no incorporation into the surrounding nucleoplasm within the bleb (Fig. 6N–Q). This suggests that the bleb microenvironment can accommodate normal RNA polymerase I (Pol I) transcription, but is nonpermissive for Pol II transcription.
During Pol II transcription, modifications of chromatin are required for the polymerase to proceed along the template. The promoter regions of transcribed genes are modified by acetylation of histone H3 as well as trimethylation of histon H3 at Lys 4 (H3K4me3) (Berger 2007). Both of these marks are present throughout the nucleus, including the blebs, in silenced (Figs. 5N–Q, 6R–U) and nonsilenced cells (Supplemental Fig. S4B–E). Transcript elongation also requires the transcribed chromatin regions to become trimethylated on histone H3K36 (Berger 2007). Immunostaining for histone H2 trimethylated on Lys 36 (H3K36me3) showed its absence in ∼90% of the blebs compared with the nuclear body in LB1 silenced cells (Fig. 6V–Y). The absence of H3K36me3 in the blebs is consistent with the failure to incorporate BrU into these structures and suggests that transcription elongation is inhibited in the blebs.
Gene-rich chromatin is concentrated in LA/C-rich, LB2-deficient nuclear blebs
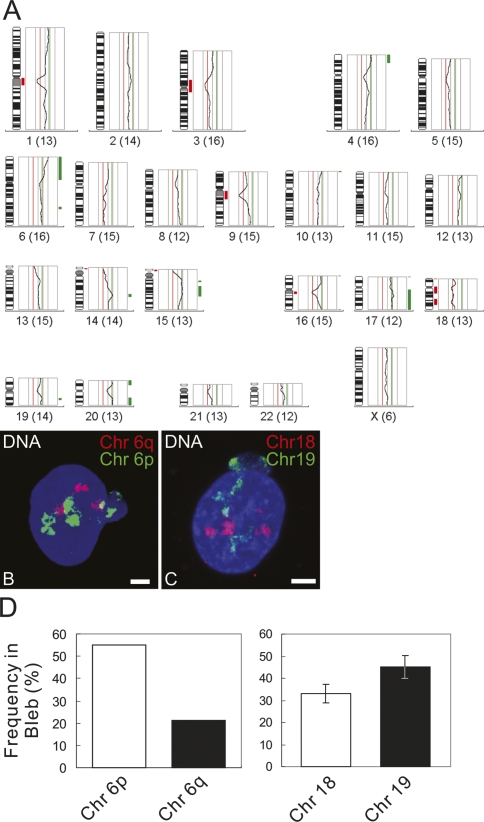
Chromosome territories (CTs) within spherical nuclei are distributed according to their gene density, with gene-rich chromosomes more likely to be in the interior and gene-poor chromosomes at the periphery (Croft et al. 1999). Because the blebs were highly enriched in euchromatin relative to the nuclear body and individual chromosomes contain different proportions of euchromatin and heterochromatin, we examined whether specific chromosomes or subregions of chromosomes were preferentially located in the blebs of LB1 silenced cells due to their higher gene density. Nuclei containing one bleb (that stained for LA/C, but not LB2) were microdissected using a Zeiss laser microdissection system (PALM) equipped with fluorescence optics (Supplemental Fig. S5A–D). The isolated DNA was amplified and characterized by comparative genomic hybridization (CGH) with pools of 20–30 blebs each to determine if there is a general over- or underrepresentation of certain chromosomal regions (Geigl and Speicher 2007). Control DNA for the CGH experiments was isolated from microdissected whole nuclei of LB1 silenced cells. In an additional experiment, DNA isolated and amplified from one large bleb was analyzed by CGH. The results showed that there was an overrepresentation of several gene-rich chromosomal regions in blebs. Most striking was a consistent overrepresentation of a region of chromosome 6p, 6p21 (Fig. 7; Supplemental Fig. S5E,F). Regions of the gene-rich chromosomes 17 and 19 were also overrepresented in bleb pools and present in the single bleb analysis, while regions of gene-poor chromosome 18 were underrepresented in the bleb pools and missing in the single bleb analysis (Fig. 7A; Supplemental Fig. S5E–G). The CGH findings were supported by interphase fluorescence in situ hybridization (FISH) experiments using specific chromosome painting probes (Fig. 7B,C). The distribution of whole chromosome paints for chromosomes 18 and 19 was compared in nuclei of LB1 silenced cells. The results demonstrated that chromosome 19 was significantly more likely to reside in a bleb (∼45% ± 5.2%) than chromosome 18 (∼33% ± 4.3%; P = 0.038). When the distribution of chromosome arms 6p and 6q was examined, it was observed that 6p was present in ∼55% of the nuclear blebs, while 6q was present in 21% (Fig. 7D). There were no changes in the radial arrangement of CTs 18 and 19 in LB1 silenced cells without nuclear blebs or in controls. As in control cells, chromosome 19 was found to be located more toward the nuclear interior and chromosome 18 more at the nuclear periphery (P < 0.001) (Supplemental Fig. S5H). Taken together, these results suggest a preference for gene-rich chromosome regions to localize in blebs that contain LA/C, but not LB2. Global changes in chromatin movements in LB1 silenced cells could explain the relocalization of a gene-rich chromosome from the nuclear interior to the blebs. However, the results of 6- to 12-h observations of LB1-silenced cells expressing GFP-histone H3 after photobleaching showed no significant long-range movements of chromosomes in nuclei with or without blebs (data not shown).
Figure 7.
(A) CGH profile from a pool of 20 blebs from LB1 silenced cells. The fluorescent intensities of the green and red channels are depicted as an intensity ratio profile next to each chromosome. Regions that are overrepresented in nuclear blebs are indicated by a green bar, and regions that are underrepresented are indicated by a red bar. Note the underrepresentation of regions of chromosome 18 and the overrepresentation of 6p, regions of chromosome 17, and regions of chromosome 19. (B,C) Chromosome distributions in LB1 silenced cells. DNA is counterstained with Hoechst 33258. DNA, chromosome 6p (or 19), and chromosome 6q (or 18) are shown in blue, green, and red, respectively. Bars, 5 μm. (D) Percentiles of the distributions of painting probes for 6q/6p and 18/19 in LB1 silenced cells with nuclear blebs. Error bars are ±SD.
Discussion
In this study, we showed by confocal microscopy that A- and B-type lamins form separate, but interconnected meshworks within the lamina and are present in less organized states in the nucleoplasm. Similar meshworks have been described recently for LB using a microscope equipped for three-dimensional (3D) structured illumination (Schermelleh et al. 2008). Selective silencing of each of the lamins has provided insights into the interactions between these different lamin meshworks. Removal of either LA/C or LB2 had no effect on the structural organization of the other lamins, but LB1 silencing dramatically changed the appearance of both the LA/C and LB2 meshworks, suggesting that LB1 provides a scaffold required for the structural integrity of the entire nuclear lamin system. The loss of LB1 also induced the formation of blebs containing LA/C, but not LB2. These blebs contained predominantly early replicating, gene-rich euchromatin. Surprisingly, the chromatin in the blebs was not actively transcribed despite the presence of activated RNA Pol II and other histone marks signifying active transcription.
Nuclear herniations that are morphologically similar to blebs have also been reported in lmna−/− embryonic fibroblasts (MEFs). These NE herniations lack LB, Nup153, and LAP2 (Sullivan et al. 1999). This is in contrast to our results in LA/C silenced HeLa cell nuclei that were devoid of blebs. In addition, ∼40% of lmnb1Δ/Δ MEFs exhibit nuclei with structures described as blebs. These blebs contain LA/C and nuclear pore complexes, whereas the blebs induced by LB1 silencing reported in this study lack pore complexes. While the blebs described in the lmnb1Δ/Δ cells have some features in common with the blebs we describe here, these cells appear to express LB1 lacking a portion of the central rod domain and the non-α-helical C terminus (Vergnes et al. 2004). Therefore, it is difficult to compare the blebs in HeLa cells in which there is silencing of LB1 with MEFs expressing a truncated form of LB1.
Our FCS data show that >90% of nucleoplasmic LA and LC exhibit rapid mobility, while LB1 and LB2 are static, further highlighting the differences in the organizational states of the two types of lamins. LB1 silencing also leads to a significant increase in the mobility of the slow nucleoplasmic LA fraction, but no measurable changes in the other LA or LC fractions. The differences in mobility between LA and LC could be explained by transient interactions with the immobile B-type lamins or perhaps other lamin-associated proteins or factors such as LAP2α, BAF, or PCNA (Schirmer and Gerace 2004; Shimi et al. 2004; Dechat et al. 2008). It should be noted that the high-molecular-weight estimates for the slowest-moving fractions are likely to be incorrect, since macromolecules larger than 2.5 MDa have been shown to be immobile in the nucleoplasm (Gorisch et al. 2005). The anomalous sizes calculated in this study for the slow fractions could be due to more stable interactions with immobile structures. It is also possible that these lamin fractions are not globular structures but are short filaments, enabling them to negotiate small channels within the chromatin network. Such filamentous structures have been described as protofilaments in in vitro lamin assembly assays (Heitlinger et al. 1991; Stuurman et al. 1998). The static B-type lamins in the nucleoplasm could be bound to immobile nuclear structures or exist in very large complexes, which could restrict their movement. Alternatively, the nucleoplasmic B-type lamins could form a polymer network in the nucleoplasm that provides a scaffold for interactions with A-type lamins and other unknown factors.
The formation of LA/C-rich blebs that do not contain LB2 in the LB1 silenced cells provides a unique opportunity to examine the formation and interrelationships of lamin networks. Our results suggest that blebs form from LA/C-rich plaques in the lamina and that the interaction of chromatin with these regions is involved in bleb formation. It is also possible that blebs are formed at the time of cell division during daughter cell nuclear assembly. In support of this latter idea, tethering of reporter genes to the nuclear periphery with lamins or lamina-associated proteins also requires a round of mitosis (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008), suggesting that interactions of the reporter gene with the lamina may be established during NE assembly. In addition, it has been shown that CTs are established during mitosis and the early G1 phase of the cell cycle (Walter et al. 2003). The formation of these LA/C-rich plaques and blebs in LB1 silenced cells suggests that, under normal conditions, the A- and B-type lamins form microdomains, each interacting with different types of chromatin. In support of this, we observed global increases in the fluorescence intensity of AcH3 and decreases in fluorescence intensity in H3K9me2. These changes were directly correlated with the increase in meshwork size. However, the most dramatic changes in AcH3 up-regulation and H3K9me2 down-regulation were observed in the blebs. These changes may be attributable to the loss of anchorage sites for peripheral heterochromatin, which in turn could affect the organization of both heterochromatic and euchromatic regions throughout the nucleus. Our immunolocalization studies of histone marks and CREST, along with double replication labeling and CGH analyses of the DNA extracted from blebs, indicate that the bleb is a unique microenvironment containing only euchromatin. The dramatic separation of LA/C and LB2 meshworks in the blebbed nucleus may reflect a LA/C association with gene-rich chromatin and LB2 (and LB1) with gene-poor chromatin. For example, the absence of CREST staining in the blebs suggests that B-type lamins are responsible for attaching centromeres to the nuclear periphery (Solovei et al. 2004). The CGH analyses of DNA associated with blebs indicate that they contain gene-rich regions of chromosomes, including those from chromosomes 6, 17, and 19, while regions of gene-poor chromosomes such as 18 were underrepresented. Most interestingly, a region of the short arm of chromosome 6, 6p21, is significantly overrepresented in blebs. With >200 gene loci, this is one of the most gene-rich regions in the human genome (Volpi et al. 2000).
Other studies have also linked significant alterations in the organization of chromatin in the nucleus with lamins. In lmnb1Δ/Δ MEFs, chromosome 18 has been shown to be positioned away from its normal location at the nuclear periphery, supporting the role of the B-type lamins in tethering gene-poor chromosomes (Malhas et al. 2007). A recent genome-wide analysis of human fibroblasts revealed that LB1 is preferentially associated with large gene-poor chromatin domains up to 10 Mb in length (Guelen et al. 2008). In another study, it has been reported that the gene-rich chromosome 19 is located closer to the nuclear periphery in LB1 silenced cells (Tang et al. 2008). However, we did not observe a change in the distribution of these CTs in the majority of LB1 silenced nuclei. We only observed changes in the positioning of whole CTs when blebs were present and there was a dramatic separation of the LA/C and LB2 meshworks.
The chromatin associated with LA/C-rich blebs and enlarged meshworks in LB1 silenced cells is primarily euchromatic. Therefore, we expected that Pol II transcription would be elevated. In support of this, immunostaining with Pol IIo antibodies increased throughout the entire nucleus of LB1 silenced cells, especially in the blebs. Enrichment of H3K4me3 in the bleb was also consistent with active transcription. However, nascent RNA labeling and H3K36me3 localization within blebs were almost undetectable. These observations are consistent with the phenomenon of promoter proximal stalling (Kumar et al. 1998). Since several Pol II transcription factors are known to promote escape from stalling (Adelman et al. 2005; Peterlin and Price 2006), it is possible that the interaction of lamins with some of these factors is disrupted in the LB1 silenced cells, leading to an increase in stalling. It is also possible that lamin binding regulates other factors necessary for transcription or recycling of the polymerase after termination. The inactivation of Pol II transcription has also been shown following disruption of lamin networks by depleting LB1 or expressing a truncated LA (Pendas et al. 2002; Spann et al. 2002; Tang et al. 2008). In contrast to our results showing an increase in Pol IIo accompanying a decrease in RNA synthesis by quantitative immunofluorescence and immunoblotting, Tang et al. (2008) found that there was a decrease in Pol IIo by immunofluorescence following LB1 silencing in HeLa cells. Although at the present time we do not understand this discrepancy, our localization of multiple factors involved in Pol II transcription and their correlation with structural changes in the lamina of LB1 silenced cells strongly suggest that inhibition of RNA synthesis is due to promoter proximal stalling, not to a decrease in the amount of enzyme. Interestingly, Pol I activity appeared to be unaffected in blebs, as nucleoli in these domains incorporated BrU (see also Spann et al. 2002). The apparent lack of nuclear pore complexes within LA/C-rich blebs is also worthy of consideration as it has been shown that some transcriptionally active genes require tethering to nuclear pores (Schneider and Grosschedl 2007). Therefore, genes that are most likely to be associated with B-type lamin microdomains may not be activated unless tethered to nuclear pore complexes. Alternatively, the lack of nuclear pore complexes in the bleb could prohibit local export of mRNA, thereby inhibiting transcription, as it has been shown that export of mRNA is coupled to splicing and transcription (Tokunaga et al. 2006; Valencia et al. 2008).
We propose that the nuclear lamina and nucleoplasmic lamin networks are organized into separate but interconnected microdomains. Our results demonstrate that when the LA/C and LB2 microdomains within the lamina are artificially expanded by silencing LB1, the separate functions of A- and B-type lamins become more obvious. These microdomains have different functions in chromatin organization and gene regulation. Our results from silencing LB1 suggest that its presence is critical for organizing both the LA/C and the LB2 microdomains. The data also show that LB1 and LB2 assemble into separate, but interconnected networks and that they each interact differently with the LA/C network. Based on the evidence presented in this study, we propose that B-type lamins within the lamina and nucleoplasm form relatively static structures that provide a scaffold for gene-poor chromosomal regions. This latter interaction is supported by the recent finding that gene-poor chromosomal regions are associated with LB1 in the lamina (Guelen et al. 2008). The results also suggest that the A-type lamins are preferentially associated with gene-rich regions. In normal cells, the close interactions of A-type and B-type lamins could provide an environment in which cross-talk between the different microdomains could finely regulate gene expression. These findings have important implications for the understanding of the laminopathies that frequently display enlarged meshworks such as in muscular dystrophies, lipodystrophies, and the LA/C-rich nuclear blebs seen in progeria (Goldman et al. 2004; Burke and Stewart 2006). In the laminopathies, significant alterations in the interrelationships between different microdomains could lead to defects in gene expression. In the future, the identification of lamin-associated factors involved in histone modification and Pol II transcription could provide new insights into their roles in chromatin organization and gene regulation.
Materials and methods
Plasmids used for live imaging and silencing of lamins
The plasmids used for live cell imaging in the FCS experiments were pEGFP-myc-HsLMNA, pEGFP-myc-HsLMNB1, and pEGFP-myc-HsLMNC (Moir et al. 2000). pEGFP-HsLMNB2 was prepared from pEYFP-HsLMNB2 (provided by Dr. Harald Herrmann, Heidelberg) (Schumacher et al. 2006).
The construction of shRNA expression plasmids were described elsewhere (Brummelkamp et al. 2002; Kojima et al. 2004). The target sequences for silencing lamin expression were as follows: LMNA T1, 5′-AGCAGTCTCTGTCCTTCGA-3′ (human LA/C); LMNA T8, 5′-ATGATCCCTTGCTGACTTA-3′ (human LA/C); LMNB1 T3, 5′- CGAGCATCCTCAAGTCGTA-3′ (human LB1); LMNB1 T4, 5′-GAATCAGAGGCGAGTAGTA3′ (human LB1); LMNB2 T7, 5′-CGGTGATGCGTGAGAAT GA-3′ (human LB2); LMNB2 T8, 5′-TCAAGAACAACTCGGA CAA-3′ (human LB2); Neg-ctrl, 5′-ATGTACTGCGCGTGGA GA-3′ (negative control, scrambled). The loop sequence is either 5′-GCTTCCTGTCAC-3′ (for LMNA T1 and T8) or 5′-TTCAA GAGA-3′ (for the others). All the constructs were verified by DNA sequencing.
Cell culture and transfection
HeLa cells were grown as described previously (Shumaker et al. 2006) and transfected using the TransIT-HeLa MONSTER transfection kit (Mirus) in accordance with the product manuals or by electroporation as described previously (Yoon et al. 2001). Transfection efficiencies were routinely ∼90%. In shRNA silencing experiments, transfected cells were assayed 3–5 d following transfection.
Gel electrophoresis and immunoblotting
HeLa cells transfected with the silencing vectors were solubilized in Laemmli sample buffer (Laemmli 1970) lacking dithiothreitol (DTT) and dyes. The protein concentration was determined by the BCA protein assay kit (Pierce Biotechnology, Inc.). Prior to electrophoresis, DTT was added to the samples to a final concentration of 0.1 M, and the proteins were separated by SDS-PAGE on 10% gels and transferred to nitrocellulose. Immunoblotting was carried out with the primary antibodies: mouse anti-LA/C (1:200; JoL2, Chemicon), rabbit anti-LB1 (1:4000) (Moir et al. 1995), mouse anti-LB2 (1:200; LN43, Abcam), mouse anti-Pol II hyperphosphorylated at Ser5 (1:500; Pol IIo, provided by Dr. David Spector, Cold Spring Harbor Laboratory, NY), mouse anti-α-tubulin (1:5000; B-5-1-2, Sigma), and mouse anti-importin β1 (1:4000; Affinity Bioreagents, Inc.). Horseradish peroxidase-conjugated secondary antibodies (KPL) were used at a dilution of 1:40,000, and the peroxidase activity was detected by the Super Signal Pico-West chemiluminescence kit (Pierce). Images were collected using a Kodak Image Station 440CF (Kodak) and quantified using Kodak Molecular Imaging software.
Indirect immunofluorescence
Cells grown on glass coverslips were fixed either in methanol for 10 min at −20°C or in 3.7% paraformaldehyde in PBS for 10 min followed by extraction in 0.1% Triton X-100 in PBS for 10 min at room temperature. Cells used for Pol IIo immunofluorescence were fixed in 3.7% paraformaldehyde on ice and permeabilized as above. Primary antibodies included mouse anti-LA/C (1:200; JoL2), goat anti-LA/C (1:500; N-18, Santa Cruz Biotechnologies), rabbit anti-LA (1:2000) (Dechat et al. 2007), rabbit anti-LB1 (1:1000) (Moir et al. 1995), goat anti-LB1 (1:300; M-20, Santa Cruz Biotechnologies), mouse anti-LB2 (1:200; LN43, Abcam), and rabbit anti-LB2 (1:1000; prepared by Cocalico using the LB2-specific peptide TVKKSSVMRENENG). Nuclear pore complexes were stained with the mouse monoclonal antibody mAb414 (1:1000; Covance). Chromatin was stained with rabbit AcH3 (1:1000; Upstate Biotechnologies), rabbit anti-histone H3K4me3 (1:1000; Abcam), rabbit antihistone H3K36me3 (1:1000; Abcam), mouse anti-Pol II hyperphosphorylated at Ser2 (H5) and Ser5 (H14) (1:100; provided by Dr. David Spector, Cold Spring Harbor Laboratory, NY), anti-Pol II (1:1000; 4H8, Abcam), human anti-CREST (1:100; provided by Dr. Bill Brinkley, Baylor College of Medicine, TX), and rabbit anti-histone H3K9me2 (1:1000; provided by Dr. Thomas Jenuwein, The Vienna Biocenter, Vienna). The secondary antibodies used were goat anti-mouse IgG-Alexa Fluor 488, goat anti-mouse IgG-Alexa Fluor 568, goat anti-mouse IgG-Cy5, donkey anti-mouse IgG-Alexa Fluor 488, donkey anti-mouse IgG-Alexa Fluor 568, donkey anti-mouse IgG-Cy5, goat anti-rabbit IgG-Alexa Fluor 488, goat anti-rabbit IgG-Alexa Fluor 568, goat anti-rabbit IgG-Cy5, donkey anti-rabbit IgG-Alexa Fluor 488, donkey anti-rabbit IgG-Alexa Fluor 568, and donkey anti-rabbit IgG-Cy5 (all 1:200; Molecular Probes, Inc., Jackson ImmunoResarch Laboratories, Inc.). DNA was stained with Hoechst 33258 (Molecular Probes, Inc.). Fixed cells were observed with either the Zeiss AxioImager.Z1 or Zeiss LSM 510 META (Carl Zeiss, Inc.) microscopes using oil immersion objective lenses (PlanApochromat, 63×, 1.40 NA; PlanApochromat 100×, 1.40 NA; Carl Zeiss, Inc.). The images were analyzed using a script, to call functions in the AutoMeasure Plus module of AxioVision 4.6 (Carl Zeiss, Inc.) to determine mesh size within the lamina in control, LA/C, LB1, and LB2 silenced cells. Briefly, mesh size was measured after converting the lamin staining picture to a thresholded binary image, and the area of each dark region within the nucleus was measured. Fluorescence intensities of the histone and Pol II staining in control and LB1 silenced cells were quantified by determining average pixel intensities over a whole nucleus. For presentation, images were adjusted for brightness, contrast, and γ settings. Statistical values were determined using the Student’s t-test.
Laser microdissection of nuclear blebs
LB1 silenced cells were grown on PEN membrane slides and prepared for immunofluorescence with antibodies directed against LA and LB2. Nuclei containing blebs that stained with anti-LA but not anti-LB were identified, imaged, and then prepared for PALM laser microdissection (Geigl and Speicher 2007). Either single blebs or pools of 20–30 blebs were collected and prepared for CGH. Whole nuclei on the same membrane were dissected and served as controls. The amplified bleb DNA was labeled with digoxigenin-11-dUTP, and the whole nuclei DNA was labeled with and biotin 16-dUTP. The labeled DNA was mixed with Cot DNA (Roche) and hybridized on 46XY metaphase spreads. The biotinylated and digoxigenin-labeled DNA probes were detected with avidin-Cy3.5 (Rockland) and mouse anti-digoxigenin-FITC (Roche). Chromosomes were counterstained with DAPI. The ratio of test DNA and control DNA was evaluated using the Zeiss Isis CGH software to identify regions that were over- or underrepresented in nuclear blebs. Three bleb pools each containing 20–30 blebs and two single blebs were analyzed by CGH.
FISH
Cells were prepared for FISH as described (Solovei et al. 2002) using DNA from flow-sorted chromosomes 18 and 19 labeled by nick translation. Chromosome 18 was labeled with biotin-dUTP, and chromosome 19 was labeled with digoxigenin-dUTP and detected with avidin-Alexa Fluor 488 (Molecular Probes, Inc.) and mouse anti-digoxigenin Cy3 (Dianova), respectively. Radial arrangements of the chromosomes were analyzed as described (Ronneberger et al. 2008) using images obtained with the Zeiss LSM510Meta.
Double replication labeling
HeLa cells were pulsed for 40 min with 0.01 mM IdU (Sigma) in complete media, chased with media without IdU for 4.5 h, incubated in complete media containing 0.01 mM CldU (Sigma) for 30 min at 37°C, then prepared for examination as described (Aten et al. 1992). IdU-labeled DNA was detected with mouse anti-BrdU (Caltag Laboratories), and CldU-labeled DNA was detected with rat anti-BrdU antibody (Abcam). Secondary antibodies were donkey anti-rat IgG-Cy3 (Jackson ImmunoResearch Laboratories, Inc.) and goat anti-mouse IgG-Alexa 488 (Molecular Probes, Inc.).
Transcription labeling
In vivo detection of transcription sites was carried out as described (Pellizzoni et al. 2001) using HeLa cells and processed for immunofluorescence as described above. Mouse monoclonal anti-BrdU (BU-33, Sigma) was used at 1:30 dilution.
FCS measurements
Live cell imaging for FCS was carried out at 24–48 h following transfection using the Zeiss LSM 510 META (Carl Zeiss, Inc.) with a water immersion objective lens (PlanApochromat, 40×, 1.45 NA). Transfected cells were maintained in Opti-MEM medium (Invitrogen) during observation. Cells with very low levels of GFP expression, <100 kcps, were used for FCS analyses as described (Pack et al. 2006). Lamin particle size was estimated using known values for nuclear viscosity as described (Pack et al. 2006).
Acknowledgments
We thank David L. Spector for the antibodies against Pol IIo. Due to the limitations on the number of references, many informative papers could not be referenced. This work was supported by grants from the National Cancer Institute (RO1 CA31760) and the National Institute on Aging (RO1 AG023776) and an Ellison Senior Scholar Award to R.D.G. K.P. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1735208.
References
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Aten J.A., Bakker P.J., Stap J., Boschman G.A., Veenhof C.H. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem. J. 1992;24:251–259. doi: 10.1007/BF01046839. [DOI] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Broers J.L., Machiels B.M., van Eys G.J., Kuijpers H.J., Manders E.M., van Driel R., Ramaekers F.C. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Burke B., Stewart C.L. The laminopathies: The functional architecture of the nucleus and its contribution to disease. Annu. Rev. Genomics Hum. Genet. 2006;7:369–405. doi: 10.1146/annurev.genom.7.080505.115732. [DOI] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Shimi T., Adam S.A., Rusinol A.E., Andres D.A., Spielmann H.P., Sinensky M.S., Goldman R.D. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc. Natl. Acad. Sci. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes & Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E., Tramier M., Coppey-Moisan M., Gaillard C., Courvalin J.C., Buendia B. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum. Mol. Genet. 2006;15:1113–1122. doi: 10.1093/hmg/ddl026. [DOI] [PubMed] [Google Scholar]
- Finlan L.E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J.R., Bickmore W.A. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/ journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigl J.B., Speicher M.R. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat. Protocols. 2007;2:3173–3184. doi: 10.1038/nprot.2007.476. [DOI] [PubMed] [Google Scholar]
- Goldman A.E., Moir R.D., Montag-Lowy M., Stewart M., Goldman R.D. Pathway of incorporation of microinjected lamin A into the nuclear envelope. J. Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorisch S.M., Wachsmuth M., Toth K.F., Lichter P., Rippe K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005;118:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Heitlinger E., Peter M., Haner M., Lustig A., Aebi U., Nigg E.A. Expression of chicken lamin B2 in Escherichia coli: Characterization of its structure, assembly, and molecular interactions. J. Cell Biol. 1991;113:485–495. doi: 10.1083/jcb.113.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S.T., Huang S., Heikal A.A., Webb W.W. Biological and chemical applications of fluorescence correlation spectroscopy: A review. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- Hilwig I., Gropp A. Staining of constitutive heterochromatin in mammalian chromosomes with a new fluorochrome. Exp. Cell Res. 1972;75:122–126. doi: 10.1016/0014-4827(72)90527-7. [DOI] [PubMed] [Google Scholar]
- Hof M., Hutterer R., Fidler V. Fluorescence spectroscopy in biology: Advanced methods and their applications to membranes, proteins, DNA, and cells. Springer; New York: 2005. [Google Scholar]
- Kojima S., Vignjevic D., Borisy G.G. Improved silencing vector co-expressing GFP and small hairpin RNA. Biotechniques. 2004;36:74–79. doi: 10.2144/04361ST02. [DOI] [PubMed] [Google Scholar]
- Krohne G., Benavente R., Scheer U., Dabauvalle M.C. The nuclear lamina in Heidelberg and Wurzburg: A personal view. Eur. J. Cell Biol. 2005;84:163–179. doi: 10.1016/j.ejcb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Kumar K.P., Akoulitchev S., Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc. Natl. Acad. Sci. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran R.I., Spector D.L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., Lee R.T. Lamin A and lamin C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- Malhas A., Lee C.F., Sanders R., Saunders N.J., Vaux D.J. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 2007;176:593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn K.J., Cabuy E., Bonne G., Levy N., Morris G.E., Novelli G., Kill I.R., Bridger J.M. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Spann T.P., Goldman R.D. The dynamic properties and possible functions of nuclear lamins. Int. Rev. Cytol. 1995;162B:141–182. doi: 10.1016/s0074-7696(08)62616-9. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Yoon M., Khuon S., Goldman R.D. Nuclear Lamins A and B1. Different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe R.T., Henderson S.C., Spector D.L. Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack C., Saito K., Tamura M., Kinjo M. Microenvironment and effect of energy depletion in the nucleus analyzed by mobility of multiple oligomeric EGFPs. Biophys. J. 2006;91:3921–3936. doi: 10.1529/biophysj.105.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Charroux B., Rappsilber J., Mann M., Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 2001;152:75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas A.M., Zhou Z., Cadinanos J., Freije J.M., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodriguez F., Tryggvason K., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Reddy K.L., Zullo J.M., Bertolino E., Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Ronneberger O., Baddeley D., Scheipl F., Verveer P.J., Burkhardt H., Cremer C., Fahrmeir L., Cremer T., Joffe B. Spatial quantitative analysis of fluorescently labeled nuclear structures: Problems, methods, pitfalls. Chromosome Res. 2008;16:523–562. doi: 10.1007/s10577-008-1236-4. [DOI] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E.C., Gerace L. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J. Biol. Chem. 2004;279:42811–42817. doi: 10.1074/jbc.M407705200. [DOI] [PubMed] [Google Scholar]
- Schneider R., Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes & Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Schumacher J., Reichenzeller M., Kempf T., Schnolzer M., Herrmann H. Identification of a novel, highly variable amino-terminal amino acid sequence element in the nuclear intermediate filament protein lamin B(2) from higher vertebrates. FEBS Lett. 2006;580:6211–6216. doi: 10.1016/j.febslet.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Shimi T., Koujin T., Segura-Totten M., Wilson K.L., Haraguchi T., Hiraoka Y. Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J. Struct. Biol. 2004;147:31–41. doi: 10.1016/j.jsb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S., et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Cavallo A., Schermelleh L., Jaunin F., Scasselati C., Cmarko D., Cremer C., Fakan S., Cremer T. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp. Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- Solovei I., Schermelleh L., During K., Engelhardt A., Stein S., Cremer C., Cremer T. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma. 2004;112:410–423. doi: 10.1007/s00412-004-0287-3. [DOI] [PubMed] [Google Scholar]
- Spann T.P., Goldman A.E., Wang C., Huang S., Goldman R.D. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 2002;156:603–608. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N., Heins S., Aebi U. Nuclear lamins: Their structure, assembly, and interactions. J. Struct. Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.W., Maya-Mendoza A., Martin C., Zeng K., Chen S., Feret D., Wilson S.A., Jackson D.A. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J. Cell Sci. 2008;121:1014–1024. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- Tokunaga K., Shibuya T., Ishihama Y., Tadakuma H., Ide M., Yoshida M., Funatsu T., Ohshima Y., Tani T. Nucleocytoplasmic transport of fluorescent mRNA in living mammalian cells: Nuclear mRNA export is coupled to ongoing gene transcription. Genes Cells. 2006;11:305–317. doi: 10.1111/j.1365-2443.2006.00936.x. [DOI] [PubMed] [Google Scholar]
- Trojer P., Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol. Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Valencia P., Dias A.P., Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L., Peterfy M., Bergo M.O., Young S.G., Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen R., Kuijpers H.J., Schlingemann R.O., Boehmer A.L., van Driel R., Brakenhoff G.J., Ramaekers F.C. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J. Cell Sci. 1989;92:123–130. doi: 10.1242/jcs.92.1.123. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Walter J., Schermelleh L., Cremer M., Tashiro S., Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.H., Yoon M., Moir R.D., Khuon S., Flitney F.W., Goldman R.D. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J. Cell Biol. 2001;153:503–516. doi: 10.1083/jcb.153.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]