Abstract
The broad cellular actions of RNase III family enzymes include ribosomal RNA (rRNA) processing, mRNA decay, and the generation of noncoding microRNAs in both prokaryotes and eukaryotes. Here we report that YmdB, an evolutionarily conserved 18.8-kDa protein of Escherichia coli of previously unknown function, is a regulator of RNase III cleavages. We show that YmdB functions by interacting with a site in the RNase III catalytic region, that expression of YmdB is transcriptionally activated by both cold-shock stress and the entry of cells into stationary phase, and that this activation requires the σ-factor-encoding gene, rpoS. We discovered that down-regulation of RNase III activity occurs during both stresses and is dependent on YmdB production during cold shock; in contrast, stationary-phase regulation was unperturbed in ymdB-null mutant bacteria, indicating the existence of additional, YmdB-independent, factors that dynamically regulate RNase III actions during normal cell growth. Our results reveal the previously unsuspected role of ribonuclease-binding proteins in the regulation of RNase III activity.
Keywords: Ribonuclease III, RpoS, cold shock, RNA decay, RNA processing
RNase III family ribonucleases, which cleave dsRNA to yield 5′ phosphate and 3′ hydroxyl termini, are remarkable for both the extent of their evolutionary conservation in prokaryotic and eukaryotic cells and the diversity of their biological roles (Court 1993; Nicholson 2003; Drider and Condon 2004; MacRae and Doudna 2007). Containing 226 amino acids, the 25.6-kDa peptide encoded by the rnc gene of Escherichia coli (EC 3.1.26.3) is structurally the least complex and functionally perhaps the most extensively studied RNase III family member. Rnc monomers, which include an N-terminal catalytic domain and a C-terminal dsRNA-binding domain (dsRBD) (Nashimoto and Uchida 1985), dimerize to form the enzymatically active protein (Dunn 1982; Nicholson 2003).
E. coli RNase III was first identified by its ability to catalyze cleavage of ribosomal RNA (rRNA) precursors during ribosome biogenesis (for review, see Dunn 1982). Subsequently, this enzyme has been shown to mediate the maturation and/or degradation of a variety of transcripts, including tRNA precursors (Régnier and Grunberg-Manago 1989), conditionally expressed small non-protein-coding RNAs (Murchison and Hannon 2004), and mRNAs that encode the exoribonuclease PNPase (Pnp) (Régnier and Portier 1986), Rnc itself (Matsunaga et al. 1996a, b), or phage and plasmid proteins (Nicholson 1996, 2003). However, notwithstanding the multiplicity and breadth of functions of RNase III, rnc-null mutant E. coli cells are viable and show no phenotypic abnormalities except for a slightly impeded rate of growth (Babitzke et al. 1993) and minimally defective translation of certain mRNAs (Talkad et al. 1978).
The first suggestion that the ribonucleolytic actions of RNase III may be regulated came from the observation by Makarov and Apirion in 1992 that extracts of E. coli cells contain an ∼17-kDa protein that can inhibit in vitro RNase III-dependent processing of p10Sa RNA (Makarov and Apirion 1992)—a 10Sa RNA precursor (also known as ssrA RNA and tmRNA) that has an important role in the recycling of ribosomes from defective mRNAs (Richards et al. 2008). However, the nature of this RNase III-inhibiting moiety and its possible biological role(s) has remained obscure for almost two decades. Using a function-based screen to identify genes encoding proteins that regulate endonucleolytic cleavages by RNase III in vivo, we discovered a highly conserved bacterial protein YmdB that inhibits RNase III actions by binding to the region required for dimerization/activation of the enzyme. We show that production of YmdB and the consequent inhibition of RNase III function are modulated in response to cellular and environmental stresses. Our results reveal a novel mechanism for the dynamic regulation of ribonuclease activity by alterations in cell physiology.
Results
Screening of RNase III regulators in E. coli
To identify trans-acting regulators of RNase III activity, we introduced a library of plasmid-borne E. coli genes [the ASKA library, which includes all known E. coli ORFs expressed from an IPTG-inducible promoter (Kitagawa et al. 2005), into an E. coli strain (RS7305) containing a single copy of a rnc′-′lacZ reporter gene fusion (Matsunaga et al. 1996a)]. Earlier work has shown that RNase III cleaves its own transcript, and consequently that β-galactosidase production from this fusion construct is increased approximately ninefold in cells containing a missense mutation that decreases RNase III activity (Matsunaga et al. 1996a, b). We reasoned that cleavage of the rnc′-′lacZ transcript would also be affected by down-regulators of Rnc activity encoded by ASKA library ORFs. Regulation of expression of β-galactosidase from the rnc′-′lacZ fusion was monitored colorimetrically on MacConkey-Lac and S-gal plates as described in the Materials and Methods. Two colonies that showed reproducibly increased color intensity on culture plates containing IPTG were obtained and the phenotype was confirmed. Sequence analysis of ASKA inserts in plasmids isolated from these colonies identified ymdB (b1045), which is located at the 23.82-min position of the E. coli chromosome and encodes a putative 177-amino-acid 18.88-kDa protein of previously unknown function (Swiss-Prot P0A8D6). Swiss-Prot database analysis of the putative YmdB protein indicates that amino acids 1–175 comprise a macro (A1pp) domain characteristic of Appr-1″-p processing family proteins, which catalyze the conversion of ADP-ribose-1″-monophosphate (Appr-1″-p) to ADP-ribose in eukaryotes (Culver et al. 1994).
Effect of YmdB on cleavage of RNase III targeted transcripts
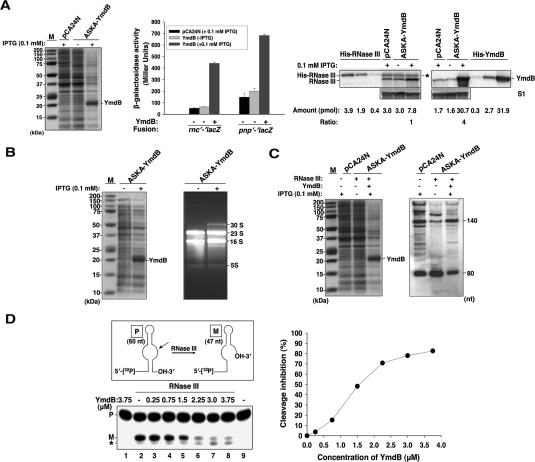
Addition of IPTG to cultures of bacteria expressing the ASKA library-derived plasmid-borne ymdB insert under control of the plac promoter increased production of a protein migrating in polyacrylamide gels at the position expected for YmdB (Fig. 1A, left). This increase was associated with reduced cleavage of transcripts containing RNase III targeted sites in rnc and pnp mRNAs fused to lacZ (Matsunaga et al. 1996a; Beran and Simons 2001) as evaluated quantitatively by measuring β-galactosidase activity encoded by lacZ (Fig. 1A, middle). Using purified RNase III and YmdB to quantify the cellular levels of these proteins in vivo by Western blotting, we determined that the ratio of YmdB to RNase III that resulted in ∼90% inhibition of RNase III activity was 4:1 (Fig. 1A, right). IPTG-induced production of YmdB in vivo also resulted in the accumulation of 30S rRNA (Fig. 1B)—which is known to increase in bacteria deficient in RNase III activity (Babitzke et al. 1993). That these effects result specifically from inhibition of Rnc activity was confirmed using PL-putL-lac fusion transcripts, which are attacked at a site immediately 3′ of putL to generate an 80-nucleotide (nt) product (Sloan et al. 2007); turn on of the YmdB expression led to accumulation of the 140-nt unprocessed transcript and concurrent reduction of the 80-nt RNase III cleavage product (Fig. 1C).
Figure 1.
Down-regulation of RNase III activity by YmdB. (A) Cleavage of RNase III targeted sites in lacZ fusions. Strains P90C containing chromosomal rnc′-′lacZ (RS7305) or pnp′-′lacZ (RS8872) fusion transformed with plasmids pCA24N or ASKA-YmdB were grown at 37°C to OD600 ∼0.2 and transcription from the plasmid-borne promoter was induced by addition of 0.1 mM IPTG for 2 h. Total cellular proteins were prepared by resuspending E. coli cells with 1× denaturing protein gel loading dye at a final concentration of OD600 = 0.1 cells per 10-μL resuspension and boiled. The suspension (20 μL) was separated on Criterion 12% Bis-Tris gel with XT-MES buffer. (Left) The gel was stained by Coomassie blue. M represents the Precision Plus Protein Standards (Bio-Rad) with the molecular masses (in kilodaltons). (Middle) β-Galactosidase production from each lacZ fusion in the absence or presence of IPTG was measured. (Right) Purified His-RNase III and His-YmdB were run relative to cell lysates of RS7305 cells (OD600 = 0.2 cells from the middle panel) on Criterion 12% Bis-Tris with XT-MOPS buffer and immunoblotted against anti-RNase III, anti-YmdB, and anti-S1. The amount of protein (pmol) was calculated using the amount of protein quantified by Bradford assays divided by molecular weight of each protein. The bands (*) migrating at the His-RNase III band indicates the nonspecific band from Western blot analysis of cellular lysates using anti-RNase III. (B) Processing of 30S RNA. Cells (SDF204; rnc+) having plasmids pCA24N or ASKA-YmdB were grown. (Left) The expression of YmdB in each sample was detected as in A. (Right) Total RNAs (5 μg) were separated by electrophoresis on 1.5% agarose gels containing ethidium bromide (0.5 μg/mL) and visualized by AlphaImager (AlphaInnotech). (C) Processing of PL-putL-lac fusion. Cells having single copy of a PL-putL-lac fusion in RW4712 (rnc+) or RW4606 (Δrnc14∷Tn10) with plasmids pCA24N or ASKA-YmdB were used. (Left) The expression of YmdB from total cellular proteins obtained as in A was shown. Total RNAs (15 μg) were separated by 10% polyacrylamide gel (+7 M Urea) and putL mRNA was probed with 5′-[32P]- putL primer by Northern blotting. The unprocessed (140-nt) and processed (80-nt) RNA bands on the blot were shown. (D, top left) In vitro processing of R1.1 RNA. Structure of R1.1 RNA (60 nt) used in RNase III cleavage. The cleavage site was indicated by arrow. P and M represent precursor and mature R1.1 RNA, respectively. (Bottom left) 5′-[32P]-labeled RNA (∼5 nM) was incubated with YmdB only (lane 1), RNase III (50 nM) plus varying concentrations of YmdB (lanes 2–8), or RNA alone (lane 9) in 20 μL of 1× cleavage buffer (10 mM Tris-HCl at pH7.5, 12 mM NH4Cl, 0.4 mM MnCl2, 0.1 mM EDTA, 0.1 mM DTT) for 10 min at 37°C. An asterisk indicates secondary cleavage product by RNase III. (Right) The percent inhibition by YmdB of cleavage of R1.1 RNA was plotted to the concentration of YmdB. The cleavage inhibition (%) of each sample was determined by using the equation, [1 − (Mi/M0)] × 100 (%), where Mi, is the intact RNA determined in the presence of both YmdB and RNase III and M0, is the intact RNA determined in reactions lacking YmdB. In the absence of YmdB, 41.6% of M was observed.
Inhibition of RNase III cleavages by purified His-tagged YmdB protein was demonstrated biochemically in vitro and the inhibition was shown to be dependent quantitatively on YmdB using R1.1 RNA (60 nt in size), a small RNase III substrate containing a sequence that is the site of RNase III-mediated processing of the phage T7 polycistronic early mRNA precursor (Fig. 1D; Amarasinghe et al. 2001). The distinctly different YmdB/RNase III ratios found to affect RNase III activity in vivo and in vitro imply that experimental conditions employed for the in vitro assays do not entirely mimic the physiological state.
YmdB interaction with RNase III
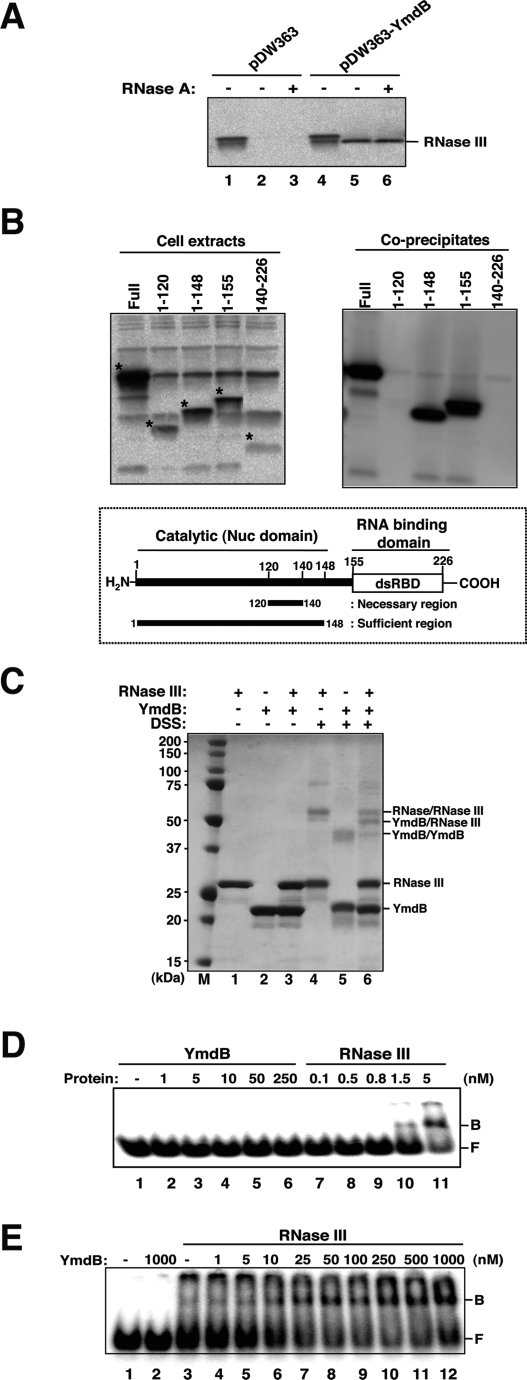
Notwithstanding the ability of YmdB to inhibit RNase III-mediated cleavage of R1.1 RNA, gel shift analysis failed to detect binding of YmdB to this substrate. Instead, multiple lines of evidence indicated that YmdB interacts with the RNase III protein. This interaction was suggested initially by our detection of an RNase III-sized ∼26 kDa protein, using rabbit polyclonal antibody generated against E. coli RNase III, in histidine-tagged YmdB preparations purified by Ni2+ column chromatography from rnc+—but not from rnc mutant—bacteria (data not shown). More direct evidence of the ability of YmdB and RNase III to interact was obtained in experiments showing that biotin-tagged YmdB preparations purified by a streptavidin bead-binding procedure (see Materials and Methods) also included an RNase III-size protein detected on Western blots by RNase III antibody (Fig. 2A). The observed copurification of this protein with biotin-tagged YmdB was unaffected by treatment of YmdB with ribonuclease A (+ vs. −), implying that interaction of this protein with RNase III does not require an RNA intermediate. Independently of these results, high-throughput MALDI-TOF screening (Butland et al. 2005) of protein complexes isolated from E. coli cells has identified YmdB in protein complexes containing RNase III (A. Emili, pers. comm.).
Figure 2.
YmdB and RNase III interactions. (A) Coprecipitation of RNase III with biotin-tagged YmdB. Streptavidin-conjugated agarose beads were added to extracts from XL1 cells containing plasmids pDW363 or pDW363-YmdB, and protein coprecipitated with beads was collected and analyzed by Western blotting. (Lanes 1,4) Total cell extracts. (Lanes 2,5) Eluates from streptavidin beads added to the cell extracts untreated with RNase A. (Lanes 3,6) Eluates from extracts pretreated with RNase A prior to bead addition. (B) Mapping the binding site of YmdB on RNase III. Total cellular extracts of NB478 (W3110, rnc <> cat) cells containing both RNase III segments (amino acids 120, 140, 148, 155, and 140–226) in plasmid pKSC1 and biotin-tagged YmdB in pDW363-YmdB were coprecipitated as in A. Total cellular extracts from RNase III segments (top left) and their coprecipitated proteins (top right) were analyzed by Western blotting with anti-RNase III. Asterisks represent the RNase III fragment bands. (Bottom) The schematic representation of a YmdB-binding site on RNase III is shown. (C) Cross-linking of RNase III and/or YmdB using DSS. Cross-linking reactions included RNase III and/or YmdB (5 μM each) and were performed at room temperature for 15 min in MOPS reaction buffer (10 mM MOPS at pH 7.5, 250 mM NaCl, 0.1 mM EDTA, and 0.1 mM DTT). Reaction products were separated by Criterion 10% Bis-Tris gel with XT-MOPS buffer after boiling the samples quenched by 2× SDS gel loading dye containing 0.5 M Tris base. Proteins were visualized by Coomassie blue staining. (Lanes 1–3) Protein incubated without DSS. (Lanes 4–6) Proteins incubated with 10 μg of DSS. (Lane M) Precision Plus protein standards with the molecular masses (in kilodaltons). (D) Binding of YmdB or RNase III to target RNA. The binding assays were performed using internally labeled R1.1 RNA with proteins that were preincubated on ice for 10 min at 37°C in the presence of 10 mM Ca2+, which enhances substrate binding but does not enable catalysis (Amarasinghe et al. 2001). F and B refer to free and bound R1.1 RNA, respectively. (Lane 1) RNA only. (Lanes 2–5) RNA with YmdB (1–50 nM). (Lanes 6–11) RNA with RNase III (0.1–5 nM). (E) Effects of YmdB on RNA-binding activity of RNase III. The binding reactions of RNase III–YmdB were carried out by increasing the amount of YmdB with or without RNase III. (Lane 1) RNA only. (Lane 2) RNA with YmdB (1000 nM). (Lanes 3–12) RNA with RNase III (1.5 nM) and YmdB (0–1000 nM). The band at the top of the gel has been routinely seen in previously published studies (Sun and Nicholson 2001; Calin-Jageman and Nicholson 2003) and as it is observed only in RNase III lanes, it appears to be aggregates of RNase III remaining in the well.
Physical interaction between YmdB and RNase III in vivo was further confirmed and the RNase III segment required for the interaction was identified by analysis, in cells mutated in the chromosomal rnc gene, of truncated RNase III proteins expressed from the pKSC1-Rnc plasmids and captured by YmdB attached to streptavidin-coated beads (Fig. 2B, top). The results of these experiments indicated that amino acids 1–148 of RNase III are sufficient for its binding to YmdB, and also that amino acids 120–140 are necessary for such binding (Fig. 2B, bottom). The required region overlaps a domain that has been reported, in the RNase III proteins of Aquifex aeolicus, Thermotoga maritima, and Mycobacterium tuberculosis, to be essential for enzymatic activity (Akey and Berger 2005; Gan et al. 2006). Analogous immunoblot experiments using full-length biotin-tagged YmdB and truncated YmdB derivatives showed interaction of RNase III with the full-length YmdB protein, but not with any of the truncated YmdB proteins tested (Supplemental Fig. S1A,B). Moreover, none of the truncated YmdB derivatives we tested regulated the activity of RNase III in vivo, as measured by the colorimetric assays described above (data not shown), implying that sequences located along most or all of the length of the YmdB protein are required for its function as an RNase III regulator. Interestingly, the macro-like domain of YmdB is congruent with virtually the full-length YmdB protein (Supplemental Fig. S1C).
Full catalytic activity of RNase III requires dimerization of the protein (Dunn 1982; Nicholson 2003; Meng and Nicholson 2008). We used the cross-linking agent DSS (Disuccinimidyl suberate; Pierce) (Li and Nicholson 1996) to experimentally test the notion that YmdB binding may interfere with RNase III dimerization. As shown in Figure 2C, RNase III bands migrating at the positions expected for RNase III monomers and dimers were observed in cross-linked reactions analyzed by SDS-PAGE (Fig. 2C, lanes 4,6), but in the absence of cross-linker, only a band migrating in the monomer position was detected (Fig. 2C, lanes 1,3). Addition of YmdB at the concentration tested (5 μM) resulted in a 33% reduction of the RNase III dimer band relative to monomer (Fig. 2C, lane 4 vs. lane 6) as shown by optical scanning of Coomassie blue stained gels and the appearance of a band that we found by MALDI-TOF analysis contains both RNase III and YmdB (Fig. 2C, lane 6). Reactions containing both the cross-linker and YmdB, but not reactions containing YmdB alone, showed a band migrating as an YmdB dimer, and the intensity of this band was decreased by 50% in the presence of 5 μM RNase III (Fig. 2C, lane 5 vs. lane 6). These results, which further confirm the ability of YmdB to interact with RNase III, strongly suggest that YmdB, as well as RNase III, exists in a dimeric form, and that interaction between the two proteins diminishes the ability of each to form homodimers.
We anticipated that the YmdB–RNase III interaction might be inhibiting RNase III activity by decreasing the ability of the endoribonuclease to interact with substrates. However, a test of the effects of YmdB on RNase III binding to R1.1 RNA in vitro showed surprisingly that the interaction increased RNase III binding to this substrate (Fig. 2E). This observation parallels the similar paradoxical finding that point mutations affecting amino acid residue 117, which is in the vicinity of the RNase III region we showed here to be required for its interaction with YmdB, enhanced the RNA-binding ability of RNase III, while rendering the enzyme catalytically inactive (Sun and Nicholson 2001). Taken together, these results raise the prospect that the monomeric form of RNase III, while being inactive catalytically (Dunn 1982; Nicholson 2003), may bind more tightly to substrates than RNase III dimers. Interestingly, neither the catalysis-nullifying effects of point mutations in RNase III amino acid 117 nor of YmdB was detectable in reactions containing Mg2+ (Sun and Nicholson 2001; our unpublished data), and consequently, the cation used in both the mutational studies and our YmdB investigations was Mn2+.
Effect of cell growth and cold shock on YmdB gene expression
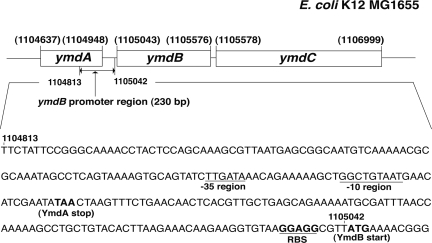
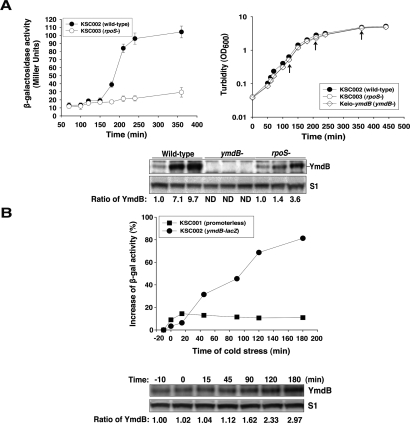
Our examination of primary microarray data obtained during studies that globally tested the effects of various physiological and environmental perturbations on E. coli mRNA abundance suggested that expression of the ymdB gene may be affected by cell physiology (Gutiérrez-Rios et al. 2003; Traxler et al. 2006; White-Ziegler et al. 2008). To further investigate this possibility, to quantify any cell growth-related changes in expression of ymdB, and to identify sites involved in the regulation of ymdB mRNA production, we made transcriptional fusions between ymdB and lacZ and inserted these into the E. coli chromosome as single-copy reporter gene constructs. Regions containing putative promoters in an operon postulated to contain the ymdA and ymdC genes (Karp et al. 2007), as well as ymdB, were identified using BPROM (Softberry, Inc.)—which detects consensus sequences for RNA polymerase σ70 recognition sites (Campbell et al. 2002). We observed that fusion to lacZ of a region containing 230 nt 5′ to the postulated ymdB protein start codon (Fig. 3) activated β-galactosidase production from the reporter gene. Moreover, lacZ expression from this construct began to increase in samples taken from bacteria in early stationary phase, and 2 h later reached a steady-state level approximately eightfold higher than was observed in logarithmically growing cells (Fig. 4A, top panels). Consistent with these findings, Western blot analysis showed that the intensity of a protein band that reacted with anti-YmdB rabbit polyclonal antibody (Fig. 4A, bottom) and that was not detected in ymdB mutant bacteria was ∼10-fold greater in stationary versus log-phase cells. An analogous increase in the abundance of the ymdB transcript during stationary phase was observed by microarray analysis (Gutiérrez-Rios et al. 2003). The observed increase in both ymdB mRNA and protein observed during stationary phase was dramatically reduced in rpoS mutant bacteria, which lack the stationary-phase σ-factor RpoS (Fig. 4A; Klauck et al. 2007), indicating its dependence on the production of RpoS. Consistent with this finding, ymdB was among the multiple genes showing altered mRNA abundance in rpoS mutant bacteria during microarray studies of global gene expression (Traxler et al. 2006). Also in agreement with earlier microarray results, which showed elevated abundance of ymdB mRNA during growth of E. coli cells at low temperature (White-Ziegler et al. 2008), we observed that shift of cultures to 13°C resulted in an approximately threefold increase in ymdB transcription, as quantified by β-galactosidase production from the ymdB-lacZ fusion construct, and YmdB protein by Western blotting (Fig. 4B).
Figure 3.
Schematic representation of ymdB gene region. DNA sequences of ymdB adjacent region extending from position 1,104,637 to position 1,106,999—numbered according to GenBank (U00096) of NCBI—including the ymdA (1,104,637–1,104,948), ymdB (1,105,043–1,105,567), and ymdC (1,105,578–1,106,999) coding regions and plausible ymdB promoter region (1,104,813–1,105,042) are shown. Promoter regions predicted by BPROM (Softberry, Inc.) are shown at the bottom of diagram and sequences of consensus sequences to σ70 (−35 and −10 regions) are underlined. “YmdA stop” and “YmdB start” represent stop codon of YmdA ORF and start codon of YmdB ORF, respectively. RBS represents putative ribosome-binding site.
Figure 4.
Transcription and translation of YmdB by stress conditions. (A) Production of β-galactosidase from ymdB-lacZ fusion. (Top left) Filled circles and empty circles represent β-galactosidase production from strains KSC002 (wild type) and KSC003 (rpoS−) harboring ymdB-lacZ transcriptional fusion, respectively. (Top right) Cell growth at 37°C was monitored by measuring turbidity (OD600) using spectrophotometer (Beckman-DU70). To measure the relative ratio of YmdB protein, total cellular proteins (OD600 = 0.2) extracted from KSC002, KSC003, and Keio-ymdB (ymdB−) cells at specific time points (indicated by arrows in the top right panel) were separated by Criterion 12% Bis-Tris gel with XT-MES buffer, immunoblotted using anti-YmdB and anti-S1, and quantified (bottom). ND indicates that band was not detected in the Western blotting. (B) ymdB transcription in cold stress. Briefly, cells were grown to OD600 ∼0.3 at 37°C, shifted to 13°C and continuously grown at 13°C, 250 rpm. The turbidity was almost unchanged (data not shown). (Top) Filled squares and empty circles represent the increase of β-gal (β-galactosidase) activity (%) by cold stress from strains containing chromosomal fusion of either promoterless-lacZ (KSC001) or ymdB-lacZ (KSC002). The value was determined by dividing the β-galactosidase activity at each time point by that of −10 min before the stress. (Bottom) YmdB and S1 proteins were immunoblotted from total cellular proteins(OD600 = 0.2) using antibodies of each protein and the ratio of YmdB protein during the time of cold stress was determined. Values shown are the relative intensities of YmdB to the value at −10 min of cold stress.
YmdB-mediated effects of stress on RNase III activity
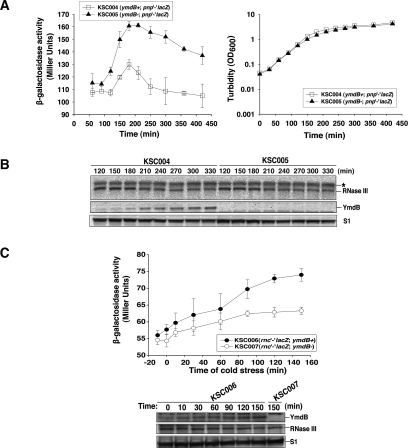
We hypothesized that inhibition of RNase III function by the observed elevation of YmdB production during stationary phase would lead to reduced cleavage of RNase III targeted transcripts encoded by a chromosomally inserted pnp′-′lacZ fusion transcript and a consequent stationary-phase elevation of β-galactosidase encoded by the fusion construct. As shown in Figure 5A, such an increase was observed. However, the increase was not sustained throughout the period of stationary-phase elevation of YmdB (Fig. 5A,B) and, importantly, also occurred in ymdB mutant bacteria—indicating its independence of YmdB. Similarly, no difference in the steady state level of the RNase III protein, which cleaves the transcripts that encode it (Matsunaga et al. 1996a, b, 1997), was detected during stationary-phase growth of ymdB+ versus ymdB mutant bacteria—notwithstanding the elevation of YmdB protein observed in these bacteria during stationary-phase cells (Fig. 5B). We conclude from these results that RNase III activity undergoes down-regulation during the stationary phase and that such down-regulation does not require the action of YmdB.
Figure 5.
Effect of ymdB knockout on gene expression in stress conditions. (A) Production of β-Gal from pnp′-′lacZ fusion in stationary phase. (Left) Empty squares and filled triangles represent β-galactosidase production from strains KSC004 (wild type) and KSC005 (Keio-ymdB) harboring pnp′-′lacZ translational fusion in the chromosome, respectively. (Right) Cell growth at 37°C was monitored by measuring turbidity (OD600) using spectrophotometer (Beckman-DU70). (B) Cellular levels of RNase III and YmdB proteins in stationary phase. Total proteins (OD600 = 0.2) from KSC004 (empty squares) and KSC005 (filled triangles) indicated time points were separated by Criterion 12% Bis-Tris gel with XT-MOPS buffer and immunoblotted with anti-RNase III, anti-YmdB, and anti-S1 protein. An asterisk indicates the nonspecific signal by immunoblotting. (C) Effect of ymdB knockout on RNase III activity in cold stress. E. coli cells grown to OD600 ∼0.3 in LB at 37°C were shifted to 13°C and continuously grown at 13°C, 250 rpm. Samples were taken by indicated time points. The turbidity was not changed by the cold stress (data not shown). (Top) Filled circles and empty circles represent β-galactosidase production from KSC006 (wild type) and KSC007 (Keio-ymdB) harboring rnc′-′lacZ translational fusion in the chromosome, respectively. (Bottom) YmdB, RNase III, and S1 proteins by cold stress were detected by immunoblotting.
We observed that RNase III activity was decreased also during cold shock, as indicated by increased production of β-galactosidase activity encoded by a rnc′-′lacZ fusion gene inserted into the E. coli chromosome. However, in contrast to the YmdB independence of stationary-phase effects on RNase III activity, down-regulation of RNase III activity during cold shock was reduced in ymdB mutant bacteria (Fig. 5C), demonstrating a role for ymdB in the regulation of RNase III during cold stress. While YmdB abundance was approximately threefold higher during stationary phase versus cold stress, the β-galactosidase activity encoded by the rnc′-′lacZ fusion was approximately the same under both conditions (Figs. 4, 5), implying that the inverse relationship between YmdB and RNase III is not linear.
Discussion
While it has long been known that the cellular actions of ribonucleases are affected by physiological conditions (Nilsson et al. 1984; Georgellis et al. 1993; Chen and Deutscher 2005; Freire et al. 2006), there has been limited understanding of the mechanisms that underlie ribonuclease regulation. Proteins that alter ribonuclease activity by interacting with substrates have been identified, and in the case of some RNase III family members, viral proteins have been shown to regulate RNA-mediated gene silencing by competing with the enzyme’s ability to bind to targets (Chao et al. 2005). Our discovery of an E. coli protein that regulates RNase III activity by interacting directly with this ribonuclease reveals a previously unsuspected mechanism for dynamic control of RNase III actions. The effects of YmdB extend beyond E. coli RNase III, as this E. coli protein inhibited the cleavages by the RNase III (AbsB protein) (Price et al. 1999; Xu et al. 2008) of the unrelated bacterial species, Streptomyces coelicolor (Supplemental Fig. S2C). YmdB homologs are present in multiple bacterial species and the actions of such YmdB homologs are likely to be specific for RNase III family members, as no inhibition of endoribonucleases RNases E or G was observed on the E. coli YmdB protein (data not shown). Conversely, recently described enzyme-binding inhibitors of E. coli RNase E (Lee et al. 2003; Gao et al. 2006; Zhao et al. 2006) did not detectably affect the actions of RNase III (our unpublished data). While the observed effect of YmdB also modulated PNPase expression (Fig. 5A), this effect was shown to be dependent entirely on YmdB inhibition of RNase III (Supplemental Fig. S3), which cleaves PNPase transcripts. YmdB was isolated using rnc′-′lacZ chromosomal fusion developed previously by Simons and colleagues (Matsunaga et al. 1996a, b, 1997) to study mutations in the rnc gene. We adapted this fusion for detection of trans-acting gene products that increased the expression of β-galactosidase encoded by a chromosomally inserted rnc′-′lacZ fusion construct. As the rnc segment of the rnc′-′lacZ transcript is cleaved by RNase III, down-regulation of RNase III resulted in elevation of β-galactosidase expression. The ASKA library of plasmid-borne ORFs that we screened expresses individual gene products at high levels under control of a lac promoter inducible by IPTG (Kitagawa et al. 2005).
The ability of YmdB to down-regulate RNase III activity has been shown by us both in vivo and in vitro. In our in vitro experiments (Fig. 1D), we observed that RNase III treatment of 5′-[32P]-R1.1 RNA, a well-characterized substrate shown in previous experiments to be cleaved at a single site within the descending arm of a hairpin structure (Amarasinghe et al. 2001), yielded a doublet band (the “M” band) when purified YmdB was added. YmdB alone showed no effect on the substrate. A similar band suggestive of a secondary cleavage site at nucleotide position U45 was observed also following treatment of R1.1 RNA by a mutant RNase III enzyme (the E65 mutation, Zhang et al. 2004). We speculate that both the E65 mutation and YmdB binding to RNase III may perturb the cleavage specificity of the endoribonuclease, leading to the targeting of R1.1 RNA at secondary cleavage site.
Our studies showed that RNase III segments in N-terminal catalytic regions containing amino acids 120–140 are necessary and amino acids 1–148 are sufficient for YmdB binding. It has been known that RNase III functions as a homodimer and contains two dsRBDs and two catalytic sites (Dunn 1982; Nicholson 2003). It also has been reported that the N-terminal portion of bacterial RNase III polypeptides includes the NucD (catalytic domain), which self-associates to form a single “processing center” at the subunit interface that contains the catalytic sites (Zhang et al. 2004). We found that like RNase III, YmdB can also form homodimers, and that interaction between these two proteins decreased homodimer formation of each. We suggest that RNase III–YmdB heterodimer formation may disrupt formation of the RNase III processing center. However, while YmdB can clearly inhibit the endoribonucleolytic activity of RNase III, interaction of the functionally inhibited RNase III with the RNA substrate was observed to be increased. Such uncoupling of binding and catalytic activity is reminiscent of the effects of mutations at amino acid residue 117, which also increased binding to R1.1 RNA while rendering the RNase III protein catalytically inactive (Sun and Nicholson 2001).
Inhibition of RNase III activity by YmdB was observed in vitro when Mn2+ was the cation cofactor, but not when Mg2+ was the cation. Interestingly, the RNase III E117 mutation identified by Sun and Nicholson (2001) was also found to abrogate endoribonuclease activity in vitro when tested using Mn2+, but not with Mg2+. Position 117, which Sun and Nicholson (2001) included in a region they designated as the “inhibitory” site of the enzyme, is also within the RNase III segment we showed to be sufficient for binding to YmdB. Collectively, these findings raise the possibility that Mn2+ binding at or near position 117 enables YmdB interaction with RNase III, and consequent inhibition of enzymatic activity.
We hypothesize that the formation of YmdB/RNase III heterodimers may inhibit RNase III activity by interfering with the formation of RNase III homodimers, which is believed to be the active form of the enzyme (Meng and Nicholson 2008). Production of YmdB was increased during stationary phase and cold shock, and our data indicate that this increase is mediated at the transcriptional level. Moreover, increased ymdB transcription during stationary phase was dependent on the stationary-phase σ factor RpoS encoded by the rpoS gene. Conversely, overexpression of YmdB during log-phase growth activated production of RpoS protein (our unpublished data), consistent with evidence that RpoS is elevated in E. coli cells mutated in RNase III, which has been shown to cleave the rpoS mRNA leader (Resch et al. 2008). As we showed that YmdB is an inhibitor of RNase III, these findings suggest an autoregulatory relationship between the ymdB and rpoS gene products.
We also screened genomic DNA libraries we constructed to express E. coli genes under control of their natural promoters. In these screenings, five additional trans-acting regulators of β-galactosidase expression from the rnc′-′lacZ fusion construct were identified (our unpublished data), suggesting that RNase III activity is controlled by other trans-acting gene products in addition to YmdB. Our experimental results (Fig. 5) showing that RNase III can be down-regulated during stationary phase by an YmdB-independent mechanism is consistent with this notion. Moreover, the actions of some of the putative regulators we identified in our screen were observed to be affected by the composition of the media on which cells were assayed—raising the further prospect that regulation by these agents is modulated by cell growth conditions. Additional experimentation is being carried out to elucidate whether cellular regulation of RNase III activity in E. coli is, in fact, controlled by a battery of trans-acting regulators that collectively mediate a dynamic response to physiological and environmental factors.
The effects of YmdB on RNase III activity in vivo also raise the question of how such an inhibitor can affect an autoregulated system as the autoregulation might be expected to compensate for the inhibitor’s actions. Similarly, how can a missense mutation increase RNase III abundance, as has been reported previously (Matsunaga et al. 1997; Wilson et al. 2002), instead of simply leading to compensation that maintains the enzyme activity at a “normal level”? Analogous findings, both for mutations and inhibitors, have been reported for another autoregulated ribonuclease, RNase E (Jain and Belasco 1995; Sousa et al. 2001; Ow et al. 2002; Lee et al. 2003) suggesting that the autoregulation of both ribonucleases is imperfect.
As observed for RNase III itself, ymdB-like genes are widely distributed in nature, and GenBank analysis by us has revealed ymdB homologs in both eukaryotes and prokaryotes. However, the structure of the operon containing YmdB (i.e., ymdA–ymdB–ymdC) is highly conserved in only Salmonella and Shigella—both close relatives of E. coli (Karp et al. 2007). As noted earlier, the YmdB protein (Swiss-Prot P0A8D6) includes a macro (A1pp) domain, a module of ∼180 amino acids that can bind ADP-ribose, an NAD metabolite or related ligands (Schreiber et al. 2006). The macro domain motif is present in both multidomain proteins and simpler proteins—where it appears to be the primary structural component (Amé et al. 2004). Given the inhibition by YmdB of endoribonucleolytic cleavages by a Streptomyces coelicolor homolog of E. coli RNase III (AbsB) (Price et al. 1999), it is worth noting that S. coelicolor encodes a 169 amino acid “stand alone” macro domain homolog of YmdB (i.e., SC6450).
Materials and methods
Medium, bacterial strains, plasmids, and phage vectors
All bacterial strains, plasmids, and phages used during this study listed in Supplemental Table S1. Bacterial strains were routinely grown in Luria-Bertani (LB) medium (Sambrook and Russell 2001). The concentrations (per milliliter) of antibiotics used were 100 μg of ampicillin (ApR), 30 μg of chloramphenicol (CmR), 50 μg of spectinomycin (SpR), and 30 μg of tetracyclin (TcR). Plasmid pKSC1 was prepared as follows. CloDF13 origin and SpR marker region was obtained from pCDuet-1 (Stratagene) cut by HpaI and EcoRV. The fragment is ligated to fragment of pBAD-myc-HisB (Invitrogen), in which both replication of origin and ApR marker are eliminated by BsaAI and ScaI.
Library screening
The ASKA library (Kitagawa et al. 2005) was transformed into RS7305 (Matsunaga et al. 1996a) and colonies (∼20,000) were separately plated onto either LB/S-Gal (50 μg/mL; Sigma-Aldrich) with 0.1 mM IPTG or MacConkey-Lac plate. After overnight incubation of the plate at 37°C, dark-black or deep-red colonies were picked. The plasmid DNAs were obtained from each colony and transformed with fresh RS7305 cells. Screenings were repeated and positive colonies were selected. As the final step, each colony was streaked onto both LB/S-Gal with 0.1 mM IPTG and MacConkey-Lac plate. Plasmid DNAs from colonies showing positive results on both plates were prepared and sequenced.
Construction of chromosomal lacZ fusions
We PCR-amplified a DNA fragment containing regions of ymdA operon—from 1104,813 to 1105,042 of E. coli K12 genome (GenBank U00096)—and cloned a fragment to lacZ transcriptional fusion vector, pSP417 (Podkovyrov and Larson 1995), generating a plasmid pKSC2 (ymdB-lacZ). The lacZ fusions in pSP417 (negative control) and pKSC2 were transferred onto the chromosome using the transducing λ phage system (Simons et al. 1987). The ymdB-lacZ fusion from pKSC2 was transferred into λRS45, whereas the control pSP417 was transferred into λRS74 via a double recombination event. We also used either λRS741 (pnp′-′lacZ) or λRS473 (rnc′-′lacZ) from RS8872 (Beran and Simons 2001) or RS7305 (Matsunaga et al. 1996a) lysogen, respectively, to transfer fusion into BW25113 strain. Plaques containing the recombinant λ phages were isolated based on their blue plaque phenotype. The recombinant λ phages were used to lysogenize BW25113, Keio-rpoS, or Keio-ymdB and generated the following strains: KSC001 (BW25113; λKSC1) with a chromosomal promoterless lacZ gene; KSC002 (BW25113; λKSC2), KSC004 (Keio-rpoS; λKSC2); KSC005 (BW25113; λRS741), KSC006 (Keio-ymdB; λRS468), and KSC007 (Keio-ymdB; λRS468). Single lysogens are verified by PCR-based method (Powell et al. 1994).
Measurement of β-galactosidase activity
β-Galactosidase activity in whole cells was determined as described by Miller (1992). Values represent the means of at least three independent experiments.
RNA extraction and Northern blotting
In general, total RNAs were extracted using the hot phenol method as described previously (Sambrook and Russell 2001), followed by RNeasy kit cleanup (Qiagen). For 30S RNA precursor detection, 5 μg of total RNA isolated from wild-type cells of uninduced or induced YmdB protein were run on 1.5% nondenaturing agarose gel containing ethidium bromide (0.5 μg/mL) and visualized by AlphaImager (AlphaInnotech). For Northern blot analyses, 15 μg of total RNAs were fractionated on a 10% polyacrylamide gel containing 7 M Urea, electrotransferred onto Hybond-N+ membrane (AP Biotech), and cross-linked the membrane using UV-cross-linker at 1250 J/cm2 (Stratagene). Primer putL (5′-CGTTCATCCTGAACCCGCCGCGC-3′; early part of the HK022 pL transcript; Sloan et al. 2007) was radiolabeled using [γ-32P]ATP and T4 Polynucleotide Kinase (New England Biolabs) and used as a probe to detect put-L-mRNA in the total RNA pool. Hybridization, washing of the membranes, and detection of signals were carried out according the RapidHyb buffer (Promega) protocol.
Protein purifications
His-tagged RNase III was purified and dialyzed from BL21(DE3) rnc105 containing pET15b-rnc as described (Amarasinghe et al. 2001) and His-tagged YmdB was purified from E. coli SDF205 (rnc105 mutant) cells having ASKA-YmdB as follows: The cultures were grown to mid-log phase, induced with 0.5 mM IPTG for 3 h and harvested. The cells were resuspended in buffer A [20 mM Tris-HCl at pH 8.0, 0.5 M NaCl, 10 mM imidazole] containing lysozyme (5 mg/mL), DNase I (10 mg/mL), Complete EDTA-free Protease Inhibitor Cocktail Tablets (Roche Molecular Biochemicals)]. After cell disruption by passing through French Press, the cell suspension was centrifuged at 30,000g, 30 min, and the crude extract was obtained. The crude extract was loaded onto Ni2+ column packed with 5 mL of Ni-Sepharose 6 fast flow resin (GE Healthcare) and equilibrated with buffer A. Unbound proteins are washed with 100 mL of 20 mM imidazole in buffer A and the protein was eluted with 15 mL of elution buffers containing 20, 50, 100, 200, or 400 mM imidazole in buffer A, respectively, with the gravity flow rate of 0.2 mL/min. YmdB protein was eluted from 50 mM imidazole fraction. Pools eluted from at 200 mM imidazole buffer were concentrated using Centriplus-YM10 (Millipore) and dialyzed against buffer B (30 mM Tris-HCl at pH 8.0, 50 mM NaCl, 1 mM EDTA, 1 mM DTT). The YmdB protein concentration was determined by Bradford assay kit (Bio-Rad) and purity was identified by SDS-PAGE gel with Coomassie blue staining. The absence of RNase III was determined by Western blotting using antibodies to RNase III.
In vitro cleavage assays
Generally, RNase III cleavage assays were performed according to a protocol (Amarasinghe et al. 2001), using 5′-[32P] R1.1 RNA in the cleavage buffer (10 mM Tris-HCl at pH 7.5, 12 mM NH4Cl, 0.1 mM EDTA, 0.1 mM DTT, 0.25 mg/mL yeast tRNA) containing 20 U of rRNasin (Promega). Specific features of the assays are detailed in the appropriate figure. Reactions were initiated by adding Mn2+ at final concentration of 0.4 mM to the protein/RNA mixtures. All reactions were carried out for 10 min at 37°C. Reactions were quenched by adding same volume of 2× loading dye that contained 20 mM EDTA (Amarasinghe et al. 2001). Aliquots were electrophoresed in 10% polyacrylamide gel containing 7 M Urea and 1× TBE buffer. The results were visualized by phosphorimaging (Typhoon Trio) and quantified by ImageQuant software. Reactions were performed in at least three independent experiments and representative data are reported.
Gel shift assays
Gel mobility-shift assays were carried out as described in previous publications (Li and Nicholson 1996; Amarasinghe et al. 2001; Sun and Nicholson 2001; Meng and Nicholson 2008), using [α-32P] UTP-labeled R1.1 RNA and proteins that were preincubated on ice for 10 min at 37°C in Ca2+ (10 mM) buffer. Electrophoresis was carried out at 4°C in a 6% (w/v) polyacrylamide gel (Acrylamide:Bisacrylamide, 80:1) containing 0.5× TBE buffer and 10 mM CaCl2. Binding reactions were visualized by phosphorimaging and quantified using ImageQuant software.
Immunodetection of YmdB, RNase III, and S1 proteins
Polyclonal YmdB antibodies were obtained from rabbits inoculated with YmdB peptide (N′-CYDEE NAHLY ERLLT QQGDE) from Proteintech Group, Inc., and were affinity-purified. RNase III antibodies raised from rabbits were obtained from Drs. R.W. Simon and K. Lee as a gift. We used S1 antibodies (Feng et al. 2001) to provide an internal standard to evaluate the amount of cell extracts in different lanes. Cellular proteins were detected by separating proteins on Criterion XT Bis-Tris gels with appropriate percentage and buffer, followed by immunoblot by antibodies of interesting proteins, anti-rabbit-IgG:HRP conjugate (Promega) and ECL detection reagents (Millipore). Specific proteins were imaged by VersaDoc 1000 (Bio-Rad) and quantified by Quantity One (Bio-Rad).
Detection of protein interactions in vivo
Full-length YmdB was cloned into XhoI and BamHI site of pDW363 vector (Tsao et al. 1996). Then, E. coli XL1 blue cells were transformed with pDW363 or pDW363-YmdB, and the resulting cells were cultured in LB media containing 8 μg/mL of D-biotin (Sigma-Aldrich) at 37°C. Expression of biotin-tagged YmdB was initiated by 1 mM of IPTG when the culture OD600 reached ∼0.4 and induced for 3 h. Cells were harvested by centrifugation at 5000 rpm for 10 min and resuspended in 1/5 volume of ice-cold NP-40 lysis buffer (150 mM sodium chloride, 1.0% NP-40, 50 mM Tris at pH 8.0). The cell lysate was obtained after passing the cells through French press (2000 psi) two times, and followed by centrifugation for 15 min at 4°C to remove intact cells and cell debris. The protein concentration was determined by BCA method (BCA Protein Assay Kit, Pierce). Aliquots (5 mg of protein) were further treated with RNase A (at a final concentration of 5 mg/mL) at 37°C for 15 min. Aliquots with or without treatment with RNase A were mixed with each 100 μL of streptavidin-agarose slurry (Invitrogen) and the volume was adjusted to 800 μL. Following 1 h rotation at 4°C, resins were centrifuged at 6000 rpm for 5 min and then washed extensively 10 times with ice-cold NP-40 buffer. Finally, the streptavidin agarose resins were resuspended with 100 μL of 2× SDS protein loading buffer (100 mM Tris-Cl at pH 6.8, 4% SDS, 200 mM β-mercaptoethanol) and boiled for 10 min. The released proteins were electrophoresed on 12% Criterion Bis-Tris gel with XT-MES buffer (Bio-Rad) and the presence of RNase III was analyzed by Western blotting using anti-RNase III.
Identification of RNase III region for YmdB interaction
Specific regions of the RNase III molecules were cloned into NcoI and HindIII site of plasmid pKSC1. E. coli NB478 (W3110, rnc <> cat) cells were transformed with both pKSC1-RNase III deletion constructs (amino acids 1–120, 1–140, 1–155, and 140–226 of Rnc) and pDW363-YmdB. Cells were cultured in LB media containing 8 μg/mL of D-biotin at 37°C. Proteins were induced by adding both 0.1% L-arabinose and 1 mM IPTG to exponentially grown cells, followed by further growth for 2 h Coprecipitation analyses were done as described in Detection of Protein Interactions in Vivo. The presence of RNase III was detected by Western blotting using RNase III antibody.
Protein cross-linking and mass mapping
To monitor the change of dimerization status of RNase III and YmdB, we used a cross-linking reaction that has been described for E. coli RNase III (Li and Nicholson 1996) using DSS, an amine cross-linking agent. Briefly, cross-linking reactions (20-μL volumes) including RNase III (5 μM monomer) and/or YmdB (5 μM monomer) were carried out for 15 min at room temperature in 10 mM MOPS (pH 7.5), containing 250 mM NaCl, 0.1 mM EDTA, and 0.1 mM DTT. Reactions were quenched by boiling after adding 20 μL of 2× SDS-gel loading buffer supplemented with 0.5 M Tris base, then analyzed by electrophoresis in 12% Criterion XT Bis-Tris gel with XT-MOPS buffer (Bio-Rad), and visualized by Coomassie blue staining. The presence of RNase III and YmdB from heterodimer band was confirmed by mass mapping analysis (PAN Facility, Stanford University). The intensity of bands on the gel was quantified by optical scanning of Coomassie stained gel, followed by analysis using the ImageQuant program.
Acknowledgments
We thank Drs. R.W. Simons, D.L. Court, R.A. Weisberg, and T.J. Larson for the gift of strains; Drs. R.W. Simons and K. Lee for polyclonal antibody to RNase III; and Dr. A. Emili for the unpublished data of protein interactions. We also thank Dr. W. Xu for providing purified AbsB protein, Dr. H. Mori from the Japanese E. coli consortium for the generous gift of the ASKA plasmid ORF library and Keio collections, and Roberta Peterson for assistance in preparing manuscript. The research was supported by NIH NIGMS Grant GM 54158 to S.N.C.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1729508.
References
- Akey D.L., Berger J.M. Structure of the nuclease domain of ribonuclease III from M. tuberculosis at 2.1 Å. Protein Sci. 2005;14:2744–2750. doi: 10.1110/ps.051665905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe A.K., Calin-Jageman I., Harmouch A., Sun W., Nicholson A.W. Escherichia coli ribonuclease III: Affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- Amé J.C., Spenlehauer C., de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Granger L., Olszewski J., Kushner S.R. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J. Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beran R.K., Simons R.W. Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 2001;39:112–125. doi: 10.1046/j.1365-2958.2001.02216.x. [DOI] [PubMed] [Google Scholar]
- Butland G., Peregírn-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Calin-Jageman I., Nicholson A.W. RNA structure-dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 2003;31:2381–2392. doi: 10.1093/nar/gkg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O., Trester-Zedlitz M.L., Darst S.A. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Chao J.A., Lee J.H., Chapados B.R., Debler E.W., Schneemann A., Williamson J.R. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chen C., Deutscher M.P. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- Court D.L. RNA processing and degradation by RNase III. In: Belasco J., Brawerman G., editors. Control of messenger RNA stability. Academic; New York: 1993. pp. 71–116. [Google Scholar]
- Culver G.M., Consaul S.A., Tycowski K.T., Filipowicz W., Phizicky E.M. tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1″, 2″-cyclic phosphate. J. Biol. Chem. 1994;269:24928–24934. [PubMed] [Google Scholar]
- Drider D., Condon C. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 2004;8:195–200. doi: 10.1159/000086700. [DOI] [PubMed] [Google Scholar]
- Dunn J.J. Ribonuclease III. In: Boyer P., editor. The Enzymes. Academic; New York: 1982. pp. 485–499. [Google Scholar]
- Feng Y., Huang H., Liao J., Cohen S.N. Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 2001;276:31651–31656. doi: 10.1074/jbc.M102855200. [DOI] [PubMed] [Google Scholar]
- Freire P., Amaral J.D., Santos J.M., Arraiano C.M. Adaptation to carbon starvation: RNase III ensures normal expression levels of bolA1p mRNA and σS. Biochimie. 2006;88:341–346. doi: 10.1016/j.biochi.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Gan J., Tropea J.E., Austion B.P., Court D.L., Waugh D.S., Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Gao J., Lee K., Zhao M., Qiu J., Saxena A., Moore C.J., Cohen S.N., Georgiou G. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol. Microbiol. 2006;61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- Georgellis D., Barlow T., Arvidson S., von Gabain A. Retarded RNA turnover in Escherichia coli: A means of maintaining gene expression during anaerobiosis. Mol. Microbiol. 1993;9:375–381. doi: 10.1111/j.1365-2958.1993.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Rios R.M., Rosenblueth D.A., Loza J.A., Huerta A.M., Glasner J.D., Blattner F.R., Collado-Vides J. Regulatory network of Escherichia coli: Consistency between literature knowledge and microarray profiles. Genome Res. 2003;13:2435–2443. doi: 10.1101/gr.1387003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C., Belasco J.G. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: Unusual sensitivity of the rne transcript to RNase E activity. Genes & Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- Karp P.D., Keseler I.M., Shearer A., Latendresse M., Krummenacker M., Paley S.M., Paulsen I., Collado-Vides J., Gama-Castro S., Peralta-Gil M., et al. Multidimensional annotation of the Escherichia coli K12 genome. Nucleic Acids Res. 2007;35:7577–7590. doi: 10.1093/nar/gkm740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Klauck E., Typas A., Hengge R. The σS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci. Prog. 2007;90:103–127. doi: 10.3184/003685007X215922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Zhan X., Gao J., Qiu J., Feng Y., Meganathan R., Cohen S.N., Georgiou G. RraA, a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- Li H., Nicholson A.W. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996;15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- MacRae I.J., Doudna J.A. Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Makarov E.M., Apirion D. 10Sa RNA: Processing by and inhibition of RNase III. Biochem. Int. 1992;26:1115–1124. [PubMed] [Google Scholar]
- Matsunaga J., Dyer M., Simons E.L., Simons R.W. Expression and regulation of the rnc and pdxJ operons of E. coli. Mol. Microbiol. 1996a;22:977–989. doi: 10.1046/j.1365-2958.1996.01529.x. [DOI] [PubMed] [Google Scholar]
- Matsunaga J., Simons E.L., Simons R.W. RNase III autoregulation: Structure and function of rncO, the posttranscriptional ‘operator.’. RNA. 1996b;2:1228–1240. [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J., Simons E.L., Simons R.W. Escherichia coli RNase III (rnc) autoregulation occurs independently of rnc gene translation. Mol. Microbiol. 1997;26:1125–1135. doi: 10.1046/j.1365-2958.1997.6652007.x. [DOI] [PubMed] [Google Scholar]
- Meng W., Nicholson A.W. Heterodimer-based analysis of subunit and domain contributions to double-stranded RNA processing by Escherichia coli RNase III in vitro. Biochem. J. 2008;410:39–48. doi: 10.1042/BJ20071047. [DOI] [PubMed] [Google Scholar]
- Miller J.H. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- Murchison E.P., Hannon G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Nashimoto H., Uchida H. DNA sequencing of the Escherichia coli ribonuclease III gene and its mutations. Mol. Gen. Genet. 1985;201:25–29. doi: 10.1007/BF00397981. [DOI] [PubMed] [Google Scholar]
- Nicholson A.W. Structure, reactivity, and biology of double-stranded RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;52:1–65. doi: 10.1016/s0079-6603(08)60963-0. [DOI] [PubMed] [Google Scholar]
- Nicholson A.W. The ribonuclease III superfamily: Forms and functions in RNA maturation, decay, and gene silencing. In: Hannon G., editor. RNAi: A guide to gene silencing. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. pp. 149–174. [Google Scholar]
- Nilsson G., Belasco J.G., Cohen S.N., von Gabain A. Growth-rate dependent regulation of mRNA stability in Escherichia coli. Nature. 1984;312:75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- Ow M.C., Liu Q., Mohanty B.K., Andrew M.E., Maples V.F., Kushner S.R. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 2002;43:159–171. doi: 10.1046/j.1365-2958.2002.02726.x. [DOI] [PubMed] [Google Scholar]
- Podkovyrov S.M., Larson T.J. A new vector-host system for construction of lacZ transcriptional fusions where only low-level gene expression is desirable. Gene. 1995;156:151–152. doi: 10.1016/0378-1119(95)00053-9. [DOI] [PubMed] [Google Scholar]
- Powell B.S., Court D.L., Nakamura Y., Rivas M.P., Turnbough C.L. Rapid confirmation of single copy λ prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price B., Adamidis T., Kong R., Champness W. A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J. Bacteriol. 1999;181:6142–6151. doi: 10.1128/jb.181.19.6142-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M. Cleavage by RNase III in the transcripts of the metY–nusA–infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol. 1989;210:293–302. doi: 10.1016/0022-2836(89)90331-8. [DOI] [PubMed] [Google Scholar]
- Régnier P., Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- Resch A., Afonyushkin T., Lombo T.B., McDowall K.J., Bläsi U., Kaberdin V.R. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA. 2008;14:454–459. doi: 10.1261/rna.603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Sundermeier T., Svetlanov A., Karzai A.W. Quality control of bacterial mRNA decoding and decay. Biochim. Biophys. Acta. 2008;1779:574–582. doi: 10.1016/j.bbagrm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schreiber V., Dantzer F., Amé J.C., de Murcia G. Poly(ADP-ribose): Novel function for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Simons R.W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sloan S., Rutkai E., King R.A., Velikodvorskaya T., Weisberg R.A. Protection of antiterminator RNA by the transcript elongation complex. Mol. Microbiol. 2007;63:1197–1208. doi: 10.1111/j.1365-2958.2006.05579.x. [DOI] [PubMed] [Google Scholar]
- Sousa S., Marchand I., Dreyfus M. Autoregulation allows Escherichia coli RNase E to adjust continuously its synthesis to that of its substrates. Mol. Microbiol. 2001;42:867–878. doi: 10.1046/j.1365-2958.2001.02687.x. [DOI] [PubMed] [Google Scholar]
- Sun W., Nicholson A.W. Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant. Biochemistry. 2001;40:5102–5110. doi: 10.1021/bi010022d. [DOI] [PubMed] [Google Scholar]
- Talkad V., Achord D., Kennell D. Altered mRNA metabolism in ribonuclease III-deficient strains of Escherichia coli. J. Bacteriol. 1978;135:528–541. doi: 10.1128/jb.135.2.528-541.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler M.F., Chang D., Conway T. Guanosine 5′,3′-bipyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. 2006;103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao K.L., DeBarbieri B., Michel H., Waugh D.S. A versatile plasmid expression vector for the production of biotinylated proteins by site-specific, enzymatic modification in Escherichia coli. Gene. 1996;169:59–64. doi: 10.1016/0378-1119(95)00762-8. [DOI] [PubMed] [Google Scholar]
- White-Ziegler C.A., Um S., Perez N.M., Berns A.L., Malhowski A.J., Young S. Low temperature (23°C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology. 2008;154:148–166. doi: 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- Wilson H.R., Yu D., Peters H.K., Zhou J.G., Court D.L. The global regulator RNase III modulates translation repression by the transcription elongation factor N. EMBO J. 2002;21:4154–4161. doi: 10.1093/emboj/cdf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Huang J., Cohen S.N. Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endonucleoribotic cleavage of absB operon transcripts. J. Bacteriol. 2008;190:5526–5530. doi: 10.1128/JB.00558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Flipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zhao M., Zhou L., Kawarasaki Y., Georgiou G. Regulation of RraA, a protein inhibitor of RNase E-mediated RNA decay. J. Bacteriol. 2006;188:3257–3263. doi: 10.1128/JB.188.9.3257-3263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]