Abstract
Division site selection in rod-shaped bacteria depends on nucleoid occlusion, which prevents division over the chromosome and MinCD, which prevent division at the poles. MinD is thought to localize MinC to the cell poles where it prevents FtsZ assembly. Time-lapse microscopy demonstrates that in Bacillus subtilis transient polar FtsZ rings assemble adjacent to recently completed septa and that in minCD strains these persist and are used for division, producing a minicell. This suggests that MinC acts when division proteins are released from newly completed septa to prevent their immediate reassembly at new cell poles. The minCD mutant appears to uncouple FtsZ ring assembly from cell division and thus shows a variable interdivisional time and a rapid loss of cell cycle synchrony. Functional MinC-GFP expressed from the chromosome minCD locus is dynamic. It is recruited to active division sites before septal biogenesis, rotates around the septum, and moves away from completed septa. Thus high concentrations of MinC are found primarily at the septum and, more transiently, at the new cell pole. DivIVA and MinD recruit MinC to division sites, rather than mediating the stable polar localization previously thought to restrict MinC activity to the pole. Together, our results suggest that B. subtilis MinC does not inhibit FtsZ assembly at the cell poles, but rather prevents polar FtsZ rings adjacent to new cell poles from supporting cell division.
Keywords: FtsZ, cell biology, cell cycle, cell division, minCD
Division site selection in rod-shaped bacteria is a highly regulated and reliable process achieved by two systems that restrict FtsZ assembly (and the subsequent assembly of a complete divisome) to mid-cell. One system, nucleoid occlusion (Noc), prevents FtsZ from assembling over the chromosome while the other, MinCD, prevents FtsZ assembly and division at the cell pole (Fig. 1A; for reviews, see Errington et al. 2003; Margolin 2005; Rothfield et al. 2005; Barak and Wilkinson 2007). Although unrelated DNA-binding proteins mediate Noc in Bacillus subtilis and Escherichia coli (Wu and Errington 2004; Bernhardt and de Boer 2005), the MinCD proteins are broadly conserved and encoded by many bacterial genomes (Rothfield et al. 2005). The simultaneous inactivation of the Noc and MinCD systems is lethal in both species, resulting in the assembly of multiple FtsZ structures that fail to coalesce into functional rings (Wu and Errington 2004; Bernhardt and de Boer 2005).
Figure 1.
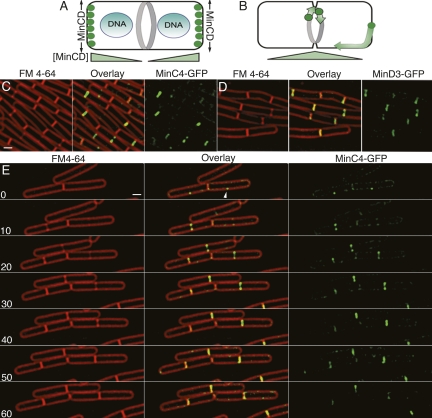
Localization of MinC4-GFP (EBS499) and MinD4-GFP (JAG118). (A) The current model for the spatial regulation of cell division in B. subtilis and E. coli indicates that the DNA-binding protein Noc blocks division over the chromosome, while MinC (green) has the highest time-averaged concentration at the poles, thereby blocking polar cell division. In E. coli, the increased concentration of MinCD at the poles is achieved by its oscillation from one cell pole to the other, while in B. subtilis MinCD were previously thought to localize to the cell poles. (B) Our studies find MinC to have the highest concentration at mid-cell, with a few molecules moving rapidly along the cell envelope. Thus, MinC has a high concentration only at the most recent cell pole (right pole), and only before it relocalizes to mid-cell. (C,D) Epifluorescence microscopy of MinC4-GFP (EBS499) (C) and MinD4-GFP (JAG118) (D) in growing B. subtilis cells. The nonfunctional MinD4-GFP fusion here complemented by amyE∷PminCD-minCD, which does not express sufficient MinC or MinD to fully complement the minCD-null mutation for minicell production (Supplemental Material), but does complement MinD4-GFP, suggesting that MinD4GFP retains some function. (E) Time-lapse microscopy showing the localization of MinC4-GFPthroughout the cell cycle. FM 4-64 and GFP images were collected every 10 min. Supplemental Movie S1 shows MinC4-GFP. Bars, 1 μm.
The MinCD system functions in many bacterial species and is therefore well suited for comparative studies of the spatial regulation of cell division. Indeed, disruption of minC or minD changes the position at which FtsZ assembles in both rod-shaped bacteria such as E. coli and B. subtilis and in cocci such as Neisseria gonorrhoeae (Ramirez-Arcos et al. 2001; Szeto et al. 2001) and the cyanobacterium Synechocystis (Mazouni et al. 2004). Furthermore, while the absence of MinC and MinD allows assembly of additional FtsZ rings (Z-rings) at aberrant positions (Levin et al. 1998), their overexpression blocks Z-ring assembly and cell division (Bi and Lutkenhaus 1993; Justice et al. 2000; Levin et al. 2001), suggesting that the Min system inhibits Z-ring formation. In keeping with this hypothesis, disruption of proteins that spatially regulate MinCD activity (MinE in E. coli and DivIVA in B. subtilis) block FtsZ assembly in a MinC-dependent manner, producing filamentous cells (de Boer et al. 1989; Zhao et al. 1995; Marston et al. 1998; Pichoff and Lutkenhaus 2001). MinC appears to be the primary division inhibitor in E. coli (de Boer et al. 1992; Hu et al. 1999), while MinD tethers MinC to the membrane, allowing it to be effective at physiological concentrations (de Boer et al. 1991). The biochemical mechanism by which MinC inhibits polar Z-ring formation is unclear, but in E. coli it appears to do so without affecting either FtsZ GTPase activity (Hu et al. 1999; Cordell et al. 2001) or the addition of FtsZ monomers to existing Z-rings (Anderson et al. 2004). Indeed recent evidence suggests that MinC inhibits lateral interactions between FtsZ protofilaments in E. coli (Dajkovic et al. 2008) and B. subtilis (Scheffers 2008), which might destabilize FtsZ rings and decrease their ability to recruit proteins involved in cytokinesis to potential division sites.
Understanding the spatial distribution of MinC and MinD within the cell is critical to understanding how they regulate Z-ring formation. Given the similar effects of MinC and MinD in B. subtilis and E. coli, one might expect them to display similar subcellular behavior. This is not the case. E. coli MinC and MinD oscillate from pole to pole with a periodicity of ∼50 sec (Hu and Lutkenhaus 1999; Raskin and de Boer 1999a), while B. subtilis MinCD is static, localizing to the septum late in division and being retained at new cell poles by DivIVA (Marston et al. 1998; Marston and Errington 1999). Current models of MinCD function in both species propose that these division inhibitors have the highest time-averaged concentration at the cell poles, thereby inhibiting polar cell division while allowing division at mid-cell (Fig. 1A; for reviews, see Ryan and Shapiro 2003; Rothfield et al. 2005; Lutkenhaus 2007). By these models, the static polar localization of B. subtilis MinCD could achieve the same biological function as the oscillation of E. coli MinCD.
We here reinvestigate the function and localization of B. subtilis MinC. First, we use a functional GFP insertion expressed from the native minC locus that maintains wild-type operon structure. We find that B. subtilis MinC moves rapidly along the cell membrane and appears to rotate around division sites, with little accumulation at the poles. Next, we investigated FtsZ dynamics and the timing of cell division in wild-type and minCD mutant cells using time-lapse fluorescence microscopy. These experiments indicate that even in wild-type cells, polar FtsZ rings often form adjacent to the recently completed septum. In the absence of MinCD, these normally transient structures are stabilized and used for cell division. We therefore propose that in B. subtilis, MinCD primarily acts at the new cell pole to prevent the maturation of transient FtsZ structures into active divisomes. Our data also suggest that MinCD affect the timing of cell division, because the interdivisional time is highly variable in minCD mutant cells.
Results
Localization of GFP insertions in MinC and MinD
We were concerned that the apparent contrast between the dynamic behavior of E. coli MinC and MinD and the stable polar localization of B. subtilis MinC and MinD might be explained by overexpression of the latter proteins from inducible promoters, because even a minor static population can mask the presence of a dynamic population of molecules using standard fluorescence microscopy. We therefore constructed and used a GFP insertion transposon (Materials and Methods) to isolate two functional gfp insertions into the chromosomal minC locus that preserve minCD operon structure. These fusions have gfp inserted after the fourth (minC4-gfp) or eighth (minC8-gfp) minC codon, produce <0.3% minicells and have wild-type cell length (Fig. 1; Supplemental Fig. S1; Supplemental Table S1). Because these insertions were generated by Tn5-mediated transposition (see Materials and Methods), there is a small duplication of the coding region. Thus, after GFP, the fusion proteins encode most of MinC, starting with the third (minC4-gfp) or seventh (minC8-gfp) amino acid. We visualized these GFP insertions in growing bacteria at 30°C. Both localized in an identical manner distinct from that reported in previous publications, forming foci and rings at new and constricting septa, with little or no MinC observed at the poles (Fig. 1C; Supplemental Fig. S1).
The septal localization of MinC was lost and replaced by cytoplasmic fluorescence when cells were immobilized on polylysine-treated coverslips (Supplemental Fig. S2A), indicating that proteins might be lost from division sites when slides are prepared by this method. Indeed, FtsZ-GFP also showed increased cytoplasmic fluorescence when visualized on polylysine coverslips rather than agarose pads (Supplemental Fig. S2B,C), and the Z-rings contained only ∼20% of the fluorescence of cells growing on agarose pads (Supplemental Table S3). Thus, there is a dramatic reduction in septal localization of both FtsZ and MinC when slides are prepared in a manner that does not support growth.
We also isolated many gfp insertions in the chromosomal minD locus, one of which (minD4-gfp) was at a location that would generate a fusion protein that is nearly identical to the previously published GFP-MinD fusion (Marston et al. 1998). Both constructs encode GFP followed by amino acids 2–268 of MinD, but minD4-gfp is integrated at and expressed from the native minCD locus rather than being integrated into an ectopic position and expressed by an inducible promoter in a minD background (Marston et al. 1998). Surprisingly, we found that MinD4-GFP produced elongated cells and minicells at a level similar to the minCD-null (Supplemental Table S2), and that it showed weak and inconsistent fluorescence intensity, and therefore did not meet our criteria for a functional insertion. We attempted to rescue MinD4-GFP by introducing a wild-type copy of minCD expressed from the minCD promoter at the amyE locus. This construct cannot rescue a minCD-null for minicell production (Supplemental Table S2), likely because minCD is also expressed from the Pmaf promoter upstream of the maf–radC–mreBCD genes that are immediately upstream of minCD (Lee and Price 1993). The strain containing both minD4-gfp and this extra copy of wild-type minCD produced fewer minicells than either alone (Supplemental Table S2) indicating that MinD4-GFP is partially functional. Unlike MinC4-GFP, MinD4-GFP appeared to be retained at some cell poles, so it is possible that MinD is retained at the cell pole to a greater extent than MinC. However, this polar localization might be caused by overexpression or the partial functionality of MinD4-GFP. We did not further characterize this fusion.
MinC is dynamic
We used time-lapse fluorescence microscopy to investigate MinC dynamics during the cell cycle. MinC localized to the division site before visible constriction, often forming a single focus at a future division site (Fig. 1E, arrowhead). MinC then assembled a ring-like structure that constricted during division and was lost from new septa with faint membrane-associated fluorescence detected throughout the cell (including the cell poles) in foci that appeared to move between time points. Thus, MinC localizes to septa immediately before the onset of septal biogenesis and disappears from completed septa, thereby behaving as expected for a late cell division protein. MinC also appeared dynamic, leaving completed septa and moving to new division sites (Supplemental Movie S1). This dynamic population of molecules was more evident using total internal reflection fluorescence (TIRF) microscopy (Supplemental Fig. S3; Supplemental Movies S2, S3), which has decreased excitation depth and increased excitation intensity, thereby allowing observation of relatively few fluorophores and rapidly moving molecules (Axelrod 2001; Sako and Uyemura 2002; Axelrod 2003; Kusumi et al. 2005). This method showed that MinC moved along the cell membrane and around the pole (Supplemental Fig. S3; Supplemental Movies S2, S3). Thus, B. subtilis MinC appears to show two distinct behaviors, a stable localization to mid-cell and a more dynamic movement throughout the cell membrane.
The poles are a secondary localization site for MinC
The previously observed polar localization of GFP-MinC (Marston et al. 1998; Marston and Errington 1999) could be a consequence of overexpression from the inducible xylose promoter (Pxyl), which could saturate septal binding sites or it could be a consequence of different slide preparation methods. We therefore compared localization of MinC4-GFP and Pxyl-GFP-MinC in growing cells (Fig. 2). Inducible Pxyl-GFP-MinC localized to division sites before and during septal biogenesis (Fig. 2B, arrows) as well as to the cell poles (Fig. 2B, arrowheads). The most intense fluorescent signal was consistently observed at the mid-cell of dividing cells (Fig. 2B), with less intense fluorescence at the cell pole. In contrast, MinC4-GFP fluorescence was largely confined to mid-cell (Fig. 2A), even when a longer exposure yielded twice the maximum pixel intensity compared with the inducible fusion. Polar localization of Pxyl-GFP-MinC depended on expression level, because decreasing the xylose concentration from the previously used concentration of 0.5% (Fig. 2B,E) to 0.05% (Fig. 2D) reduced polar but not mid-cell localization, while retaining the ability of GFP-MinC to prevent minicell formation. Thus, both MinC4-GFP, which is expressed from the native promoter, and inducible Pxyl-GFP-MinC localize to mid-cell, but the inducible fusion is stably associated with the cell poles only when substantially overexpressed. We also noted that MinC4-GFP is recruited to the cell poles during intermediate stages of FtsZ depletion (Supplemental Fig. S4; Supplemental Movie S4). These results suggest that the poles are secondary localization sites that recruit MinC when the primary site (active division sites) is either missing or saturated by overexpressed MinC.
Figure 2.
Quantitative comparison of MinC4-GFP and inducible GFP-MinC (Pxyl-GFP-minC) localization. Arrowheads indicate the cell poles; arrows indicate the mid-cell position. The pixel intensities are shown as a heat intensity plot with red being the highest, blue the lowest with colors corresponding to the scale on the right for each image. (A) MinC4-GFP (EBS499) with a 5-sec exposure, which results in a two times higher maximum pixel intensity than Pxyl-GFP-MinC. The highest fluorescence intensity is at mid-cell, with low levels of fluorescence elsewhere along the membrane including the cell pole. No polar caps of MinC are observed. (B) PxylGFP-MinC (JAG189) grown with 0.5% xylose and imaged with a 1-sec GFP exposure. The protein localizes both to septa and to mid-cell. (C–E) Pxyl-GFP-MinC was expressed at various levels by titrating the xylose concentration from 0.02% (C), a concentration that does not produce sufficient protein to block minicell formation, to 0.05% (D) and 0.5% (E), both of which produce sufficient protein to prevent minicell formation. Decreasing the xylose concentration to 0.05% decreases polar GFP-MinC localization while maintaining mid-cell localization. Bars, 1 μm.
Dynamic behavior of MinC at the septum
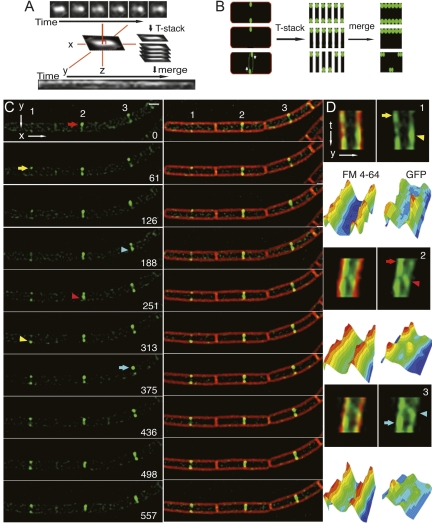
MinC initially assembles an asymmetric focus at the future division site (Fig. 1C). We noted that MinC often remained asymmetric during constriction, with one side of the septum appearing >2× brighter than the other (Fig. 3, arrows). Time-lapse epifluorescence microscopy with images collected at 1-min time intervals showed that MinC at individual septa appeared to transition between foci and symmetric or asymmetric rings and back to foci (Fig. 3C; Supplemental Movie S5, S6). In addition, the region of highest fluorescence intensity appeared to move from one side of the septum to the other (Fig. 3C, arrows and arrowhead). To quantitatively view this data, we devised a kymogram-like method that projects the GFP fluorescence at the septum through time (Fig. 3A). Briefly, the Y-axis was manually aligned to each septum, the septum was cropped and plotted against time giving a two-dimensional projection of each septum through time. After projection, a uniform static ring would show two parallel lines of equal intensity, whereas an asymmetric static ring would show one brighter line (Fig. 3B). The pixel intensity data of these projections clearly showed that the maximum pixel intensity of MinC moved from one side of the septum (Fig. 3D, arrows) to the other (Fig. 3D, arrowheads) in many cells, including each of the three shown in Figure 3C (cells 1, 2, and 3). The apparent movement across the septum required ∼4 min for the cells shown in Figure 3, and was sometimes visualized as a diagonal line in the projections (cells 2 and 3). In contrast, variation in FM 4-64 staining intensity was always symmetric (likely due to fluctuations in excitatory light intensity).
Figure 3.
Movement of MinC4-GFP at division sites visualized by epifluorescence microscopy. To quantify changes in the appearance of MinC4-GFP at septa over time, we employed a kymogram-like method to follow individual septa through time, together with an analysis of the pixel intensities over time. (A) Briefly, the septa were rotated to align the septa with the Y-axis, cropped, stacked, and the Y-axis projected through time. (B) An example of the expected kymograms for symmetric rings (top), for an asymmetric assembly (middle), and for an asymmetric assembly that rotates about the septum (bottom). (C) Time-lapse microscopy of MinC4-GFP (EBS499) at three adjacent division sites (numbers 1–3). Images were taken at ∼1-min intervals; the time in seconds is shown on the right. MinC appeared to translocate from one side of the septa (arrows) to the other (arrowhead). See Supplemental Movies S5 and S6 for more examples. (D) Kymograms of each septum (labeled 1–3) showing GFP (right) and FM 4-64 fluorescence (left) and the corresponding 3D pixel intensity plots. The number in the top right indicates the septum in C; the arrows and arrowheads also correspond to those in C for each cell. The FM 4-64 intensity fluctuates evenly across the septum, in contrast to the GFP intensity, in which one side can gain or lose intensity. Bar, 1 μm. Time is indicated in seconds.
Thus, MinC appears dynamic even at the septum, moving from one side of the septum to the other. This behavior could represent the circumferential movement of MinC foci or arcs around the septum, or it could represent the assembly and disassembly of MinC complexes on different sides of the septum. At this time, we cannot distinguish between these two possibilities. However, if MinC is rotating around the septum, then its calculated velocity around the septum is 0.25–0.34 μm/min for slightly constricted septa.
MinC localizes to the septum after the early division proteins
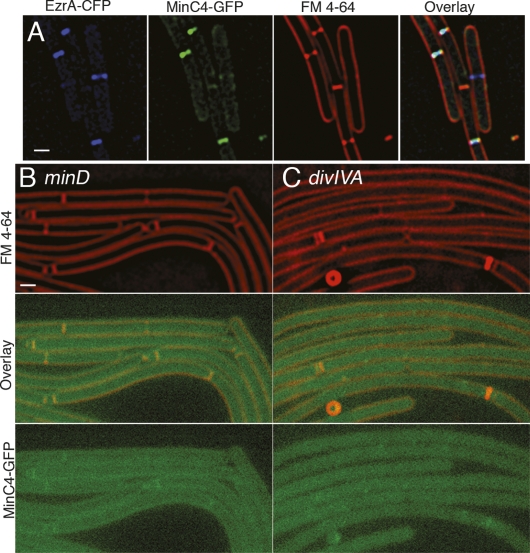
MinC localization is consistent with that of a late division protein, as the protein appears to arrive at mid-cell just prior to septal biogenesis, which is readily visualized at early stages by two spots of increased fluorescence after staining with the vital membrane stain FM 4-64 (Fig. 1E, 10 min). Indeed, quantification of MinC in growing cells revealed that it was present at mid-cell in 12% of predivisional cells (in which FM 4-64 staining showed no evidence of septal biogenesis) compared with 93% of dividing cells (Supplemental Table S4). To confirm that MinC arrives after the early division proteins, we colocalized MinC4-GFP with EzrA fused to CFP in growing cells (Fig. 4A; Supplemental Table S5). We found that 87% of predivisional cells showed localization of EzrA but not MinC at mid-cell, while 13% showed MinC and EzrA at mid-cell. In contrast, 92% of cells that were actively dividing showed both EzrA and MinC localized at the nascent septum. Thus, EzrA arrives at mid-cell prior to MinC and both proteins remain at the septum during constriction. Septal localization of MinC4-GFP was dependent on MinD and DivIVA (Fig. 4B,C), which were shown previously to be required for polar localization of MinC (Marston et al. 1998; Marston and Errington 1999). These results indicate that MinC is a late recruit to the septum as it arrives after the early division protein EzrA, and that DivIVA localizes MinC to the septum, rather than stably sequestering it at the cell pole.
Figure 4.
MinC localizes to division sites late in division in a manner dependent on DivIVA and MinD. (A) Colocalization of EzrA-CFP (blue) and MinC4-GFP (green) with FM 4-64n-stained membranes (red) in strain JAG226. (B,C) MinC4-GFP fails to localize in the absence of MinD (JAG79) (B) or DivIVA (JAG67) (C), and shows low fluorescence that is barely above background. Thus, DivIVA and MinD appear to be required for septal localization of MinC, rather than for retention at the cell pole. Bars, 1 μm.
FtsZ dynamics visualized by time-lapse microscopy
The results described above indicated that MinC is recruited to nascent division sites after the early division proteins and immediately before septal biogenesis, and that it leaves the new cell pole to relocalize to the mid-cell of the newborn daughter cells. Thus, MinC spends little time associated with the cell poles, where it must act to prevent minicell formation. We therefore hypothesized that MinC might prevent minicell formation by inhibiting the assembly of a mature divisome immediately adjacent to the most recently completed septum (or more specifically in the space between the pole and the chromosome where Noc is not inhibiting Z-ring formation). This hypothesis predicts that in minCD mutant cells, polar FtsZ rings would frequently assemble adjacent to the recently completed septum, and that minicells would most frequently form at these sites, rather than at the old cell pole where FtsZ polymers are not present.
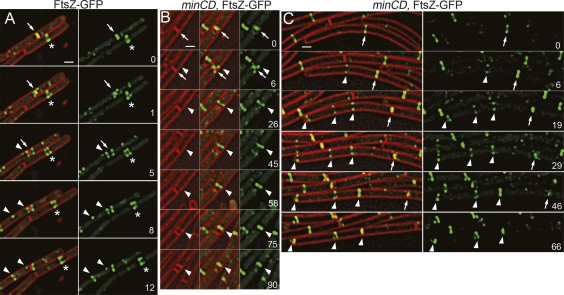
To test this hypothesis and to gain further insight into FtsZ dynamics during the cell cycle, we used time-lapse fluorescence microscopy to watch FtsZ-GFP relocalization in wild-type and minCD mutant cells. We first used FtsZ-GFP, which expresses ftsZ-gfp as the only copy of ftsZ (Fig. 5A), and is somewhat temperature sensitive for growth, but at higher temperatures than used in our time-lapse experiments (Levin et al. 1999). Similar fusions are routinely used for studies of FtsZ localization and dynamics. We noted two behaviors of FtsZ-GFP in this strain. First, the FtsZ ring often assembles at mid-cell by the collapse of a dynamic spiral-like shape into a ring (Fig. 5A, asterisk; Supplemental Movie S7), as previously suggested by other studies (Thanedar and Margolin 2004; Peters et al. 2007). Second, when FtsZ leaves a recently completed septum (Fig. 5A, arrows), it often resulted in the transient assembly of FtsZ structures near the cell pole that fall apart and relocalize to mid-cell within ∼10 min (Fig. 5A, arrowheads). Similar minor populations of FtsZ are detected in some time-lapse images of FtsZ-YFP (Peters et al. 2007). We suspect that these transient structures are labile and not readily visualized without time-lapse microscopy, given our finding that the amount of FtsZ present even in medial rings decreases fivefold in nongrowing cells.
Figure 5.
Comparison of FtsZ-GFP dynamics in wild-type and minCD cells. Images of FtsZ-GFP (green) and FM 4-64 (red) were collected every 1–3 min for several cell division events. Times were selected for display that best showed the illustrated behavior; additional time points and examples of wild-type and minCD mutant cells can be found in Supplemental Movies S7 and S8). Time is indicated in minutes in the right panel of each set. (A) Relocalization of FtsZ-GFP (JAG212) from a completed septum (arrows), showing the transient assembly of an FtsZ-ring adjacent to the new pole (arrowheads) that disappears while an FtsZ ring assembles at mid-cell (arrowheads). The assembly of an FtsZ ring at mid-cell often entails stabilization of a dynamic structure into a single, brighter ring (asterisk). (B) In the minCD strain JAG252, FtsZ-GFP assembles structures (arrowhead) adjacent to the recently completed septum (arrow); these ultimately constrict to generate a minicell. This pathway accounts for 75% of all minicells. (C) The remaining minicells are produced in long cells that assemble several FtsZ rings. The arrow indicates the first FtsZ ring to assemble, arrowheads subsequent rings. The leftmost ring constricts to generate a minicell. Bars, 1 μm.
We hypothesized that the transient polar FtsZ rings we observed in wild-type cells might mature into active divisomes in a minCD mutant. We therefore analyzed FtsZ-GFP dynamics in a minCD mutant (Supplemental Movie S8). In a minCD mutant, FtsZ-GFP assembled polar FtsZ structures adjacent to the recently completed septum, but unlike wild-type cells, these structures persist and eventually support cell division (Fig. 5B, arrowheads; Supplemental Movie S8). Indeed, 73% of the minicells were the result of FtsZ-rings that assembled adjacent to recently completed septa (here defined as those that contained FtsZ-GFP in previous time points) rather than at the older cell pole (here defined as those that lacked FtsZ-GFP in previous time points) (Fig. 5B; Supplemental Table S6). The remaining minicells were typically formed from long cells that assembled multiple FtsZ rings, some of which were at the cell poles (Fig. 5C, arrowheads) and which could constrict nearly simultaneously to give a cell with two poles of equal age. These results suggest that MinC and MinD ensure that polar FtsZ structures visualized in wild type are transient and unable to support cell division. This view is consistent with recent biochemical evidence that MinC does not inhibit FtsZ polymerization, but rather the lateral interactions between protofilaments that are likely necessary for septation (Dajkovic et al. 2008; Scheffers 2008) We propose that MinCD act immediately after cell division, when the high concentration of FtsZ and other cell division proteins at the new cell pole would favor the immediate reassembly of the cell division proteins adjacent to the new pole, a hypothesis consistent with the MinC localization data presented above.
Increasing the FtsZ/MinCD ratio causes the assembly of Z-rings near the new cell pole
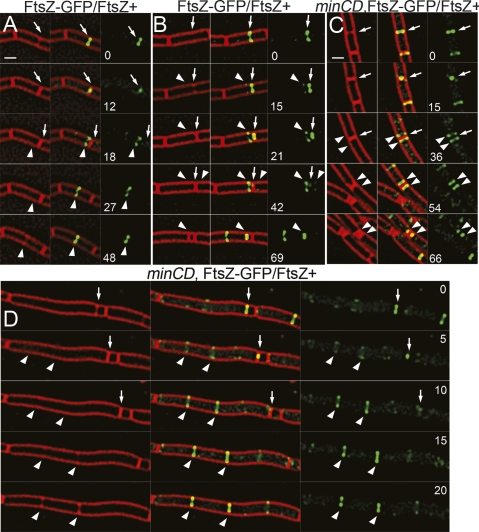
It has previously been shown that overexpressing FtsZ overcomes Min inhibition and produce minicells (Ward and Lutkenhaus 1985; Weart and Levin 2003). If MinC acts primarily at new cell poles, then under these conditions, minicells should also be assembled from FtsZ rings that assemble adjacent to recently completed septa. We therefore examined FtsZ dynamics in a strain that expresses gfp-ftsZ in addition to a wild-type copy of ftsZ (denoted FtsZ-GFP/FtsZ+). This strain makes slightly shorter cells than wild type, likely because it produces more FtsZ (Supplemental Table S6). The extra copy of ftsZ rescues the temperature sensitivity of the GFP fusion and also causes the production of ∼7% minicells in the presence of wild-type MinCD (Supplemental Table S6; Supplemental Movie S8). This is likely due to the increased levels of cytoplasmic FtsZ overcoming Min inhibition, as supported by our observation that overexpression of MinCD in the FtsZ-GFP/FtsZ+ strain reduces the number of minicells from 9.9% to 2.6% (Supplemental Table S6). Polar FtsZ assemblies were readily visualized in this strain (Fig. 6A; Supplemental Movie S9), and 79% of the minicells were formed adjacent to a recently completed septum (again defined as those that contained FtsZ-GFP in previous time points), from these polar FtsZ-GFP structures.
Figure 6.
FtsZ dynamics in strains coexpressing FtsZ-GFP and FtsZ. Images of FtsZ-GFP (green) and FM 4-64 (red) were collected every 3 min for several cell division events. (A) Relocalization of FtsZ-GFP/FtsZ+ (AD3007) from a completed septum (arrow) to a transient polar site and to mid-cell (arrowhead). (B) Relocalization of FtsZ-GFP/FtsZ+ (AD3007) from completed septum (arrow) to a polar site that constricts to form a minicell (arrowhead). (C) In the minCD strain JAG135, FtsZ-GFP/FtsZ+ assembles polar FtsZ rings (arrowheads) adjacent to completed septa (arrow), which constrict to form minicells. (D) This strain also forms long cells that assemble multiple FtsZ-rings that can constrict nearly simultaneously (arrowheads). Bars, 1 μm.
As expected, the absence of MinCD in the FtsZ-GFP/FtsZ+ strain exacerbated this defect and caused additional polar FtsZ rings to persist and mature into active division sites, producing minicells (Fig. 6B,C, arrowheads; Supplemental Movie S10). Again, most minicells were assembled adjacent to the recently completed septum, but some were also assembled in long cells with multiple FtsZ rings (Fig. 6C; Supplemental Table S6). These results suggest that reducing the MinC/FtsZ ratio below that needed to prevent polar cell division allows minicells to form, most adjacent to the most recently completed septum.
The minCD mutant affects the coupling between FtsZ ring assembly and cell division
In wild-type cells, a single stable FtsZ ring assembles at mid-cell and is used for septal biogenesis in each cell cycle, so septation and FtsZ ring assembly are both temporally and spatially correlated. However, our studies of FtsZ dynamics in the minCD mutant revealed an uncoupling of FtsZ ring assembly and septation, so that septation no longer provided an accurate indicator of the timing of FtsZ ring assembly. For example, multiple FtsZ rings assembled at approximately the same time often constricted at different times (Fig. 5C, arrowheads), so that an FtsZ ring assembled in the parent cell could be used in a subsequent division event in the daughter cell. Thus, an FtsZ-ring could assemble and persist at a new pole until after another FtsZ ring in the cell had divided, ultimately producing a minicell at the old cell pole, but one cell cycle after the FtsZ ring was assembled. Our analysis also revealed that in the minCD mutant multiple cell division events could occur in rapid succession or even simultaneously producing cells that did not have a clear old and new pole. This defect made it impossible to use a simple lineage analysis to determine if minicells generally formed from FtsZ assembled adjacent to a recently completed septum. Our results suggest that the absence of MinCD affects either the time required for Z-ring assembly or the time required for the onset of septal biogenesis, or both.
The absence of MinCD alters the timing of cell division
Time-lapse observation of minCD mutant cells stained with the fluorescent membrane stain FM 4-64 revealed that they appeared defective in the timing of cell division (Supplemental Movie S11). First, unlike the wild type (Fig. 7A), newly formed sister cells produced by a medial division rapidly became asynchronous (Fig. 7B). Second, the strain often produced abnormally long cells, even in the absence of a minicell division (Fig. 7B). These results were surprising because it has been proposed that the longer than normal cells observed in minCD mutants are the result of an asymmetric division that gives rise to a mini cell and a long cell. Third, individual cells often initiated multiple cell division events simultaneously (Fig. 7B, division #4), rather than having only a single division event per cell as observed in wild type (Fig. 7A). Together, these data suggest that minCD cells have a defect in the timing of cell division in addition to an inability to prevent maturation of FtsZ-rings at the new cell pole.
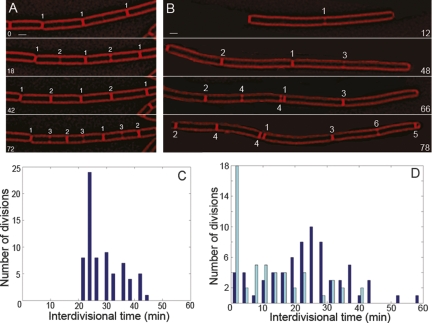
Figure 7.
Comparison of interdivisional time in wild-type and minCD cells. Time-lapse microscopy of FM 4-64-stained cells was performed with images collected every 6 min. Numbers near septa indicate the order they were formed. Identical numbers in the same time point denote septa that formed simultaneously. (A) The wild-type strain PY79 shows well-synchronized daughter cells. (B) The minCD strain MDS642 shows asynchronous division in the daughter cells (divisions 2 and 3), and simultaneous divisions (division 4). The cell shown has been cropped out of a field for clarity. Bar, 1 μm. (C,D) The time between the completion of one septum and the start of the next (described in the Materials and Methods) was plotted for PY79 (C) and MDS642 (D). Dark-blue lines denote mid-cell to mid-cell interdivisional time and light-blue lines denote that for any division involving a minicell.
To more quantitatively investigate this possibility, we used time-lapse microscopy of wild-type and minCD strains, collecting images of FM 4-64-stained membranes every 3 min at 30°C. We noted the time at which each division event was completed (with uniform FM 4-64 staining across the cell) and the time at which septal biogenesis commenced in each daughter cell (two spots of FM 4-64 staining on each side of the cell). The difference between these times (the interdivisional time) was plotted on a histogram (Fig. 7C,D). Wild-type cells have a well-coordinated cell cycle during which the time between completion of division and subsequent formation of a new septum is never shorter than 20 min, with an average of 29 ± 6 min. We then repeated this using minCD cells and separated the mid-cell to mid-cell divisions (Fig. 7C,D, dark blue, 25 ± 12 min) and those that form a minicell (Fig. 7C,D, light blue, 13 ± 12 min). Neither population maintained the 20 min minimum interdivisional time, as cells were able to initiate cell division events simultaneously (see Fig. 6B, division 4). There was also a population of cells that had long interdivisional times, which would result in the production of cells that are longer than wild type and capable of assembling multiple FtsZ rings.
Discussion
Most current models for MinC function indicate that this division inhibitor is restricted to the cell poles, where it inhibits FtsZ assembly (Ryan and Shapiro 2003; Rothfield et al. 2005; Margolin 2006; Barak and Wilkinson 2007; Lutkenhaus 2007). The activity of E. coli MinC is restricted to the cell poles by oscillation from one pole to the other (together with MinD), while B. subtilis MinC is thought to be stably tethered to the cell pole by DivIVA (again together with MinD). Surprisingly, several of our observations (discussed below) contradict this view and suggest a modified model for Min function.
First, functional GFP fusions to MinC expressed from the native chromosomal locus do not stably localize to the cell poles, but rather are primarily associated with sites of active cell division, to which they are recruited late in divisome assembly and prior to septation. Our data suggest that the prominent bipolar localization of MinC observed previously is due to protein overexpression, which causes MinC to localize to the cell poles as a secondary localization site that is also observed when division is blocked by FtsZ depletion. Thus, rather than being stably localized to both cell poles, MinC is only transiently associated with the new cell pole, remaining at this site for a short amount of time after constriction and then moving to mid-cell.
Second, B. subtilis MinC is dynamic, although in a manner distinct from that of E. coli. Specifically, we find that the net movement of B. subtilis MinC is from the new pole to mid-cell, rather than oscillating from pole to pole. B. subtilis MinC relocalizes from the new cell pole to mid-cell immediately prior to division via membrane associated foci that move at rates of ∼3 μm/min. During division, it also appears to move from one side of the invaginating septum to the other once every ∼4 min (for a predicted rate of ∼0.3 μm/min). Both of these latter rates of movement are slower than the measured diffusion rate of cytoplasmic or membrane proteins of similar sizes (∼500 μm2/min, ∼10 μm2/min, respectively) (Elowitz et al. 1999; Mullineaux et al. 2006), and more similar to proteins in which movement is mediated by polymerization (0.2–2 μm/min) (Perez et al. 1999; Waterman-Storer et al. 1999; Watanabe and Mitchison 2002), and similar to the rate of MinC oscillation in E. coli (Hu and Lutkenhaus 1999; Raskin and de Boer 1999a, b). This suggests that MinC might be moved by a cytoskeletal protein, such as MinD, which polymerizes in a membrane and ATP-dependent manner (Hu et al. 2002; Suefuji et al. 2002) and is required for the oscillatory movement of E. coli MinC (Hu and Lutkenhaus 1999; Raskin and de Boer 1999a, b). Thus, B. subtilis MinC shows two distinct dynamic behaviors, movement of smaller populations along the cell envelope that result in a net relocalization from the new pole to mid-cell and its apparent rotation around the septum (Fig. 8A).
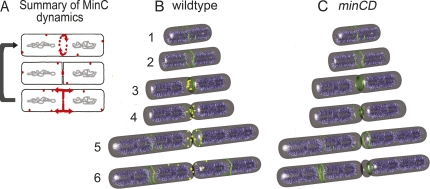
Figure 8.
Summary of MinC dynamics in B. subtilis and a proposed role for MinC in relocalization of FtsZ from new cell poles. (A) MinC (red) localizes to septa before septal biogenesis, rotating around sites of active cell division (top cell) and constricting as septal biogenesis completes (middle cell). After septation is complete, it leaves the new pole and moves to mid-cell. (B) Model for the role of MinC (yellow) in relocalization of FtsZ (green filaments) during the cell cycle. Chromosomes are shown in cyan. Cells 1 and 2 show the formation of an FtsZ ring at mid-cell from short FtsZ protofilaments (green filaments) (Li et al. 2007; Osawa et al. 2008), cells 3 and 4 the onset of septal biogenesis and the recruitment of MinC (yellow) to the active divisome (solid green ring). As septation completes (cells 4 and 5), FtsZ is released from the new cell pole and accumulates in ring-like structures near the cell pole that MinC either disrupts prevents from recruiting the late division proteins. (C) In the absence of MinC, these normally transient structures persist and are used for cell division.
Finally, we were surprised to observe that polar FtsZ rings (or ring-like structures) assemble in wild-type cells adjacent to recently completed septa, as most models propose that MinC prevents the formation of these structures. This indicates that MinC does not prevent FtsZ from assembling at the poles, but rather interacts with and destabilizes polar FtsZ polymers or newly formed Z-rings before they can become active sites of cell division. Time-lapse fluorescence microscopy demonstrated that in the minCD mutant, 75%–83% (Supplemental Table S6) of minicells are produced from these polar FtsZ structures; the remaining minicells are produced in long cells that result from a defect in the initiation of cell division (discussed below).
A refined model for Min function
Our results suggest a refined model of MinCD function in B. subtilis (Fig. 8B). First, FtsZ coalesces into a ring at mid-cell, recruiting the early division proteins FtsA, EzrA and ZapA, then the late cell division proteins, followed by DivIVA, and ultimately MinD and MinC (Fig. 8B, cell iii). Septal biogenesis begins and as it completes, FtsZ leaves the old division site to assemble ring-like structures adjacent to the new cell pole (cells iv and v). After septation, the affinity of MinC for the pole is reduced, and it moves away from the pole, probably together with MinD (cells v and vi). This movement from the newly completed septum effectively scans the new cell pole for FtsZ polymers that could assemble divisomes in the chromosome free space adjacent to the cell pole. We propose that MinC and MinD interact with these FtsZ polymers and either prevent them from associating with the late division proteins or directly destabilize the FtsZ assemblage, thereby allowing FtsZ to relocalize to mid-cell more efficiently. These possibilities are consistent with recent biochemical data demonstrating that E. coli and B subtilis MinC do not inhibit FtsZ polymerization, but rather inhibit lateral interactions between FtsZ protofilaments (Dajkovic et al. 2008; Scheffers 2008). Such interactions are likely necessary to stabilize transient FtsZ rings and to allow their association with the late division proteins. Interestingly, E. coli MinC is capable of disassociating previously assembled FtsZ networks (or bundles) (Dajkovic et al. 2008); if this is also true of B. subtilis MinC, then it could directly dissociate the FtsZ rings that assemble adjacent to the new cell pole.
The mechanism by which MinC is kept inactive and unable to block cell division upon recruitment to mid-cell remains unclear, but we can envision two possibilities. First, the different protein–protein interactions that mediate these events could regulate MinC activity: Recruitment of MinC to mid-cell depends on DivIVA and the late cell division proteins (Edwards and Errington 1997; Marston et al. 1998), whereas MinC is capable of directly inhibiting FtsZ bundling both in vitro and in vivo (de Boer et al. 1992; Marston et al. 1998; Dajkovic et al. 2008; Scheffers 2008). It is therefore possible that DivIVA and/or the late division proteins inhibit MinC activity. Second, it is possible that MinC interacts with transient FtsZ assemblages before the late division proteins, and that it cannot disrupt the activity of the complete divisomes to which it is recruited at mid-cell. Additional experiments are required to discriminate between these models, but our studies clearly demonstrate that in B. subtilis, MinC activity is not restricted to the poles by localization, but rather is regulated by an unknown mechanism.
The absence of MinC and MinD causes a cell cycle timing defect
The B. subtilis minCD mutant also has a striking defect in the timing of cell division. Whereas wild-type B. subtilis shows well-synchronized chains of daughter cells and regular interdivisional times, minCD daughter cells (even those produced by medial division) are not synchronized and the strain shows highly variable interdivisional times. We find that it is the longer interdivisional times, rather than the production of minicells, that produces the long cells that are characteristic of the minCD strain, because long cells are often produced either before or without a minicell division. Such longer cells are capable of assembling multiple FtsZ rings that constrict nearly simultaneously, resulting in very short interdivisional times and often producing a polar minicell. Similar observations have been made regarding the timing of cell division in E. coli minCD mutants (Akerlund et al. 1992), suggesting that the role of the Min system in controlling the timing of cell division is conserved. One possible model for this timing defect is that in the absence of MinC, FtsZ polymers, perhaps together with other division proteins, accumulate at the new cell pole as well as the mid-cell, thus splitting these proteins between two sites instead of a single mid-cell site as in wild type. This might cause the cell to grow longer until both sites accumulate enough FtsZ and other proteins to support cell division, resulting in an extended latent period between assembly of the FtsZ ring and the onset of constriction. This model is in keeping with the observation that minCD mutant cells often make two concurrent or nearly concurrent divisions (Fig. 7; Akerlund et al. 1992).
The rotation of MinC at active division sites suggests it might associate with another septal component that rotates. Indeed, time-lapse microscopy indicates that FtsZ itself is highly dynamic within the cell and appears to rotate around mid-cell before coalescing into a more stable ring (Fig. 5; Supplemental Movies S5, S6). It is possible that this rotation persists during division, and that we are unable to observe it in our time-lapse experiments because the Z-ring is relatively uniform. Previous photobleaching experiments have indicated that the Z-ring is dynamic and exchanging subunits from a nonlocalized pool (Stricker et al. 2002). It is therefore possible that rotation is a general characteristic of the divisome, rather than a unique property of MinC.
Materials and methods
Strains and reagents
Strains (Table 1) are derived from PY79 (Youngman et al. 1984) except PAL1213 and PL642 (which are 0168 derivatives), using standard methods (Hoch 1991). The methods used to construct the minD deletion (JAG79), EzrA-CFP (JAG151), amyE∷Pxyl-gfp-minCD (PAL1213), and FtsZ-GFP/FtsZ (AD3007) are described in the Supplemental Material.
Table 1.
Strains used in this study
Isolation of GFP insertions in minCD
We wanted to localize functional GFP fusions to MinC and MinD expressed at native levels, to minimize potential artifacts. We therefore constructed the GFP insertion transposon TAGIT (to be described in detail elsewhere) (J.A. Gregory, E.C. Becker, I.P. Tuwatananurak, J. Jung, and K. Pogliano, in prep.) to mediate the random insertion of GFP into the chromosomal minCD locus, without disrupting normal transcriptional regulation or operon structure. Briefly, TAGIT is a Tn5-based transposon into which we cloned lacZ without translational initiation sites to allow identification of in frame insertions in an expressed target gene, a kanR gene to allow selection for insertions in both E. coli and B. subtilis, and gfp without translational initiation sites. We flanked lacZ and kan with loxP sites to allow Cre-mediated excision of lacZ and kan. TAGIT generates insertions of the GFP protein into the middle of a target protein, similar to Alkaline Phosphatase “sandwich” fusions that generate bifunctional fusion proteins (Ehrmann et al. 1990). As described below, we integrated TAGIT insertions into B. subtilis chromosome at minCD and expressed Cre in vivo to excise lacZ and kan from the in-frame and expressed fusions.
We used in vitro transposition (Goryshin et al. 1998) to insert TAGIT into pJAG1, a TOPO plasmid (pCR2.1-topo, Invitrogen) containing a 3.2-kb fragment encoding minCD genes plus ∼600 nucleotides upstream and downstream amplified with primers JG3 and JG4 (Supplemental Material). To allow selection for TAGIT insertions, the kanR gene in the pCR2.1 vector was inactivated by digesting the plasmid with NarI, which cuts twice within the gene. After in vitro transposition, the resulting pool was transformed into XL-10 Gold competent cells (Stratagene), and the plasmid DNA isolated and transformed into wild-type B. subtilis strain EBS42 (amyE∷PspoIIQ-cre). The transformation was plated on LB containing 10 μg/mL kanamycin to select for integration of the plasmid into the chromosome and 10 μg/mL X-Gal to screen for insertions in which TAGIT had inserted into the same reading frame as minCD, allowing expression of β-galactosidase. Strain EBS42 expresses the Cre recombinase from the sporulation-specific spoIIQ promoter, so to excise the lacZ and kan genes we sporulated the cells in DSM for 24 h, heated the culture for 20 min at 80°C to kill vegetative cells. The cells were plated on LB and patched to LB kanamycin to identify KanS colonies in which lacZ and kan had been excised to leave GFP in the same reading frame as upstream and downstream coding regions. PCR was used to ensure that the plasmids had integrated by the desired double recombination event (replacing minCD with minCD∷TAGIT from the plasmid) rather than a single recombination event (duplicating minCD and integrating the entire plasmid into the chromosome).
We initially used fluorescence microscopy of FM 4-64-stained cells to screen for functional GFP insertions, but noted that at 42°C on DSM plates the minCD-null strain made irregular star-shaped colonies with projecting tendrils, while the wild-type strain made smooth edged colonies. This allowed us to more rapidly screen for functional GFP insertions. In total, ∼500 colonies with in-frame GFP insertions were screened for minCD function by this assay, and 100 by microscopy. Only two functional insertion sites (minC4-GFP and minC8-GFP) were isolated. To assess the randomness of TAGIT transposition, 100 randomly selected insertions were sequenced; these were distributed throughout the gene (Supplemental Fig. S5), although the only functional insertions were at the N terminus of MinC. In other experiments, we used TAGIT to isolate functional GFP insertions in the middle of LacI (J.A. Gregory, E.C. Becker, I.P. Tuwatananurak, J. Jung, and K. Pogliano, in prep.), suggesting that the failure to isolate functional GFP sandwich fusions in these studies is specific to MinC.
Time-lapse microscopy
Time-lapse microscopy used 1.2% agarose pads (Becker and Pogliano 2007) prepared as follows. FM 4-64 (Molecular Probes) was added to a final concentration of 0.5 μg/mL in a 1.2% solution of molten agar/media (CH or LB diluted 1:5 in water) and added to the well of a culture slide and covered with a glass slide. To induce Pxyl-GFP-MinC, xylose was added to the indicated concentration. After cooling, the slide was removed and two air pockets were cut out of the agar leaving a 3- to 5-mm agar bridge in the center of the well. Cells were grown overnight on the appropriate solid media at 30°C and applied to the agar bridge and covered by a glass cover slip. To prevent drying during the experiment, 50% glycerol was applied to the region of contact between the slide and the coverslip. The slide was then allowed to equilibrate in an environmentally controlled chamber at 30°C (Precision Control Weather Station) for at least 30 min prior to visualization. Exposure times were minimized to reduce phototoxicity. These slides supported growth at nearly the same rate as in liquid (a doubling time of ∼50 min on slides versus ∼40 min in liquid). Images were acquired using an Applied Precision Spectris microscope and deconvolved using softWoRx version 3.3.6 software (Applied Precision). Figures were assembled with Photoshop version 7.0 software (Adobe).
TIRF microscopy
TIRF experiments were performed using the same microscope and a 488-nm argon laser installed in a TIRF module supplied by Applied Precision and a Plan Apochromat 100× 1.40 NA objective. GFP was visualized with using FITC excitation and emission filters and 0.3-sec pulses at 35% power. FM 4-64 was imaged once using the 488-nm laser (with 0.1-sec pulses at 35% power) to excite and the rhodamine emission filter (control experiments demonstrated that no FM 4-64 fluorescence was visualized with the GFP emission filter). Summation and maximum intensity projections were constructed using the Quick Projection tool (softWoRx 3.3.6).
Quantitative image analysis:two-dimensional (2D), three-dimensional (3D), and kymogram-like plots
3D and 2D pixel intensity plots were created using the Data Inspector tool in the softWoRx version 3.3.6 software (with the heat intensity setting), with plots exported to Photoshop. To create kymogram-like plots of EBS499, the septum was cropped and rotated to align with the Y-axis using the Resample 2D function. The Volume Viewer function normally creates 3D projections of 2D images by using several Z-sections of one sample. This function was utilized to project 2D images through time by converting time points into Z-sections in the file header. Care was taken to align (Align Image tool) and crop (Resample 2D tool) each septum to the smallest area possible to faithfully represent the movement of GFP molecules through time. If images could not be aligned for any reason (focal plane and X/Y drift, rapid movement of cells due to growth, etc.) they were not used to create kymogram-like projections.
Quantification of interdivisional time
Cells were grown according to time-lapse microscopy methods and images were taken every 3 min. The interdivisional time was defined as the amount of time that elapsed between the completion of division in the parent cell and beginning of constriction in each daughter cell. The interdivisional time for each daughter cell/parent cell pair was recorded and plotted in a histogram using 20 bins (giving each time point its own bin; 3-min time points for 60 min). The histogram was created using Matlab and edited in Adobe Photoshop.
Acknowledgments
We thank Eric Stewart for advice on time-lapse fluorescence microscopy, William Reznikoff for Tn5 transposase, Richard Losick and Petra Levin for strains, and Alan Derman for constructing the FtsZ-GFP/FtsZ+ fusion. This research was supported by the National Science Foundation (NSF0135955), the National Institute of Health (GM57045), and by the University of California at San Diego Academic Senate.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1732408.
References
- Akerlund T., Bernander R., Nordstrom K. Cell division in Escherichia coli minB mutants. Mol. Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- Anderson D.E., Gueiros-Filho F.J., Erickson H.P. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Methods Enzymol. 2003;361:1–33. doi: 10.1016/s0076-6879(03)61003-7. [DOI] [PubMed] [Google Scholar]
- Barak I., Wilkinson A.J. Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Rev. 2007;31:311–326. doi: 10.1111/j.1574-6976.2007.00067.x. [DOI] [PubMed] [Google Scholar]
- Becker E.C., Pogliano K. Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol. Microbiol. 2007;66:1066–1079. doi: 10.1111/j.1365-2958.2007.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E., Herrera F.G., Gunderson F., Derman A.I., Dance A.L., Sims J., Larsen R., Pogliano J. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt T.G., de Boer P.A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell S.C., Anderson R.E., Lowe J. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 2001;20:2454–2461. doi: 10.1093/emboj/20.10.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A., Lan G., Sun S.X., Wirtz D., Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 2008;18:235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Rothfield L.I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Hand A.R., Rothfield L.I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Rothfield L.I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D.H., Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- Ehrmann M., Boyd D., Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc. Natl. Acad. Sci. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M.B., Surette M.G., Wolf P.E., Stock J.B., Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J., Daniel R.A., Scheffers D.J. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryshin I.Y., Miller J.A., Kil Y.V., Lanzov V.A., Reznikoff W.S. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J.A. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z., Mukherjee A., Pichoff S., Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Gogol E.P., Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice S.S., Garcia-Lara J., Rothfield L.I. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol. 2000;37:410–423. doi: 10.1046/j.1365-2958.2000.02007.x. [DOI] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- Lee S., Price C.W. The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division. Mol. Microbiol. 1993;7:601–610. doi: 10.1111/j.1365-2958.1993.tb01151.x. [DOI] [PubMed] [Google Scholar]
- Levin P.A., Shim J.J., Grossman A.D. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J. Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A., Kurtser I.G., Grossman A.D. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P.A., Schwartz R.L., Grossman A.D. Polymer stability plays an important role in the positional regulation of FtsZ. J. Bacteriol. 2001;183:5449–5452. doi: 10.1128/JB.183.18.5449-5452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Trimble M.J., Brun Y.V., Jensen G.J. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Bacterial division: Another way to box in the ring. Curr. Biol. 2006;16:R881–R884. doi: 10.1016/ j.cub.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Thomaides H.B., Edwards D.H., Sharpe M.E., Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes & Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazouni K., Domain F., Cassier-Chauvat C., Chauvat F. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 2004;52:1145–1158. doi: 10.1111/j.1365-2958.2004.04042.x. [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W., Nenninger A., Ray N., Robinson C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J. Bacteriol. 2006;188:3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Anderson D.E., Erickson H.P. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Diamantopoulos G.S., Stalder R., Kreis T.E. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Peters P.C., Migocki M.D., Thoni C., Harry E.J. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol. Microbiol. 2007;64:487–499. doi: 10.1111/j.1365-2958.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S., Lutkenhaus J. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 2001;183:6630–6635. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Arcos S., Szeto J., Beveridge T., Victor C., Francis F., Dillon J. Deletion of the cell-division inhibitor MinC results in lysis of Neisseria gonorrhoeae. Microbiology. 2001;147:225–237. doi: 10.1099/00221287-147-1-225. [DOI] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 1999a;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. 1999b;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Taghbalout A., Shih Y.L. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 2005;3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- Ryan K.R., Shapiro L. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 2003;72:367–394. doi: 10.1146/annurev.biochem.72.121801.161824. [DOI] [PubMed] [Google Scholar]
- Sako Y., Uyemura T. Total internal reflection fluorescence microscopy for single-molecule imaging in living cells. Cell Struct. Funct. 2002;27:357–365. doi: 10.1247/csf.27.357. [DOI] [PubMed] [Google Scholar]
- Scheffers D.J. The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett. 2008;582:2601–2608. doi: 10.1016/j.febslet.2008.06.038. [DOI] [PubMed] [Google Scholar]
- Stricker J., Maddox P., Salmon E.D., Erickson H.P. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suefuji K., Valluzzi R., RayChaudhuri D. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl. Acad. Sci. 2002;99:16776–16781. doi: 10.1073/pnas.262671699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto J., Ramirez-Arcos S., Raymond C., Hicks L.D., Kay C.M., Dillon J.A. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 2001;183:6253–6264. doi: 10.1128/JB.183.21.6253-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanedar S., Margolin W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.E., Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Mitchison T.J. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C., Desai A., Salmon E.D. Fluorescent speckle microscopy of spindle microtubule assembly and motility in living cells. Methods Cell Biol. 1999;61:155–173. doi: 10.1016/s0091-679x(08)61980-9. [DOI] [PubMed] [Google Scholar]
- Weart R.B., Levin P.A. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J., Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Youngman P., Perkins J.B., Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- Zhao C.R., de Boer P.A., Rothfield L.I. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. 1995;92:4313–4317. doi: 10.1073/pnas.92.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]