Abstract
Mixed-lineage leukemia (MLL) fusion proteins are potent inducers of leukemia, but how these proteins generate aberrant gene expression programs is poorly understood. Here we show that the MLL-AF4 fusion protein occupies developmental regulatory genes important for hematopoietic stem cell identity and self-renewal in human leukemia cells. These MLL-AF4-bound regions have grossly altered chromatin structure, with histone modifications catalyzed by trithorax group proteins and DOT1 extending across large domains. Our results define direct targets of the MLL fusion protein, reveal the global role of epigenetic misregulation in leukemia, and identify new targets for therapeutic intervention in cancer.
Keywords: Chromatin, epigenetic, genome-wide, leukemia, stem cell, transcription
Chromosomal translocations involving the mixed-lineage leukemia gene (MLL) are a frequent occurrence in human acute leukemias of both children and adults (Eguchi et al. 2005). In over half of all infant acute leukemias, the MLL protein fuses to one of >50 identified partner genes, resulting in a MLL fusion protein that acts as a potent oncogene (Krivtsov and Armstrong 2007). While extensive gene expression signatures have been determined for primary human leukemia samples (Armstrong et al. 2002; Yeoh et al. 2002; Ferrando et al. 2003; Ross et al. 2003; Rozovskaia et al. 2003; Haferlach et al. 2005), the direct genomic targets of MLL fusion proteins remain unknown. This information is essential to determine how MLL fusion proteins impose oncogenic transcriptional programs and to identify targets for therapeutic intervention in human disease.
Distinct chromatin-modifying complexes and histone modifications are associated with distinct phases of transcription (Li et al. 2007). The trithorax group proteins, including MLL, catalyze histone H3-Lys-4 trimethyl (H3K4me3) modifications at the start sites of transcriptionally engaged genes (Ruthenburg et al. 2007). These H3K4me3-modified regions are largely constrained to the transcription start site regions of genes that are transcriptionally initiated, but not necessarily fully transcribed (Bernstein et al. 2006; Barski et al. 2007; Guenther et al. 2007). As a gene becomes fully transcribed, elongating RNA Polymerase II (Pol II) molecules proceed through gene coding regions along with associated elongation factors including DOT1, which catalyzes dimethylation of histone H3-Lys-79 (H3K79me2) (Li et al. 2007). Physical interactions between the most common MLL partner proteins and transcriptional elongation components suggest that defects in H3K4 and H3K79 methylation might be a key factor in MLL leukemogenesis (Erfurth et al. 2004; Milne et al. 2005b ; Okada et al. 2005; Zeisig et al. 2005; Bitoun et al. 2007; Mueller et al. 2007), but the mechanism and extent of H3K79 methylation targeting throughout the genome is poorly understood in human cancer cells.
In order to define the portion of gene regulatory circuitry that is controlled directly by MLL fusion proteins in human leukemia, we determined the binding patterns of an MLL fusion protein and chromatin modifications across the entire human genome. We performed this mapping in leukemic cells harboring the MLL-AF4 fusion gene, because this rearrangement is the most common among MLL fusions and is associated with an extremely poor prognosis in infants and adults (Eguchi et al. 2005). Our results reveal that the MLL-AF4 oncogene produces gross defects in chromatin structure at a newly defined set of hematopoietic stem cell genes.
Results and Discussion
Identification of MLL-AF4-occupied regions of the genome in human leukemia cells
We used chromatin immunoprecipitation (ChIP) coupled to massively parallel sequencing (ChIP-seq) to determine how the MLL-AF4 fusion protein was distributed across the entire genome in human leukemia cells. This was performed using two acute lymphoblastic leukemia (ALL) cell lines (Fig. 1). The SEM cell line, which was derived from precursor B-cell ALL patient blast cells (Greil et al. 1994) harbors a t(4;11) chromosomal translocation and expresses MLL-AF4 fusion protein, endogenous MLL and endogenous AF4. The REH precursor B-cell ALL patient-derived cell line (Rosenfeld et al. 1977), which expresses only wild-type MLL and wild-type AF4, served as a control to identify regions bound by normal MLL and AF4, but not by the MLL-AF4 fusion protein. Because REH cells were derived from patients with B-cell ALL, these cells also served to control for general effects of B-cell-type leukemia.
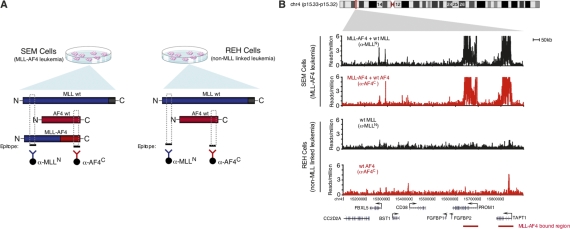
Figure 1.
Mapping MLL-AF4 fusion protein-binding sites in human leukemia cells. (A) Schematic diagram of strategy for mapping MLL-AF4 fusion protein-binding sites. SEM precursor B acute leukemia cells express the MLL-AF4 fusion protein. REH precursor B acute leukemia cells express only endogenous AF4 and MLL1. The N terminus of MLL (blue) is recognized by ChIP antibody anti-MLL-N (blue) and immunoprecipitates both wild-type MLL and MLL-AF4 fusion protein in SEM cells. The C terminus of AF4 (red) is recognized by ChIP antibody anti-AF4-C and immunoprecipitates both wild-type AF4 and MLL-AF4 fusion protein in SEM cells. Wild-type AF4 and MLL-N are immunoprecipitated by anti-AF4-C and anti-MLL-N, respectively. (B) Binding of AF4 (red) and MLL-N (black) in SEM cells (top panels) and REH cells (bottom panels) as determined by ChIP-seq. Binding profiles are shown across an 800-kb portion of the genome surrounding the PROM1 gene (gene models shown in black below graph; a black arrow indicates transcription start sites). MLL-AF4 fusion protein binding is indicated by a red bar.
We mapped protein–DNA interactions for the MLL-AF4 protein in SEM cells using antibodies to the C terminus of AF4 (anti-AF4C) and the N terminus of MLL (anti-MLLN) (Fig. 1A). Since REH cells do not express MLL-AF4 fusion protein, the sites of MLL and AF4 occupancy that were highly similar in SEM and REH cells are most likely due to binding of normal MLL or AF4 proteins. We expected large numbers of regions in both SEM and REH to be occupied by either AF4 or MLL, since these proteins are widely involved in activated transcription by Pol II (Guenther et al. 2005; Milne et al. 2005a; Bitoun et al. 2007). Indeed, we found that MLL or AF4 localized near the transcription start sites of several thousand genes in both SEM and REH cells (Supplemental Table S1). These bound regions tended to be relatively small, extended 1–3 kb into the 5′ end of the transcribed portion of the gene (Supplemental Fig. S1), and were associated with transcriptionally active genes (>80% of transcripts called present by array-based methods in each cell type) (Supplemental Table S2). These data identified the set of genes occupied by MLL and/or AF4 in precursor B ALL cells and confirmed that these genes tend to be transcriptionally active.
We next examined the genome for evidence of binding events specific to MLL fusion protein. To identify MLL-AF4 fusion protein targets, we applied an algorithm that identifies coincident ChIP-seq signals for MLLN and AF4C (Supplemental Material). We identified 226 regions of MLL-AF4 co-occupancy across the genome in SEM cells. This co-occupancy pattern did not occur in REH cells. Many of the MLL-AF4 target regions showed a striking behavior; the signal spanned regions of 5–100 kb (Fig. 1B). This binding pattern of MLL-AF4 occurred at genes encoding a variety of important developmental regulators (Fig. 2). For example, it occurred at the PROM1 (prominin-1/CD133) gene (Fig. 1B), which encodes a defining antigen of hematopoietic stem cells and is purported to play a role in asymmetric cell divisions in adult stem cells (Wagner et al. 2004; Toren et al. 2005), and HOXA7, HOXA9, HOXA10 (Fig. 2A), whose overexpression are hallmarks of ALLs carrying the t(4;11) chromosome translocation (Rozovskaia et al. 2001, 2003; Armstrong et al. 2002; Yeoh et al. 2002; Ferrando et al. 2003). Overall, MLL-AF4 occupied large domains of 169 known RefSeq genes that were previously unknown as direct targets (Fig. 2B,C; complete gene list in Supplemental Table S2).
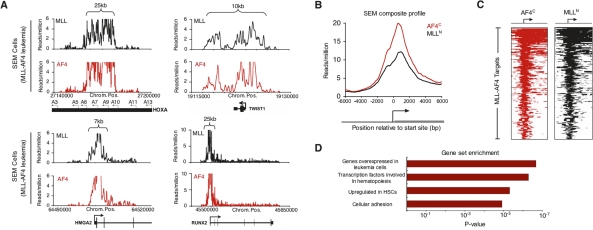
Figure 2.
MLL-AF4 target genes are enriched for early developmental regulators. (A) Signals for AF4 (red) and MLL-N (black) reflecting binding of the presumptive MLL-AF4 fusion protein in SEM cells as determined by ChIP-seq. Binding profiles are shown across a 15- to 150-kb portion of the genome surrounding the HOXA9, TWIST1, HMGA2, and RUNX2 genes (gene models shown in black below graph; a black arrow indicates transcription start sites). Size of MLL-AF4-enriched region is indicated by top brackets. A detailed description of data analysis methods is provided in the Supplemental Material. (B) Composite AF4-C terminus (red) and MLL-N terminus (black) binding profiles for all MLL-AF4 target genes. The start site and direction of transcription of the average gene are indicated by an arrow. (C) ChIP-seq density heat map of AF4-C terminus (red) and MLL-N terminus (black) for all MLL-AF4 target genes. The genomic region from −5kb to +10kb relative to the transcription start site of each gene is shown. Gene order is determined by highest average MLL/AF4 read density from top to bottom. The start site and direction of transcription of the genes are indicated by an arrow. (D) Selected results of GSEA (http://www.broad.mit.edu/gsea) of MLL-AF4 target genes .
MLL-AF4 target genes encode hematopoietic stem cell developmental regulators
MLL-AF4 target genes were analyzed by Gene Set Enrichment Analysis (GSEA, htttp://www.broad.mit.edu/gsea) (Subramanian et al. 2005) to determine whether any known phenotypes were associated with the gene set. The most highly enriched subsets were genes overexpressed in leukemia cells and genes that encode transcription factors involved in hematopoiesis (Fig. 2D). Genes from the transcription factor group included HOXA9, RUNX1, and ETV6, all of which displayed large areas of MLL-AF4 binding that extended well into the coding regions of these important proleukemia genes. We also discovered other developmental regulators not previously associated with ALL, including the TWIST1 gene. The TWIST1 transcription factor is an essential mediator of metastatic growth in human breast cancer cells and also plays a role in nephroblastomas, neuroblastomas, and gastric cancer progression (Pajer et al. 2003; Yang et al. 2004). The TWIST1 protein has also been shown to inhibit apoptosis (Puisieux et al. 2006). Other targets, including the RUNX2 transcription factor and JMJD1C histone demethylase, were previously shown to be up-regulated in MLL-AF9-induced leukemic stem cells (Krivtsov et al. 2006), but were not known to be direct targets of MLL fusion proteins. Interestingly, UTX is a chromatin modifier responsible for activating HOX loci (Cloos et al. 2008), suggesting a possible reinforcement of HOXA locus overexpression in leukemia. Another category of genes observed in the GSEA analysis was a set of genes differentially expressed in hematopoietic stem cells (Fig. 2D; Ramalho-Santos et al. 2002). These included PROM1/CD133, which encodes a surface antigen that is a defining marker of hematopoietic stem and progenitor cells and is highly expressed in tumor-initiating cells of the colon and brain (Toren et al. 2005; O’Brien et al. 2007), and the FLT3 signaling mediator. The enrichment for developmental regulatory factors indicates that the MLL-AF4 oncogene activiates specialized transcriptional programs in cancer cells.
MLL-AF4 targets predict leukemia subclass in human patients
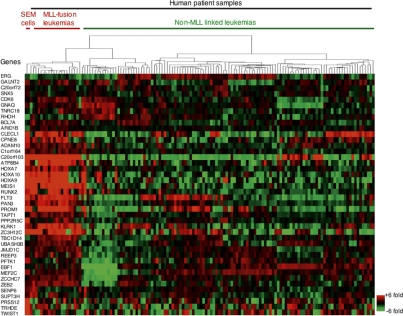
We next tested whether MLL-AF4 target genes identified in SEM cells had altered gene expression patterns in human patients with MLL-associated leukemia. If MLL-AF4 target genes specify the MLL-associated leukemia subclass in human cancer, we would expect up-regulation of these genes in patients exhibiting MLL-associated acute leukemia, but not in patients exhibiting non-MLL-linked leukemia. RNA transcript levels from leukemic blasts of 132 pediatric ALL patients of B and T lineages (Ross et al. 2003) were compared for expression of the MLL-AF4 target genes discovered in SEM cells (Fig. 3; Supplemental Fig. S4). Significantly, about two-thirds of MLL-AF4 targets in SEM cells were at least 50% overexpressed in patients with MLL-associated leukemia, but not in non-MLL-associated leukemia. This overexpression of MLL-AF4 targets was evident not only in MLL-AF4-derived patients’ samples, but also in MLL-AF9 and other MLL-derived leukemias (Ross et al. 2003), suggesting a central role of this core gene set in the most common MLL-associated leukemias. The concordance of our MLL-AF4 targets discovered in SEM cells and gene expression signatures in human leukemia patients indicates that the MLL-AF4 target genes discovered in vitro are important for disease progression in vivo.
Figure 3.
MLL-AF4 target genes define MLL-linked leukemia in vivo. Hierarchical clustering of relative expression levels of 42 genes occupied by MLL-AF4 fusion protein target regions >10 kb. Comparisons were made across the SEM and REH cell lines and 132 peripheral blood samples of patients diagnosed with leukemia. Each row corresponds to a gene that is bound by MLL-AF4 for which expression data were available. Each column corresponds to a single gene expression microarray. For each gene, expression is shown relative to the average expression level of that gene across all samples, with shades of red indicating higher than average expression and green lower than average expression. Columns and rows were ordered by unsupervised hierarchical clustering. A detailed description of data analysis methods is provided in the Supplemental Material.
Aberrant chromatin domains occur at regions of MLL-AF4 occupancy
Aberrant modification of chromatin is linked to disease progression in leukemia and other cancers (Jones and Baylin 2007). Among these modifications is the methylation of histone H3 at Lys-79 (H3K79me2), which occurs at the 5′ coding regions of genes that are experiencing productive transcriptional elongation (Steger et al. 2008), and is a critical checkpoint in transcriptional control (Peterlin and Price 2006; Saunders et al. 2006). Many common MLL partner proteins have been shown to interact with transcriptional elongation components, suggesting that H3K79 methylation might be a key factor in MLL leukemogenesis (Erfurth et al. 2004; Milne et al. 2005b; Okada et al. 2005; Zeisig et al. 2005; Bitoun et al. 2007; Mueller et al. 2007), but the mechanism and extent of H3K79me2 targeting throughout the genome is poorly understood in human cancer cells.
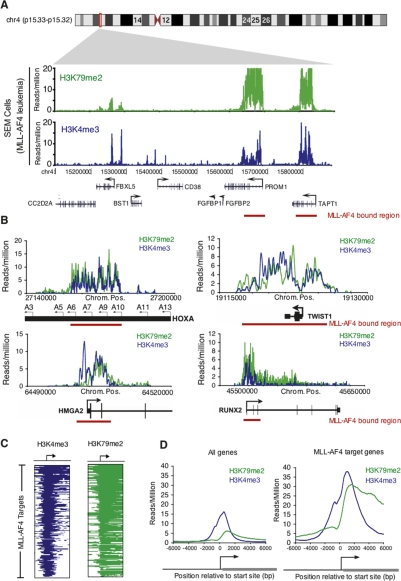
We used ChIP-seq to determine how the H3K79me2 chromatin modification was distributed across the genome in SEM cells and to ascertain whether this modification was associated with all of the MLL-AF4 target regions (Fig. 4). We found ∼8000 regions of H3K79me2 enrichment in MLL-AF4 leukemia cells or control cells, with the vast majority (95%) mapping to known transcripts (Supplemental Tables S14, S19). As expected, most genes (∼95%) marked by H3K79me2 in MLL-AF4 leukemia and in control cells produced transcripts that were detectable by microarray-based methods (Supplemental Table S2), with peak enrichment occurring downstream from the transcription start sites (Fig. 4D; Supplemental Fig. S5). We next extracted the set of H3K79me2-enriched regions in SEM cells and compared them with genomic regions enriched for MLL-AF4 fusion protein. This analysis revealed that ∼98% of MLL-AF4 targets were enriched for the H3K79me2 elongation mark. Strikingly, most of these H3K79me2 enrichments formed abnormal domains spanning 5–100 kb extending upstream of and/or downstream from target gene transcriptional start sites in a highly similar pattern to the MLL-AF4 fusion protein (Fig. 4A,B). In fact, there was a 92% overlap between the MLL-AF4 target regions and H3K79me2-enriched regions at the DNA base-pair level. Based on the induction of these chromatin modifications at MLL-AF4 target regions, we speculate that the MLL-AF4 fusion protein is directly involved in establishing these aberrant chromatin domains in MLL-linked cancer cells.
Figure 4.
Mistargeting of chromatin modifications occur as epigenetic lesions at MLL-AF4 target regions. (A) Binding of H3K79me2 (green) and H3K4me3 (blue) in SEM cells as determined by ChIP-seq. Binding profiles are shown across an 800-kb portion of the genome surrounding the PROM1 gene. Gene models shown in black below the graph; a black arrow indicates transcription start sites. MLL-AF4 fusion protein-binding regions are indicated by red bars. (B) Binding of H3K79me2 ChIPs (green) and H3K4me3 ChIPs (blue) in SEM cells as determined by ChIP-seq. Binding profiles are shown across 15- to 150-kb portions of the genome surrounding the HOXA9, TWIST1, HMGA2, and RUNX2 genes. Gene models are shown in black (below graph). A black arrow indicates transcription start sites. MLL-AF4 fusion protein binding is indicated by a red bar. (C) H3K79me2 (green) and H3K4me3 (blue) binding profiles for all MLL-AF4 target genes in SEM cells. Genes are ordered as in Figure 2C. (D) Composite H3K79me2 ChIP enrichments (green) and H3K4me3 ChIP enrichments (blue) for all genes (left) and all MLL-AF4 target genes (right). The start site and direction of transcription of the average gene are indicated by an arrow.
Since the elongation-linked H3K79me2 modification forms across large domains with MLL-AF4 (Fig. 4A–C), we asked whether regulators of the elongation checkpoint behaved similarly. Indeed, elongation proteins were present across aberrant H3K79me2 domains as determined by ChIP–chip and coimmunoprecipitation experiments. The ENL elongation factor (eleven-nineteen leukemia) bound across aberrant H3K79me2 domains at the HOXA and MEIS1 loci (Supplemental Fig. S6; data not shown) and MLL-AF4 associated with ENL and the pTEFb elongation factor in SEM cells (Supplemental Fig. S7; data not shown). Together, these results indicate that the MLL-AF4 protein, the elongation-associated H3K79me2 modification, and additional elongation factors are mistargeted to regions of the genome encoding key developmental regulators.
We next carried out ChIP-seq experiments in MLL-AF4 leukemia cells using an antibody directed against the histone H3K4me3 modification. This modification is a mark of transcriptional initiation that can be deposited by the MLL complex near the start sites of genes in normal cells (Ruthenburg et al. 2007). Since MLL suffers a monoallelic deletion of its SET-containing the H3K4 methyltransferase domain in MLL-AF4 leukemia, we asked whether MLL-AF4 leukemia cells were able to deposit this histone modification normally across the genome. We found that H3K4me3 modification occurred normally at the start sites of most active genes (∼90%) in MLL-AF4 leukemia cells (Supplemental Table S2). Strikingly, at many areas of H3K79me2 mistargeting, H3K4me3 was not only present, but also extended across broad domains of similar size (Fig. 4A–C). These chromatin modifications overlapped highly with the MLL-AF4 fusion and occurred at key leukemia and stem cell-associated genes including HOXA7, HOXA9, PROM1, and HMGA2 (Fig. 4; Supplemental Table S2). This mislocalization of both H3K79me2 and H3K4me3 modifications in MLL-AF4-induced leukemia suggests that the MLL-AF4 fusion protein, a strong transcriptional activator, may be acting at target loci by directly coupling transcriptional initiation and elongation machinery.
A model for MLL-AF4-mediated activation of stem cell-like transcriptional program in leukemia
We describe here the first genome-wide assessment of chromatin modifications and oncogene binding in the most common form of human MLL-associated acute leukemia. The results indicate that MLL-AF4 fusion proteins selectively occupy regions of the genome that contain developmental regulators important for stem cell identity and self-renewal. Our results also show that abnormal patterns of chromatin modifications, including histone H3K79 and histone H3K4 hypermethylation, occur within large domains of MLL-AF4 occupancy. The observation that MLL-AF4 occupies regions of H3K79 hypermethylation and evidence that MLL-AF4 is physically associated with elongation factors including DOT1 strongly suggests that the fusion protein is responsible for generating large domains of H3K79 hypermethylation. The presence of these aberrant chromatin domains demonstrates that chromatin mistargeting to key regions across the genome is a feature of leukemogenesis.
The presence of aberrant chromatin domains in MLL-linked cancer cells suggests that an abnormal “epigenetic” state exists in these cells. Unlike “genetic lesions” that involve changes in genome sequence in disease cells, “epigenetic lesions” include changes to histone modification states, DNA methylation states, or distribution of chromatin-modifying enzymes (Esteller 2007; Feinberg 2007). By this definition, the aberrant chromatin domains associated with MLL-AF4 binding may thus be considered epigenetic lesions. While it has not been established that these epigenetic lesions cause disease, the H3K79 methylation within these domains likely contributes to oncogenesis since H3K79 methyltransferase activity is required for transformation in MLL-AF10-induced leukemia (Okada et al. 2005). The mechanism of how MLL-AF4 and epigenetic lesions are targeted in the genome remains a central question in leukemia biology.
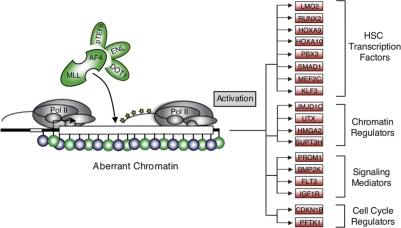
The binding of the MLL-AF4 fusion protein to a distinct set of developmental genes is of particular interest. Rather than associating with most cellular genes that are engaged in the act of transcription, the fusion protein imposes a more specialized gene expression program. Components of this program include genes not associated previously with MLL-AF4/H3K79me2 mistargeting in leukemia patient-derived cells. These include developmental transcription factors, chromatin regulators, and signaling proteins that are central to leukemia stem cell identity, hematopoietic stem cell identity, and self-renewal (Fig. 5). Our findings suggest that MLL-AF4 directly activates a partial hematopoietic stem cell-like transcriptional program found in leukemia stem cells (Krivtsov et al. 2006; Barabe et al. 2007) in concert with an underlying gross defect in chromatin structure.
Figure 5.
Model of MLL-AF4-mediated gene activation in leukemia. Schematic diagram of transcriptional misregulation in MLL-AF4-induced leukemia. MLL-AF4 associates with elongation proteins (pTEFb, ENL, and DOT1) at sites of epigenetic lesions. Phosphorylated and elongating RNA Pol II shown in yellow. Blue (H3K4me3) and green (H3K79me2) circles represent histone modifications at aberrant chromatin domains. Genes annotated as bound by MLL-AF4 and playing a role in hematopoietic stem cell function are shown to the right and subdivided into functional categories.
Materials and methods
A detailed description of all experimental procedures and data analysis methods can be found in the Supplemental Material.
Cells and cell culture
Human SEM cells with the t(4;11) translocation and REH control cells were purchased from the American Type Culture Collection (ATCC). All cell lines were maintained in RPMI medium 1640 supplemented with 10% FBS. Cells were cross-linked with 1% formaldehyde as described in the Supplemental Material.
ChIP-seq
ChIP was combined with direct sequencing as described in detail (Supplemental Material). Briefly, DNA from 1 × 108 cells was immunoprecipitated with epitope-specific antibody. Amplified DNA was gel purified and prepared for sequencing using Illumina’s Genomic DNA sample kit. Clustering and 26-cycle sequencing were performed using an Illumina Cluster station and 1G analyzer as per the manufacturer’s instructions. ChIP-seq reads were aligned to the human genome and analyzed as described in the Supplemental Material.
Gene expression analysis
Total RNA was isolated from 5 × 106 REH or SEM cells by TRIzol extraction. One microgram of total RNA was labeled according to Affymetrix protocols and hybridized to Affymetrix HG-U133 2.0 plus arrays. The data were analyzed by using Affymetrix Gene Chip Operating Software using default settings. Additional gene expression data from human leukemia patient samples was collated from Ross et al. (2003). Gene expression data and analysis results are provided in the Supplemental Material.
ChIP-seq, ChIP–chip, and microarray gene expression data and analysis
Complete ChIP-seq, ChIP–chip, and microarray gene expression data, analysis methods, and results are provided in the Supplemental Material section and in the Gene Expression Omnibus (http://www.ncbi.nlm. nih.gov/geo) database under accession number GSE13313.
Acknowledgments
We thank Y. Zonis, T. Lee, A. Marson, F. Camargo, S. Gupta, and J. Love for help and discussions. We thank the National Cell Culture Center (NCCC) for REH and SEM cells. This work was supported by grants from the Israel Cancer Research Fund and the Wolfson Foundation (to E.C.), the US-Israel BSF (to E.C. and C.M.C.), NIH grant CA 128609 (to C.M.C. and T.N.), and NIH grant HG002668 (to R.Y.). L.N.L. was supported by individual NRSA fellowship from NHLBI (grant no. F32HL082448).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1741408.
References
- Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., Korsmeyer S.J. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Barabe F., Kennedy J.A., Hope K.J., Dick J.E. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bitoun E., Oliver P.L., Davies K.E. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes & Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi M., Eguchi-Ishimae M., Greaves M. Molecular pathogenesis of MLL-associated leukemias. Int. J. Hematol. 2005;82:9–20. doi: 10.1532/IJH97.05042. [DOI] [PubMed] [Google Scholar]
- Erfurth F., Hemenway C.S., de Erkenez A.C., Domer P.H. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia. 2004;18:92–102. doi: 10.1038/sj.leu.2403200. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: The DNA hypermethylome. Hum. Mol. Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Armstrong S.A., Neuberg D.S., Sallan S.E., Silverman L.B., Korsmeyer S.J., Look A.T. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: Dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- Greil J., Gramatzki M., Burger R., Marschalek R., Peltner M., Trautmann U., Hansen-Hagge T.E., Bartram C.R., Fey G.H., Stehr K., et al. The acute lymphoblastic leukaemia cell line SEM with t(4;11) chromosomal rearrangement is biphenotypic and responsive to interleukin-7. Br. J. Haematol. 1994;86:275–283. doi: 10.1111/j.1365-2141.1994.tb04726.x. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Jenner R.G., Chevalier B., Nakamura T., Croce C.M., Canaani E., Young R.A. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T., Kohlmann A., Schnittger S., Dugas M., Hiddemann W., Kern W., Schoch C. Global approach to the diagnosis of leukemia using gene expression profiling. Blood. 2005;106:1189–1198. doi: 10.1182/blood-2004-12-4938. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Armstrong S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Milne T.A., Dou Y., Martin M.E., Brock H.W., Roeder R.G., Hess J.L. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. 2005a;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T.A., Martin M.E., Brock H.W., Slany R.K., Hess J.L. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005b;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Mueller D., Bach C., Zeisig D., Garcia-Cuellar M.P., Monroe S., Sreekumar A., Zhou R., Nesvizhskii A., Chinnaiyan A., Hess J.L., et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Pajer P., Pecenka V., Karafiat V., Kralova J., Horejsi Z., Dvorak M. The twist gene is a common target of retroviral integration and transcriptional deregulation in experimental nephroblastoma. Oncogene. 2003;22:665–673. doi: 10.1038/sj.onc.1206105. [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Puisieux A., Valsesia-Wittmann S., Ansieau S. A twist for survival and cancer progression. Br. J. Cancer. 2006;94:13–17. doi: 10.1038/sj.bjc.6602876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rosenfeld C., Goutner A., Choquet C., Venuat A.M., Kayibanda B., Pico J.L., Greaves M.F. Phenotypic characterisation of a unique non-T, non-B acute lymphoblastic leukaemia cell line. Nature. 1977;267:841–843. doi: 10.1038/267841a0. [DOI] [PubMed] [Google Scholar]
- Ross M.E., Zhou X., Song G., Shurtleff S.A., Girtman K., Williams W.K., Liu H.C., Mahfouz R., Raimondi S.C., Lenny N., et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T., Feinstein E., Mor O., Foa R., Blechman J., Nakamura T., Croce C.M., Cimino G., Canaani E. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T., Ravid-Amir O., Tillib S., Getz G., Feinstein E., Agrawal H., Nagler A., Rappaport E.F., Issaeva I., Matsuo Y., et al. Expression profiles of acute lymphoblastic and myeloblastic leukemias with ALL-1 rearrangements. Proc. Natl. Acad. Sci. 2003;100:7853–7858. doi: 10.1073/pnas.1132115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A.J., Allis C.D., Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Saunders A., Core L.J., Lis J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Steger D.J., Lefterova M.I., Ying L., Stonestrom A.J., Schupp M., Zhuo D., Vakoc A.L., Kim J.E., Chen J., Lazar M.A., et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toren A., Bielorai B., Jacob-Hirsch J., Fisher T., Kreiser D., Moran O., Zeligson S., Givol D., Yitzhaky A., Itskovitz-Eldor J., et al. CD133-positive hematopoietic stem cell “stemness” genes contain many genes mutated or abnormally expressed in leukemia. Stem Cells. 2005;23:1142–1153. doi: 10.1634/stemcells.2004-0317. [DOI] [PubMed] [Google Scholar]
- Wagner W., Ansorge A., Wirkner U., Eckstein V., Schwager C., Blake J., Miesala K., Selig J., Saffrich R., Ansorge W., et al. Molecular evidence for stem cell function of the slow-dividing fraction among human hematopoietic progenitor cells by genome-wide analysis. Blood. 2004;104:675–686. doi: 10.1182/blood-2003-10-3423. [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yeoh E.J., Ross M.E., Shurtleff S.A., Williams W.K., Patel D., Mahfouz R., Behm F.G., Raimondi S.C., Relling M.V., Patel A., et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Zeisig D.T., Bittner C.B., Zeisig B.B., Garcia-Cuellar M.P., Hess J.L., Slany R.K. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene. 2005;24:5525–5532. doi: 10.1038/sj.onc.1208699. [DOI] [PubMed] [Google Scholar]