Abstract
Cytoplasmic polyadenylation element-binding protein (CPEB) stimulates polyadenylation and translation in germ cells and neurons. Here, we show that CPEB-regulated translation is essential for the senescence of human diploid fibroblasts. Knockdown of CPEB causes skin and lung cells to bypass the M1 crisis stage of senescence; reintroduction of CPEB into the knockdown cells restores a senescence-like phenotype. Knockdown cells that have bypassed senescence undergo little telomere erosion. Surprisingly, knockdown of exogenous CPEB that induced a senescence-like phenotype results in the resumption of cell growth. CPEB knockdown cells have fewer mitochondria than wild-type cells and resemble transformed cells by having reduced respiration and reactive oxygen species (ROS), normal ATP levels, and enhanced rates of glycolysis. p53 mRNA contains cytoplasmic polyadenylation elements in its 3′ untranslated region (UTR), which promote polyadenylation. In CPEB knockdown cells, p53 mRNA has an abnormally short poly(A) tail and a reduced translational efficiency, resulting in an ∼50% decrease in p53 protein levels. An shRNA-directed reduction in p53 protein by about 50% also results in extended cellular life span, reduced respiration and ROS, and increased glycolysis. Together, these results suggest that CPEB controls senescence and bioenergetics in human cells at least in part by modulating p53 mRNA polyadenylation-induced translation.
Keywords: CPEB, senescence, polyadenylation, translation, bioenergetics, p53
Initiation is the rate-limiting step for translation of most eukaryotic mRNAs and requires both a 5′-methylated guanosine cap (m7G[5′]ppp[5′]N) and a 3′ poly(A) tail. The ends of the RNA are brought into close proximity by a protein–protein–protein bridge composed of eIF4E (the cap-binding factor), eIF4G, and poly(A)-binding protein (PABP) (Tarun and Sachs 1996; Tarun et al. 1997; Wells et al. 1998). PABP may facilitate the interaction of eIF4G with eIF4E, which is necessary for initiation since eIF4G, via the multisubunit eIF3, positions the 40S ribosomal subunit on the 5′ end of the mRNA (Sonenberg and Hinnebusch 2007). One mode of translational control that is particularly important for germ cell development is the abrogation of this 5′–3′ protein bridge by the near absence of the poly(A) tail (and hence PABP) and by the association of a specialized eIF4E-binding protein with some mRNAs. These inert (masked) mRNAs are activated by subsequent poly(A) tail growth, the binding of PABP to poly(A), and the replacement of the eIF4E-binding protein with eIF4G. The cytoplasmic polyadenylation element (CPE) controls poly(A) tail length; it resides in mRNA 3′ untranslated regions (UTRs) and serves as the binding site for CPE-binding protein (CPEB), a factor that associates with Gld2, a poly(A) polymerase (Barnard et al. 2004); PARN, a deadenylating enzyme (Kim and Richter 2006); ePAB, a PABP (Kim and Richter 2007); Maskin, an eIF4E-binding protein (Stebbins-Boaz et al. 1999; Cao et al. 2006); CPSF, a multisubunit RNA-binding complex (Mendez et al. 2000; Dickson et al. 2001); and symplekin, a probable scaffold or assembly protein (Barnard et al. 2004). In response to developmental cues, CPEB becomes phosphorylated, causing the expulsion of PARN from the RNP complex and results in Gld2-catalyzed polyadenylation (Mendez et al. 2000; Kim and Richter 2006). The newly elongated poly(A) tail is bound by ePAB that in turn binds eIF4G; this complex displaces Maskin from eIF4E, thus circularizing the RNA and promoting initiation (Barnard et al. 2005; Cao et al. 2006; Kim and Richter 2006).
CPEB-controlled translation has also been found to modulate neuronal synaptic plasticity (Klann and Richter 2007; Richter 2007) and cellular senescence in murine embryonic fibroblasts (MEFs) (Groisman et al. 2006). Like apoptosis, senescence is a mechanism that prohibits unrestricted cell proliferation. DNA damage, nutrient deprivation, improper cell contacts, and oncogenic signaling all converge on the p53 and/or retinoblastoma (Rb) tumor suppressor pathways to initiate cell cycle arrest and entry into senescence (Lowe et al. 2004; Campisi and d’Adda di Fagagna 2007). While senescence is usually examined in cultured cells, recent studies in animals have demonstrated that it is an important barrier to malignant transformation (Braig et al. 2005; Chen et al. 2005; Collado et al. 2005; Michaloglou et al. 2005).
The bypass of senescence, or immortalization, is required for, but does not necessarily lead to, cellular transformation. Although both phenomena are often studied in mouse and human cells, there are a number of differences between the two organisms, indicating the complexity of molecules that influence these processes. For example, cultured mouse cells are relatively easy to transform, requiring the addition of only one or two oncogenes (Drayton and Peters 2002). Normal diploid human cells, however, are more refractory and require the inactivation of the p53 and Rb proteins, as well as the activation of the catalytic subunit of the telomerase, hTERT. In addition, mouse telomeres are typically 25–60 kb in length while those in humans are much shorter, ∼10–15 kb; thus, telomere erosion is not necessarily essential for senescence in mouse cells while it is in human cells (Chin et al. 1999). Moreover, murine cells senesce abruptly in culture five to 10 times faster than human cells. These and other differences between mouse and human cells notwithstanding, it is increasingly evident that in both species, cellular and replicative senescence are not only triggered by cellular perturbations that generate a DNA damage response, such as oncogene expression and cell culture stress (Lee et al. 1999; Wu et al. 2004), but that senescence onset and maintenance requires a sustained DNA damage response (Hemann and Narita 2007).
While comparing rates of mitosis between MEFs derived from wild-type and CPEB knockout (KO) mice, we made the startling observation that the KO MEFS did not senesce as did wild-type MEFs, but instead were immortal (Groisman et al. 2006). Reintroduction of CPEB into early passage KO MEFs induced them to senesce; reintroduction of CPEB into late passage KO MEFs (i.e., those that had gone through several passages beyond when wild-type MEFs senesce) did not. CPEB-mediated senescence required the tumor suppressors p53 and p19ARF; conversely, Ras-induced senescence required CPEB. Myc protein levels were elevated in the KO MEFs and more myc mRNA was found on polysomes compared with wild-type MEFs. Most importantly, shRNA knockdown of myc in the KO MEFs caused them to become senescent. These data indicated that in mouse cells, the removal of CPEB-inhibited myc RNA translation led to immortalization.
Here, we investigate the importance of CPEB for senescence of normal human diploid fibroblasts. While wild-type and mock-infected human foreskin fibroblasts senesced after ∼70 population doublings, those infected with a lentivirus harboring a shRNA against CPEB did not; the life span of these cells was extended by nearly fivefold. A second infection of these knockdown cells with a retrovirus expressing murine CPEB restored a senescence-like phenotype in early but not late passage cells. Human WI-38 lung fibroblasts also bypassed senescence when CPEB levels were reduced. As expected, CPEB knockdown cells that bypassed senescence retained long telomeres. Interestingly, promotion of the senescence-like phenotype by over expressed CPEB could be reversed when the levels of this protein were reduced. CPEB knockdown cells had abnormally low levels of mitochondria; they also resemble transformed cells in that they had reduced respiration, but an elevated rate of glycolysis presumably to maintain homeostatic ATP levels. CPEB knockdown cells contained ∼50% reduction of p53, and p53 mRNA had a shorter poly(A) tail and a reduced translational efficiency compared with p53 mRNA in wild-type cells. An ∼50% reduction of p53 levels in cells containing normal levels of CPEB also bypassed senescence and had reduced mitochondrial mass and respiration. We propose that the senescence bypass and change in energy metabolism observed in CPEB knockdown cells is due at least in part to dysfunctional p53 mRNA polyadenylation and translation.
Results
CPEB is required for senescence in primary human cells
To determine whether primary human cells require CPEB to become senescent, human diploid foreskin fibroblasts were, in two separate occasions, infected at passage eight with a lentivirus expressing one of two different shRNAs against CPEB RNA (shCPEB) as a control for possible off-target effects. As an additional control, the cells were also infected with an empty lentivirus or one expressing shRNA against mRNA encoding the tetracycline resistance gene (shTETR). The lentivirus vector also expressed GFP, which indicated that ∼70% of cells were infected with the virus. The cells were analyzed for growth and morphology without drug selection.
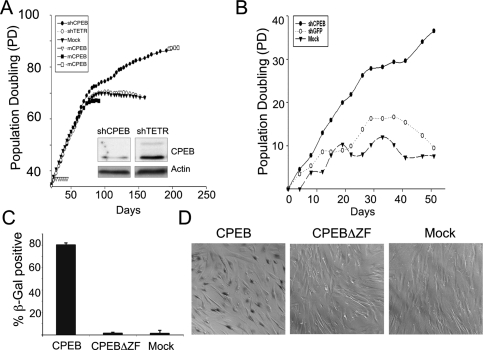
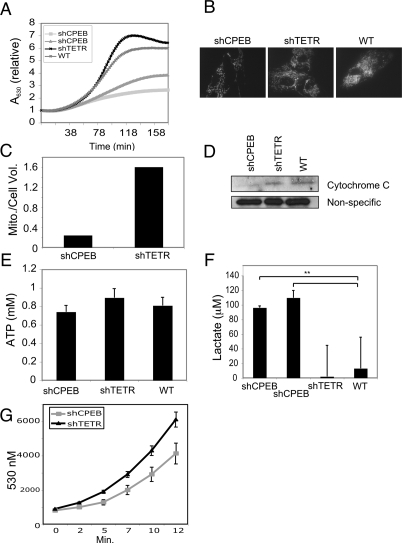
The efficiency of the CPEB knockdown was monitored by both RT–PCR of RNA (data not shown) and by Western analysis (Fig. 1A). CPEB was reduced by >80% compared with the control shTETR knockdown; the top band in Figure 1A might represent phosphorylated CPEB. After ∼68 population doublings, the mock-infected and shTETR-infected cells stopped dividing and assumed a flat senescent-like morphology. These cells also stained for β-galactosidase activity at acidic pH, a common marker for senescence (Fig. 1D; Itahana et al. 2007; data not shown). The shCPEB-infected cells, however, continued to grow, did not undergo a morphology change, and did not stain for β-galactosidase activity (Fig. 1A,D). Moreover, while mock-infected and shTETR-infected cells expressed high levels of p21CIP1 and p16INK4A, which is consistent with entry into senescence, the cells infected with shCPEB did not (Fig. 5, below; data not shown).
Figure 1.
CPEB is necessary for cellular senescence. (A) Human foreskin fibroblasts were infected with a lentiviruses encoding shRNA targeting CPEB mRNA (shCPEB), or the tetracycline resistance mRNA (shTETR), as a control. Additional cells were mock infected with an empty lentivirus. Population doublings were then determined by counting cells with a hemocytometer. Some cells were also infected with a retrovirus expressing mouse CPEB (because of differences in the primary sequence, mouse CPEB mRNA is not a target of the shCPEB), followed by cell counting. The inset shows a Western blot probed for CPEB and actin; these extracts were prepared from cells 40 d post-lentivirus infection. (B) Human lung fibroblasts (WI-38) were infected with lentiviruses targeting CPEB or, as a control GFP; mock refers to infection with an empty virus. The cells were counted as in A. (C,D) Foreskin fibroblasts previously infected with shCPEB were also infected with retroviruses expressing wild-type CPEB or a CPEB lacking a zinc finger, which renders the protein incapable of RNA binding. Some cells were also mock infected. The cells were then stained for b-galactosidase at acidic pH and were counted (C), and visualized by bright-field microscopy 3 d after retrovirus infection (D).
Figure 5.
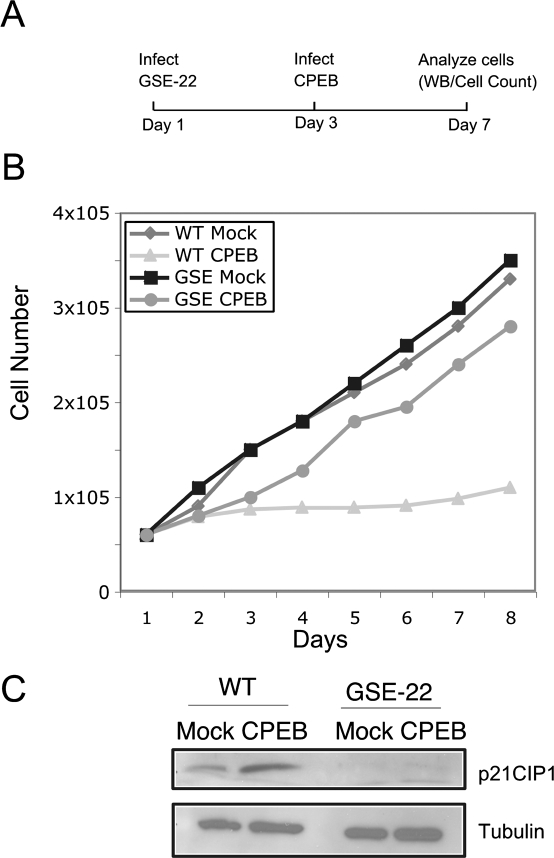
CPEB induced senescence requires p53. (A) Fibroblasts were infected with a retrovirus expressing GSE-22, a p53 dominant-negative peptide. Two days later, the cells were infected with a virus harboring CPEB; examination of the cells began on day 7. (B) Growth curves of GSE-22 or CPEB-infected or mock-infected cells. (C) Western blot analysis of p21 and tubulin in infected or mock-infected cells.
Cells expressing shCPEB were infected with a retrovirus expressing mouse CPEB, which, while >95% identical to human CPEB, would not be a target of the human shCPEB. When the cells were infected with the retrovirus after 20 or 68 doublings, they quickly entered a senescence-like state as indicated by the cessation of cell division, assumption of flattened morphology, and staining for β-galactosidase activity. However, when the cells were infected with the retrovirus after 87 population doublings, none of these events took place. Thus, the reintroduction of CPEB into knockdown cells induces a senescence-like state at early and middle, but not late, passages.
We also assessed whether CPEB is required for senescence in WI-38 human lung fibroblasts. These cells were infected with the same lentiviruses used for the foreskin fibroblasts; while the mock-infected or control-infected cells entered senescence after an additional 10 population doublings, those with reduced levels of CPEB did not and indeed reached at least 38 doublings (Fig. 1B). Thus, at least two human cell types require CPEB to enter senescence.
Finally, the zinc finger of CPEB was deleted, which in addition to containing two RNA recognition motifs, is required for CPE binding (Hake et al. 1998). This mutant protein was unable to restore senescence (a mock infection also did not rescue senescence) (Fig. 1C–E). Therefore, CPEB must bind RNA to induce the senescence-like phenotype.
CPEB is required for the suppression of telomere maintenance
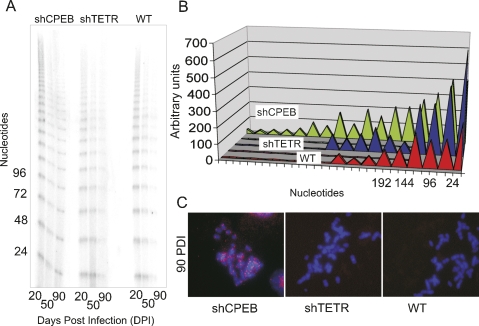
Because senescence in human cells is generally accompanied by telomere loss, we measured telomere length in cells infected with shCPEB by a telomere oligonucleotide ligation assay (T-OLA) (Stewart et al. 2003) and by fluorescence in situ hybridization (FISH) (Henegariu et al. 2001). The T-OLA assay (Fig. 2A,B) shows that, as expected, the telomeres of wild-type or shTETR-infected cells eroded as the cells entered senescence. While the shCPEB-infected cells also underwent some telomere shortening (e.g., at 90 d post-infection), the erosion was not as severe as with the control cells. This maintenance of telomere length was also evident by FISH for the telomeric region (Fig. 2C); that is, at 90 d post-infection with shCPEB, there was clearly hybridization to the telomeric region. In contrast, cells infected with shTETR underwent extensive telomere erosion as evidenced by the lack of a FISH signal to this same region. As expected, wild-type cells also lacked a FISH signal due to telomere shortening. These data indicate that CPEB is required, directly or indirectly, for telomere erosion.
Figure 2.
CPEB knockdown cells retain long telomeres. (A) DNA was extracted from skin fibroblasts infected with shCPEB and shTETR-containing lentiviruses 20, 50, or 90 d post-infection and used to determine telomere length by the T-OLA. (B) The relative telomere lengths derived from the analysis in A were quantified by scanning desitometry. (C) shCPEB, shTETR, and noninfected wild-type (WT) cells were fixed at 90 d post-infection and used for FISH to telomeric regions (telo-FISH) using a Cy3-conjugated locked nucleic acid oligonucleotide (LNA). The DNA was stained with DAPI.
The CPEB-induced senescence-like arrest is reversible
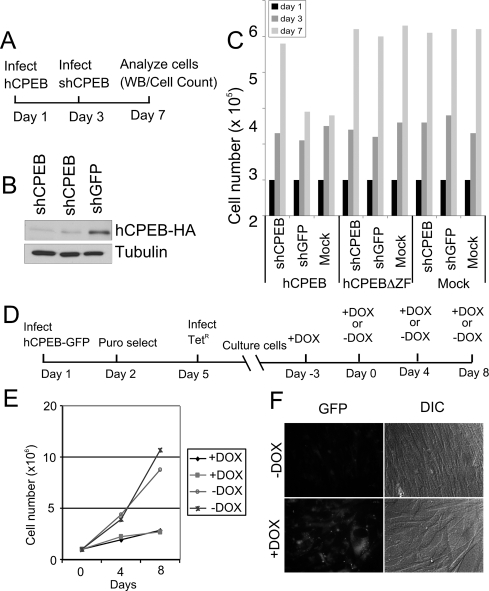
Two approaches were used to determine whether the CPEB-induced senescence-like phenotype is reversible. First, wild-type fibroblasts were infected with lentivirus expressing human HA-CPEB on day one, which was followed 2 d later by the infection of another lentivirus expressing shRNA against this same CPEB; the cells were analyzed 4 d later (Fig. 3A). Western analysis shows that the shRNA efficiently knocked down HA-CPEB (Fig. 3B). As expected, shCPEB, shGFP (another control), or mock lentivirus infections had no effect on cell growth when the cells were also infected with a retrovirus expressing CPEB lacking a zinc finger (hCPEBΔZF), which is unable to bind RNA, or with a retrovirus that expressed no heterologous protein (mock) (Fig. 3C). However, while retrovirally transduced CPEB efficiently attenuated cell growth as shown previously (see Fig. 1), the cells returned to prolific cell division once CPEB was knocked down by the lentivirus-expressed shCPEB (Fig. 3C).
Figure 3.
CPEB-induced senescence-like phenotype is reversible. (A) Schematic representation of one sequence of events used to examine the reversibility of the CPEB-induced senescence-like phenotype. (B) Western blot for CPEB and tubulin derived from fibroblasts that were sequentially infected with a retrovirus encoding HA-tagged human CPEB (hCPEB-HA) followed by a lentivirus encoding shCPEB (two different sequences) or shGFP. (C) Cell numbers were determined following sequential infection of the viruses noted in A. (D) Schematic representation of the experimental timeline when cells were infected with a retrovirus expressing CPEB under the control of the tetracycline repressor. Skin fibroblasts were infected with a virus expressing a CPEB-GFP fusion protein under the control of the tetracycline response element (TET-ON), followed the next day by puromycin selection. Three days later, the cells were infected with a virus encoding the tetracycline repressor (TETR); the cells were then cultured for several days in the presence of DOX, which allows GFP-CPEB expression to remain high. Some cells were then cultured in medium lacking DOX, which will repress GFP-CPEB transcription. (E) Growth chart of two cell lines containing exogenous CPEB-GFP under control of the TETr cultured in the absence or presence of DOX. (F) Live cell images of GFP-CPEB containing cells cultured in the absence or presence of DOX.
In a second approach to address reversibility, sequences encoding a CPEB-GFP fusion protein were cloned into a vector containing the tetracycline response element. Following infection and antibiotic selection, the cells were infected with a lentivirus encoding the tetracycline repressor (TetR); several days later, the cells were incubated with doxycyclin (DOX), which represses the expression of CPEB. The cells were then split; some were further incubated with DOX, while others where transferred to DOX-free medium (−DOX). The cells were analyzed 4 and 8 d later (Fig. 3D). Figure 3E demonstrates that CPEB (−DOX) inhibited cell division; conversely, the subsequent down-regulation of CPEB expression (+DOX) allowed the cells to continue to divide. Finally, CPEB-GFP was readily detected in cells incubated in the presence, but not the absence, of DOX (Fig. 3F). These results demonstrate that the CPEB-induced senescence-like phenotype is reversible.
CPEB is necessary for stress-induced cellular senescence
Cellular senescence is generally thought to be a response to stresses that induce telomere shortening, oncogene activation, reactive oxygen species (ROS), etc. (Ben-Porath and Weinberg 2005). In MEFs, at least one inducer of senescence, constitutively active Ras, requires CPEB (Groisman et al. 2006). This is also the case with human fibroblasts that have reduced CPEB levels (Supplemental Fig. 1). Also, as in CPEB KO MEFs, Ras failed to induce senescence in human fibroblasts with reduced CPEB (Supplemental Fig. 1). Because Ras is involved in modulating levels of ROS (Irani et al. 1997; Archer and Bar-Sagi 2002), we used two approaches to examine whether ROS was also involved in the CPEB-induced senescence-like phenotype. First, wild-type fibroblasts were treated with 200 μM N-acetyl-cysteine (NAC), an oxygen free-radical scavenging agent, followed by infection of a retrovirus expressing CPEB. While CPEB strongly induced senescence in untreated cells, its ability to do so in NAC-treated cells was substantially reduced (Supplemental Fig. 2A). Moreover, fibroblasts treated with hydrogen peroxide to increase the ROS concentration readily senesced, but did not do so when they had reduced CPEB levels (Supplemental Fig. 2B).
While indirect, these results suggest some connection between CPEB and ROS, and possible changes in cellular bioenergetics. To examine bioenergetics directly, we measured mitochondrial respiration in two CPEB knockdown cell lines; Figure 4A shows that relative to wild-type or shTETR control cells, mitochondrial respiration in these cells was reduced by ∼50%. To investigate the origin of this reduced respiration, we stained wild-type, shTETR, and shCPEB cells with Mitotracker, which reflects mitochondrial mass, followed by stacking of confocal images. These images indicate that shCPEB caused a reduction of mitochondrial number (Fig. 4B); the quantification of the Mitotracker fluorescent signal indicates that there was nearly eightfold fewer mitochondria (Fig. 4C). This decrease was confirmed by a Western blot for cytochrome C, a mitochondrial marker (Fig. 4D).
Figure 4.
Reduced respiration and mitochondrial number in CPEB knockdown cells. (A) Fibroblasts infected with shCPEB (targeted to two different sequences), shTETR, or empty virus (mock) were cultured for ∼47 d when they were used to measure oxygen consumption. (B) Z-plane stacks of confocal images obtained from live cells, some of which were infected with shCPEB or shTETR, stained with MitotrackerRed to visualize mitochondria. (C) Quantification of MitotrackerRed fluorescence from B. (D) Immunoblot of cytochrome C from wild-type, shTETR-, and shCPEB-infected cells. A nonspecific immuno-reactive band served as a loading control. (E) Wild-type, shCPEB, and shTETR cells were used to measure ATP concentration. (F) Lactate, an indicator of glycolysis, was determined in shCPEB (targeting two different sequences) and control cells. (G) Determination of relative ROS levels in cells expressing shCPEB or shTETR.
Because mitochondrial oxidative phosphorylation generates most of the cell’s ATP, it might be inferred that the CPEB knockdown fibroblasts contain less ATP than wild-type cells. However, the level of ATP was nearly identical between wild-type and shCPEB cells (Fig. 4E). Transformed cells also have reduced respiration but maintain relatively normal levels of ATP by increasing glycolysis (Bensaad and Vousden 2007). To determine whether this is also the case with CPEB knockdown cells, the amount of lactate, an indicator of glycolysis, was determined. Indeed, the CPEB knockdown cells produced about five times more lactate than did wild-type cells (Fig. 4F), indicating a substantial up-regulation of glycolysis. Finally, CPEB knockdown cells were also found to have a decrease in ROS levels (Fig. 4G), as might be expected from the reduction in mitochondrial respiration.
CPEB-induced senescence requires p53
In MEFs, CPEB requires p53 to induce senescence (Groisman et al. 2006). To determine whether this is also the case with human cells, fibroblasts were infected with a retrovirus expressing GSE-22, a p53 dominant-negative peptide that inhibits p53 activity (Beausejour et al. 2003). Two days later, the cells were infected with a virus expressing CPEB; the cells were then analyzed for growth and p21, a target gene of p53 (Figs. 5A–C). While CPEB induced senescence in cells lacking GSE-22, they were unable to do so if they contained the inhibitory peptide. Moreover, GSE-22 prevented p21 expression, thus demonstrating that it indeed inhibited p53 activity. These results indicate that CPEB-induced senescence requires p53 in human cells.
CPEB control of p53 mRNA translation
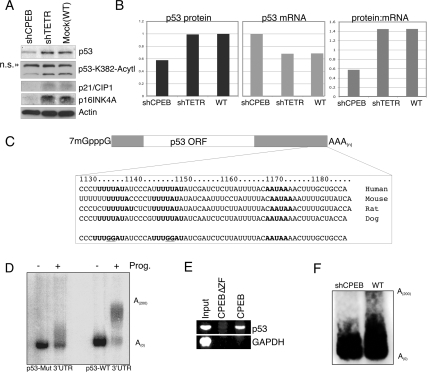
Several senescence-related proteins were analyzed in extracts derived from wild-type, shTETR, and shCPEB cells. p53 as well as K382-acetylated p53 were reduced by ∼50%–60% in shCPEB-containing cells; the cell cycle inhibitory proteins p21CIP1 and p16INK4A were also reduced, consistent with the bypass in senescence (Fig. 6A). Because p53 mRNA levels were not commensurately lower with the p53 protein levels (Fig. 6B), we inferred that CPEB might control, directly or indirectly, the post-transcriptional regulation of p53. One indication that this regulation might be direct is the fact that two conserved CPEs are found in the 3′ UTRs of p53 mRNA from several mammalian species (Fig. 6B). To examine whether these CPEs are functional, radiolabeled RNA corresponding to the p53 3′ UTR was injected into Xenopus oocytes, followed by incubation with progesterone, which stimulates oocyte maturation and CPE-dependent polyadenylation. Indeed, the p53 3′ UTR underwent robust polyadenylation. Moreover, a p53 3′ UTR with mutated CPEs exhibited little polyadenylation (Fig. 6C), demonstrating that the p53 3′ UTR has the proper sequences to promote cytoplasmic polyadenylation. In transfected fibroblasts, HA-tagged wild-type CPEB, but not a CPEB with deleted zinc fingers, efficiently coprecipitated p53 mRNA; GAPDH mRNA, a negative control was not precipitated under any condition (Fig. 6E). Finally, we investigated whether a knockdown of CPEB would alter the poly(A) tail length of p53 mRNA. Figure 6F shows a PCR-based PAT assay (Salles and Strickland 1995) for poly(A); while the poly(A) tail of p53 RNA reached nearly 200 nucleotides (nt) in wild-type cells, it was only ∼50 nt in shCPEB knockdown cells. Thus, CPEB controls p53 RNA poly(A) tail length in human fibroblasts.
Figure 6.
CPEB promotes cytoplasmic polyadenylation of p53 mRNA. (A) Extracts from shCPEB, shTETR, or mock-infected cells were probed with antibodies specific for p53, K382 acetylated p53, p21CIP1, p16INK4A, and actin. (B) The amount of p53 protein in A was quantified by densitometry, and the amount of p53 RNA in other infected cells was quantified by qRT–PCR; the ratios of these values were then plotted. (C) Diagram of the salient features of the p53 mRNA 3′ UTRs from several mammals; the CPEs and AAUAAA hexanucleotide are in bold. (Bottom) Sequence denotes dinucleotide substitutions in each of the two CPEs. (D) The human wild-type and mutated p53 3′ UTRs were radiolabeled and injected into Xenopus oocytes, which were then treated with progesterone to induce meiotic maturation and cytoplasmic polyadenylation. (E) Fibroblasts infected with a retrovirus encoding HA-CPEB or HA-CPEB lacking the zinc finger were used for HA antibody coimmunoprecipitation and RT–PCR detection of p53 mRNA and the non-CPE-containing GAPDH mRNA. (F) Ligation-mediated polyadenylation test (LM-PAT) assay was used to estimate the poly(A) tail length of p53 mRNA in wild-type and shCPEB knockdown cells.
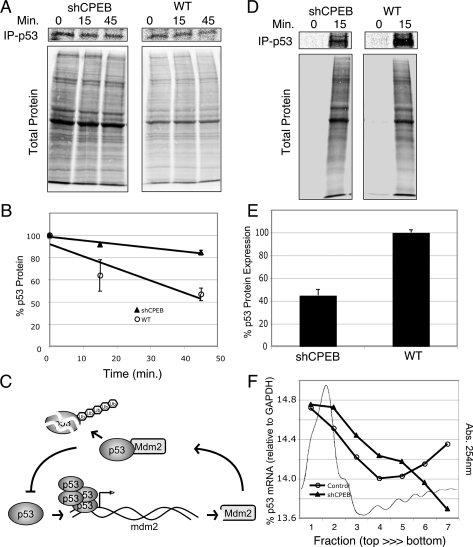
Because p53 protein levels are maintained by a balance of synthesis and destruction, we sought to determine which of these processes was controlled by CPEB. Wild-type and shCPEB-infected fibroblasts were first starved of methionine and cysteine, then pulsed with 35S-methoinine and 35S-cysteine, followed by a chase with radio-inert methionine and cysteine. p53 was then immunoprecipitated and the decay of this protein was monitored by SDS-PAGE and phosphorimaging. While shCPEB had no effect on the decay rate of general cellular proteins, there was a stabilizing effect on p53 (Fig. 7A,B). This result, however, is complicated by the fact that p53 is a positive regulator of the human homolog of mdm2, the E3 ligase that controls ubiquitin-mediated p53 destruction (Fig. 7C). In other words, elevated p53 levels induce mdm2 transcription, which in turn leads to p53 destruction. Consequently, we used alternative approaches to determine why p53 levels are reduced in CPEB knockdown cells. First, wild-type and shCPEB-infected cells were pulsed with 35S-methonine and 35S-cysteine in the presence of the proteasome inhibitor MG132, followed by p53 immunoprecipitation. Compared with total protein, the rate of p53 synthesis was ∼50% lower in the shCPEB-infected cells compared with wild type (Fig. 7D,E).
Figure 7.
CPEB controls p53 mRNA translation. (A,B) Wild-type and shCPEB-infected cells were incubated in methoinine and cysteine-free medium for 30 min, followed by an incubation with 35S-methionine and 35S-cysteine for 30 min, and then an incubation of up to 45 min with excess radioinert methionine and cysteine. p53 was immunoprecipitated from the extracts at 0, 15, and 45 min of the amino acid chase and resolved by SDS-PAGE, as was total cell protein. (C) Diagram illustrating a feedback loop where high levels of p53 induce transcription of the E3 ligase mdm2, which in turn induces p53 destruction. Ub refers to ubiquitin. (D,E) wild-type and shCPEB-infected cells were incubated in methionine and cysteine-free media for 1 h followed by incubation in 35S-methionine and 35S-cysteine for 15 min. p53 was then immunoprecipitated and analyzed by SDS-PAGE and phosphorimaging. The quantification of p53 levels in three different experiments is shown. (F) Extracts from wild-type and shCPEB-infected fibroblasts were centrifuged through 15%–50% sucrose gradients, fractionated, and scanned with 254-nm light. The relative amounts of p53 and GAPDH mRNAs were quantified by qRT–PCR. A representative polysome profile (absorbance at 254-nm light) from wild-type cells is shown.
The translational efficiency of p53 mRNA was determined by polysome sucrose density centrifugation of extracts from wild-type and shCPEB-infected cells. The gradients were fractionated and following RNA extraction, p53 mRNA was assayed by quantitative RT–PCR (qRT–PCR) using GAPDH mRNA as an internal standard. Figure 7F demonstrates that in shCPEB-infected cells, there was a shift in the sedimentation profile of p53 mRNA from heavy to lighter polysomes, consistent with a reduction in translational efficiency.
Senescence bypass and alteration in bioenergetics in p53 knockdown cells
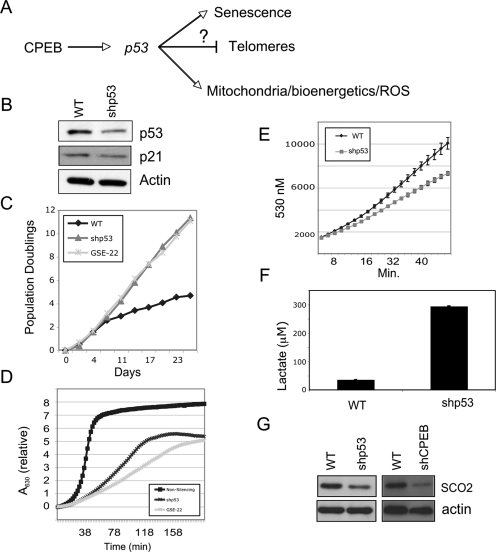
We next investigated whether the ∼50% reduction in p53 in CPEB knockdown cells is sufficient for the senescence bypass and change in bioenergetics (Fig. 8A). To do so, we stably expressed a p53 shRNA via lentiviral gene transfer, which reduced p53 protein levels by ∼50% (Fig. 8B). As expected, expression of p21, a p53 target gene, was also inhibited (Fig. 8B). The 50% reduction of p53 induced senescence bypass, as did expression of the GSE-22 dominant-negative p53 peptide (Fig. 8C). The 50% reduction of p53 also resulted in reduced mitochondrial respiration (Fig. 8D) and ROS (Fig. 8E), and stimulated a greater than sixfold increase in lactate production, indicating substantial up-regulation of glycolysis (Fig. 8F). To assess what factors might be downstream from CPEB and p53 that influences the change in bioenergetics, we determined the levels of synthesis of cytochrome oxidase 2 (SCO2), which has been reported to modulate the Warburg effect in a p53-dependent fashion (Matoba et al. 2006). Figure 8F demonstrates that SCO2 levels were reduced in cells in which either CPEB or p53 were knocked down, suggesting that the influence of CPEB on bioenergetics occurs via p53 mRNA translation and SCO2.
Figure 8.
p53 regulation of senescence and bioenergetics. (A) Proposed pathway in which CPEB, at least in part, influences senescence, telomere maintenance, and bioenergetics. (B) Western blot showing a shRNA-directed ∼50% knockdown of p53, which inhibits the expression of p21. (C) Growth curves of wild-type cells or cells infected with shp53 or GSE-22. (D) Oxygen consumption in cells infected with a nonsilencing shRNA, shp53 RNA, or GSE-22, a p53 inhibitory peptide. (E) Relative ROS levels in wild-type or shp53-infected cells. (F) Levels of lactate in wild-type and shp53 knockdown cells. (G) Western blot of SCO2 in CPEB and p53 knockdown cells.
Response to chemical carcinogen in CPEB KO mice
Based on the results of Groisman et al. (2006) and this study that describe the importance of CPEB in cell growth and metabolism in vitro, we initiated studies to examine the relative importance of CPEB in aging and malignant transformation in mice. CPEB KO mice have ∼2- to 2.3-yr life spans that are virtually indistinguishable from those of wild-type animals; they also display no unusual proclivity for tumor formation (data not shown). However, CPEB KO mice do form papillomas at a significantly faster rate than wild-type animals in a two-step DMBA-TPA carcinogenesis assay (Supplemental Fig. 3). Thus, CPEB does appear to offer some protection against at least one type of induced tumor formation and perhaps other induced stresses as well.
Discussion
We demonstrate that reduced levels of CPEB cause a bypass of senescence in primary human cells. While these cells have a nearly fivefold extended life span, their rate of cell division eventually begins to slow and they cease to divide after ∼93 population doublings. While mouse cells (MEFs) that lack CPEB also bypass senescence, they do not begin to slow even after >40 passages and are immortal (Groisman et al. 2006). In both mouse and human cells lacking (or with reduced) CPEB, the reintroduction of CPEB at early passages restores a senescence-like phenotype; a similar reintroduction of CPEB into late passage cells, however, has little effect on cell division. While it is unclear why only early passage cells respond to exogenous CPEB, such experiments do demonstrate that it is CPEB and not another factor that is responsible for the senescence bypass. In both MEFs and human fibroblasts, p53 is required for CPEB-induced senescence, while CPEB is required for Ras-induced senescence (Fig. 5; Supplemental Fig. 1; Groisman et al. 2006). These similarities between mouse and human cells notwithstanding, CPEB-controlled senescence in MEFs is mediated at least in part by myc, while in human cells one key factor is p53. In MEFs containing or lacking CPEB, there is no detectable change in the amount of p53 over many cell passages, while myc protein is elevated in CPEB KO MEFs (Groisman et al. 2006). Myc mRNA is also translated more efficiently in the KO MEFs and is at least one factor that mediates immortalization when CPEB is absent since a knockdown of myc in CPEB KO MEFs causes a cessation of cell division (Groisman et al. 2006). In human skin fibroblasts, there is no evidence for CPEB control of myc translation that contributes to senescence. On the other hand, CPEB mediates poly(A) tail length and translational control of p53 mRNA. Without the translational enhancement of p53 mRNA by CPEB, p53 levels are reduced to less than half their normal levels, which leads to senescence bypass (Fig. 8; see also Rogan et al. 1995; Wei et al. 2003; Lynch and Milner 2005) and changes in bioenergetics (see below).
Our results indicate that CPEB controls p53 mRNA polyadenylation and translation, possibly in a manner similar to that which occurs in vertebrate germ cells (Tay and Richter 2001; Tay et al. 2003; Barnard et al. 2004; Kim and Richter 2006, 2007; Richter 2007). If this is the case, then other factors such as Gld2 and PARN, which mediate CPEB-directed polyadenylation in oocytes, may have the same function in human fibroblasts, and a reduction in their steady state levels might modulate senescence.
In response to DNA damage, ribosomal protein L26 (rpL26) has been reported to stimulate p53 mRNA translation, while nucleolin inhibits it (Takagi et al. 2005). p53 mRNA translation in also enhanced by the RNA-binding protein HuR in response to UV irradiation (Mazan-Mamczarz et al. 2003). While we have no evidence that CPEB regulates p53 mRNA translation in response to DNA damage, it appears to regulate steady state translation by insuring that the p53 poly(A) tail is the proper length. Polyadenylation is a complex process regulated in both the nucleus and cytoplasm. For example, both nuclear pre-mRNA and cytoplasmic mRNA polyadenylation can be regulated during the cell cycle (Colgan et al. 1996; Groisman et al. 2006) and in response to certain signaling events (Wu et al. 1998; Mellman et al. 2008). Moreover, poly(A) dynamics are also regulated by environmental stress (Hilgers et al. 2006) and by miRNAs (Wu et al. 2006). We speculate that CPEB activity may be necessary for maintaining steady state p53 levels under normal conditions where it has important functions in cellular aging and energy metabolism (Bensaad and Vousden 2007).
Reversible CPEB-induced senescence-like phenotype
The observation that the CPEB-induced senescence-like phenotype was reversible was surprising because senescence is generally considered to be an irreversible process. On the other hand, there have been reports of senescence reversibility under particular circumstances. For example, Macip et al. (2006) have shown that in p53-null cells, a ROS-induced senescence-like phenotype is reversible. This type of senescence reversibility may not be directly related to that described here since oxidative stress of CPEB knockdown cells does not readily induce senescence (Supplemental Fig. 2). Moreover, Beausejour et al. (2003) have shown that senescence accompanying telomere shortening can also be reversed. While the nature of the CPEB reversibility of senescence requires further investigation, it is clear that cells’ entry into senescence is particularly sensitive to the amount of this protein; that is, in human cells, a knockdown of CPEB, either endogenous or exogenous to ∼20% of normal levels results in senescence bypass. In MEFs, cells heterozygous for CPEB also bypass senescence (Groisman et al. 2006). Conversely, cells containing relatively low amounts of exogenous CPEB require several passages before they senesce, whereas cells containing high levels senesce much faster. We speculate that the amount of CPEB is important for the relative translational efficiency of p53 mRNA, which in turn could be responsible for the rate, or timely onset, of senescence.
CPEB, p53, and energy metabolism
CPEB knockdown fibroblasts have nearly eightfold fewer mitochondria compared with wild-type cells and overall, respire about half as well. This startling observation suggests that these cells have reduced ATP levels and thus would probably divide more slowly than wild-type cells. Such is not the case, however, because knockdown cells have a very high rate of glycolysis; normal levels of ATP are therefore generated and the cell division rate at early passages is indistinguishable from wild type. The so-called “Warburg effect” of reduced oxygen consumption and elevated glycolysis was recognized many years ago as a hallmark of cancer cells (Shay and Wright 2000; Gatenby and Gillies 2004; Bensaad and Vousden 2007). Based on the Warburg effect, it might be inferred that CPEB knockdown skin fibroblasts are transformed; this is unlikely to be the case, however, because they are contact inhibited (data not shown). Moreover, the CPEB KO MEFs do not grow in reduced serum, do not show significant anchorage independent growth, and do not form tumors when injected into nude mice (Groisman et al. 2006). In the human CPEB knockdown fibroblasts, it seems likely that the Warburg effect is due to reduced levels of p53 (Fig. 8; Matoba et al. 2006). In p53-null or even hypomorphic cells, aerobic respiration is reduced and glycolysis is elevated. One downstream gene whose expression was recently reported to be regulated by p53, synthesis of cytochrome oxidase 2 (SCO2), may be responsible for several of these changes in energy metabolism (Matoba et al. 2006). This protein is necessary for assembly of the multiprotein cytochrome C oxidase (COX) complex, which forms the molecular foundation for oxidative phosphorylation. Indeed, SCO2 is reduced in CPEB as well as p53 knockdown cells (Fig. 8), which probably is responsible for the lowered respiration and elevated glycolysis. However, there are likely to be a number of mRNAs whose translation is misregulated in cells that lack CPEB, some of which are likely to contribute to the senescence bypass and/or the Warburg effect. In this vein, Wajapeyee et al. (2008) recently identified a growth factor, IGFBP7, and 16 additional factors that control BRAF-induced senescence. In MEFs, CPEB is necessary for Ras and probably BRAF-induced senescence, suggesting that mRNAs encoding some of these factors might be under the translational control of CPEB. We are presently investigating whether the translational regulation of other mRNAs modulates senescence.
Materials and methods
Cells and culture conditions
Primary human foreskin fibroblasts were obtained from the Cell Culture Core Facility of the Yale University Skin Disease Research Center and cultured as described (Rangarajan et al. 2004) in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal calf serum. About 106 cells, counted with a hemocytometer following treatment with trypan blue, were passaged every 3 d. Human lung WI-38 fibroblasts were cultured in a similar manner.
Virus production, infection, and cell lines
Amphotropic retroviruses and lentiviruses were produced by transient transfection of 293T cells with a transfer vector and amphotropic packaging plasmids encoding VSV-G and gag-pol using Lipofectamine 2000 (Invitrogen). Human cells at 50% confluency were infected for 8–12 h with viral supernatants containing 7 μg/mL polybrene. Typically 70%–90% infection efficiency was achieved as assessed by using a GFP-encoding viral gene or by immunostaining cells using anti-HA (Covance). After infection, fresh media was added to the infected fibroblasts.
Some cells were analyzed by Western blotting for p16 (BD Biosciences), p21 (Santa Cruz Biotechnologies), p53 (DO-1, Neomarkers), CPEB (Affinity BioReagents), cytochrome C (BD Pharmingen), SCO2 (Dr. Paul M. Hwang, NHLBI-NIH), and β-actin (Abcam). Other cells were fixed with 0.2% glutaraldehyde and stained for b-galactosidase activity at acidic pH according to Dimri et al. (1995).
Telomere analysis
A T-OLA assay was performed as described previously (Stewart et al. 2003). Briefly, DNA extracted from wild-type, shTETR-, and shCPEB-infected cells (5 mg of each) was added to a 20-mL reaction volume containing 0.5 pmol 32P-end-labeled (CCCTAA)4 oligonucleotide and hybridized for 12–15 h at 50°C, followed by addition of 20 U of Taq DNA ligase (New England Biolabs) for 5 h at 50°C. After ligation, the DNA was precipitated, dried, resuspended and applied to a 5% polyacrylamide-6 M urea gel. To ensure equal DNA loading of T-OLA gels, 10 ng of each T-OLA sample was analyzed by qPCR for the GAPDH gene. Primer sequences are presented in the supplemental Material.
Metaphase chromosomes from wild-type, shTETR-, and shCPEB-infected cells were prepared as described (Henegariu et al. 2001) and blocked with COT1 DNA and hybridized with 50 ng of a locked nucleic acid (LNA) probe (TTAGGGTTAGGGT TAGGG; locked nucleotides are underlined) that was 3′-end-conjugated with Cy3.
RNA analysis
To examine the p53 mRNA poly(A) tail, total RNA (300 ng) was preannealed with 5′-phosphorylated oligo d(T)18 (20 ng/mL) followed by RT with an oligo d(T)18 anchor primer (5′-GCTTCA GATCAAGGTGACC-d(T)18). Subsequent PCR used 2 mL from the previous RT with primers specific for p53 RNA and the linker in the presence of dNTPs plus 32P-ATP. Details can be found in the supplemental Material.
Immunoprecipitation of mRNA–protein complexes was performed on early passage fibroblasts infected with virus encoding HA-CPEB (Peritz et al. 2006), followed by RT–PCR for p53 and GAPDH mRNAs.
Analysis of p53
Control and shCPEB-infected fibroblasts were cultured in methionine and cysteine-free media (Invitrogen) for 45 min and then cultured in media containing 140 μCi 35S methionine and 35S cysteine (ProMix, Amersham) for 30 min. The cells were then washed and cultured in fresh DMEM supplemented with 2 mM each methionine and cysteine for the times indicated. The cells were then frozen and stored until they were lysed and used to immunoprecipitate p53 (DO-1 antibody, Neomarkers), which was analyzed by SDS-PAGE and PhosphorImaging. Details of the procedure can be found in the supplemental Material.
Some cells were cultured in methionine/cysteine free media as noted above in the presence of MG132, a proteasome inhibitor, for 1 h, followed by a 15-min culture in 100 μCi 35S methionine and cysteine; p53 was then immunoprecipitated and analyzed as noted above.
Bioenergetics
To measure oxygen consumption, ∼4 × 105 cells were washed and resuspended in 200 mL of Krebs-Ringers solution plus HEPES (125 mM NaCl, 1.4 mM KCl, 20 mM HEPES at pH 7.4, 5 mM NaHCO3, 1.2 mM MgSO4, 1.2 mM KH2,PO4, 1 mM CaCl2) containing 1% BSA. Cells from each condition were aliquoted into a BD Oxygen Biosensor System plate (BD Biosciences) in triplicate. Plates were assayed on a SAFIRE multimode microplate spectrophotometer (Tecan) at 1-min intervals for 60 min at an excitation wavelength of 485 nm and emission wavelength of 630 nm.
Lactate assay
The concentration of lactate in 40,000 cells was measured as described by Chang et al. (1992); the NADH produced by the conversion of lactate to pyruvate by lactate dehydrogenase was measured at 340 nm using a SAFIRE multimode spectrophotometer (TECAN).
ATP
ATP concentrations were determined using a CellTiter-Glo Luminescent Cell viability assay kit (Promega) by first plating ∼40,000 cells in 96-well format plate and following the manufacturer’s instructions.
Mitochondria
Images were obtained from live cells plated on coverslips after incubating them with 500 nM Mitotracker-Red (Molecular Probes). Cells were stained cells for 20 min in 10% FBS, DMEM, 5% CO2 using a Zeiss AxioVert 200M Confocal with a PerkinElmer UltraView Spinning Disc. Images were analyzed with Metamorph and Imarus software, similar to Kang et al. (2007).
ROS
ROS levels in 80,000 cells were determined by using 10 μM CM-H2DCFDA (dichlorodihydrofluorescein diacetate) (Invitrogen) in Krebs-Ringer bicarbonate buffer (Sigma-Aldrich). Using a Tecan plate reader, CM-H2DCFDA was excited at 485 nm and detected at 530 nm with a 4 × 4 pattern reader.
Detailed procedures
Details of all procedures an be found in the Supplemental Material.
Acknowledgments
We thank Richter laboratory members—in particular, Stephanie Nottrott, Kiran Padmanabhan, Jong-Heon Kim, and Maryia Ivshina—for discussions and technical advice. Additionally, we thank Ittai Ben-Porath, Kenkichi Masutomi, Jack Rosa, Chun-Ting Chen, Sambra Redick, Xiarong Shi, Pedja Sekaric, and the Craig Peterson and Silvia Corvera laboratories for sharing equipment and protocols. Special thanks are given to Eric Campeau for his expert technical advice and generosity in sharing reagents. We thank Paul Hwang for the SCO2 antibody, Barbara Walker and Waldemar Racki for help with the two-step carcinogenesis assay, and Dafna Bar-Sagi for suggesting its use. This work was supported by NIH grant AG30323. Additional core support from the Diabetes and Endocrinology Research Center Program Project (DK32520) is gratefully acknowledged.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1697808.
References
- Archer H., Bar-Sagi D. Ras and Rac as activators of reactive oxygen species (ROS) Methods Mol. Biol. 2002;189:67–73. doi: 10.1385/1-59259-281-3:067. [DOI] [PubMed] [Google Scholar]
- Barnard D.C., Ryan K., Manley J.L., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Barnard D.C., Cao Q., Richter J.D. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell. Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausejour C.M., Krtolica A., Galimi F., Narita M., Lowe S.W., Yaswen P., Campisi J. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I., Weinberg R.A. The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Bensaad K., Vousden K.H. p53: New roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Campisi J., di d’Adda Fagagna F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Cao Q., Kim J.H., Richter J.D. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 2006;13:1128–1134. doi: 10.1038/nsmb1169. [DOI] [PubMed] [Google Scholar]
- Chang L.H., Shimizu H., Abiko H., Swanson R.A., Faden A.I., James T.L., Weinstein P.R. Effect of dichloroacetate on recovery of brain lactate, phosphorus energy metabolites, and glutamate during reperfusion after complete cerebral ischemia in rats. J. Cereb. Blood Flow Metab. 1992;12:1030–1038. doi: 10.1038/jcbfm.1992.140. [DOI] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Murthy K.G., Prives C., Manley J.L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguría A., Zaballos A., Flores J.M., Barbacid M., et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Dickson K.S., Thompson S.R., Gray N.K., Wickens M. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 2001;276:41810–41816. doi: 10.1074/jbc.M103030200. [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Testori A., Acosta M., Campisi J. Replicative senescence, aging and growth-regulatory transcription factors. Biol. Signals. 1996;5:154–162. doi: 10.1159/000109185. [DOI] [PubMed] [Google Scholar]
- Drayton S., Peters G. Immortalisation and transformation revisited. Curr. Opin. Genet. Dev. 2002;12:98–104. doi: 10.1016/s0959-437x(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Groisman I., Ivshina M., Marin V., Kennedy N.J., Davis R.J., Richter J.D. Control of cellular senescence by CPEB. Genes & Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake L.E., Mendez R., Richter J.D. Specificity of RNA binding by CPEB: Requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Narita M. Oncogenes and senescence: Breaking down in the fast lane. Genes & Dev. 2007;21:1–5. doi: 10.1101/gad.1514207. [DOI] [PubMed] [Google Scholar]
- Henegariu O., Artan S., Greally J.M., Chen X.N., Korenberg J.R., Vance G.H., Stubbs L., Bray-Ward P., Ward D.C. Cryptic translocation identification in human and mouse using several telomeric multiplex fish (TM-FISH) strategies. Lab. Invest. 2001;81:483–491. doi: 10.1038/labinvest.3780256. [DOI] [PubMed] [Google Scholar]
- Hilgers V., Teixeira D., Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani K., Xia Y., Zweier J.L., Sollott S.J., Der C.J., Fearon E.R., Sundaresan M., Finkel T., Goldschmidt-Clermont P.J. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Itahana K., Campisi J., Dimri G.P. Methods to detect biomarkers of cellular senescence: The senescence-associated β-galactosidase assay. Methods Mol. Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- Kang B.H., Plescia J., Dohi T., Rosa J., Doxsey S.J., Altieri D.C. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131:257–270. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Richter J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Richter J.D. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes & Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E., Richter J.D. Translational control in biology and medicine. (eds. M. Mathews et al.); Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 2007. Translational control of synaptic plasticity and learning and memory; pp. 485–506. [Google Scholar]
- Lee A.C., Fenster B.E., Ito H., Takeda K., Bae N.S., Hirai T., Yu Z.X., Ferrans V.J., Howard B.H., Finkel T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J. Biol. Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Lynch C.J., Milner J. Loss of one p53 allele results in four-fold reduction of p53 mRNA and protein: A basis for p53 haplo-insufficiency. Oncogene. 2006;25:3463–3470. doi: 10.1038/sj.onc.1209387. [DOI] [PubMed] [Google Scholar]
- Macip S., Kosoy A., Lee S.W., O’Connell M.J., Aaronson S.A. Oxidative stress induces a prolonged but reversible arrest in p53-null cancer cells, involving a Chk1-dependent G2 checkpoint. Oncogene. 2006;25:6037–6047. doi: 10.1038/sj.onc.1209629. [DOI] [PubMed] [Google Scholar]
- Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Galban S., de Lopez Silanes I., Martindale J.L., Atasoy U., Keene J.D., Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman D.L., Gonzales M.L., Song C., Barlow C.A., Wang P., Kendziorski C., Anderson R.A. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Mendez R., Murthy K.G., Ryan K., Manley J.L., Richter J.D. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell. 2000;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S. BRAFE600 associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Peritz T., Zeng F., Kannanayakal T.J., Kilk K., Eiríksdóttir E., Langel U., Eberwine J. Immunoprecipitation of mRNA–protein complexes. Nat. Protocols. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Rangarajan A., Hong S.J., Gifford A., Weinberg R.A. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Richter J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Rogan E.M., Bryan T.M., Hukku B., Maclean K., Chang A.C., Moy E.L., Englezou A., Warneford S.G., Dalla-Pozza L., Reddel R.R. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol. Cell. Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles F.J., Strickland S. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl. 1995;4:317–321. doi: 10.1101/gr.4.6.317. [DOI] [PubMed] [Google Scholar]
- Shay J.W., Wright W.E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 2000;1:72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B., Cao Q., de Moor C.H., Mendez R., Richter J.D. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Stewart S.A., Ben-Porath I., Carey V.J., O’Connor B.F., Hahn W.C., Weinberg R.A. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 2003;33:492–496. doi: 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- Takagi M., Absalon M.J., McLure K.G., Kastan M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z., Sachs A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Wells S.E., Deardorff J.A., Sachs A.B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J., Richter J.D. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- Tay J., Hodgman R., Sarkissian M., Richter J.D. Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes & Dev. 2003;17:1457–1462. doi: 10.1101/gad.1071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N., Serra R.W., Zhu X., Mahalingam M., Green M.R. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Herbig U., Wei S., Dutriaux A., Sedivy J.M. Loss of retinoblastoma but not p16 function allows bypass of replicative senescence in human fibroblasts. EMBO Rep. 2003;4:1061–1066. doi: 10.1038/sj.embor.7400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wu L., Wells D., Tay J., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Miloslavskaya I., Demontis S., Maestro R., Galaktionov K. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature. 2004;432:640–645. doi: 10.1038/nature03173. [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]