Abstract
Many steps in gene expression and mRNA biosynthesis are coupled to transcription elongation and organized through the C-terminal domain (CTD) of the large subunit of RNA polymerase II (RNAPII). We showed recently that Spt6, a transcription elongation factor and histone H3 chaperone, binds to the Ser2P CTD and recruits Iws1 and the REF1/Aly mRNA export adaptor to facilitate mRNA export. Here we show that Iws1 also recruits the HYPB/Setd2 histone methyltransferase to the RNAPII elongation complex and is required for H3K36 trimethylation (H3K36me3) across the transcribed region of the c-Myc, HIV-1, and PABPC1 genes in vivo. Interestingly, knockdown of either Iws1 or HYPB/Setd2 also enhanced H3K27me3 at the 5′ end of the PABPC1 gene, and depletion of Iws1, but not HYPB/Setd2, increased histone acetylation across the coding regions at the HIV-1 and PABPC1 genes in vivo. Knockdown of HYPB/Setd2, like Iws1, induced bulk HeLa poly(A)+ mRNAs to accumulate in the nucleus. In vitro, recombinant Spt6 binds selectively to a stretch of uninterrupted consensus repeats located in the N-terminal half of the CTD and recruits Iws1. Thus Iws1 connects two distinct CTD-binding proteins, Spt6 and HYPB/Setd2, in a megacomplex that affects mRNA export as well as the histone modification state of active genes.
Keywords: Transcription elongation, RNAPII CTD, Spt6, Iws1, H3K36me3, mRNA export
The C-terminal domain (CTD) of the RNAPII Rbp1 subunit contains 52 heptapeptide repeats of a consensus sequence, YSPTSPS, that are differentially phosphorylated during the transcription cycle (Meinhart et al. 2005; Chapman et al. 2008). After initiation, Ser5P RNAPII complexes frequently pause in the vicinity of the promoter (Core and Lis 2008; Price 2008) to await mRNA 5′-end capping and the arrival of the Ser2-specific CTD kinase, P-TEFb (CycT1:Cdk9) (Peterlin and Price 2006). The Ser2P CTD serves as a scaffold for pre-mRNA splicing, polyadenylation, mRNA export, and chromatin modification factors that are required for mRNA biosynthesis and chromatin transactions during elongation (Aguilera 2005; Phatnani and Greenleaf 2006; de Almeida and Carmo-Fonseca 2008; Egloff and Murphy 2008). Structural, functional, and evolutionary studies suggest that the mammalian CTD is composed of three segments: a short N-terminal linker region (R1), a region defined by a contiguous stretch of identical consensus heptad repeats (R2), and a diverse C-terminal region (R3) containing predominantly nonconsensus repeats (Chapman et al. 2008). Although the importance of heptad sequence variation is unclear, genetic studies indicate that consensus repeats cannot be functionally replaced with noncanonical repeats (Bentley 2005; Chapman et al. 2005). The overall length of the CTD is also important for various functions. For example, only ∼15 repeats are required for 5′-end capping, whereas 20–25 repeats are needed for splicing and 3′-end processing (Fong and Bentley 2001; Fong et al. 2003), and greater than 30 repeats are necessary for efficient mRNA release from the site of transcription (Custodio et al. 2007; de Almeida and Carmo-Fonseca 2008). The terminal CTD repeat protects RNAPII from proteolytic degradation in vivo (Chapman et al. 2005).
The Ser5P RNAPII CTD is bound selectively by the 5′ end capping complex and the MLL-1/Setd1 H3K4me3 histone methyltransferase, and pauses after promoter clearance due to the actions of the Spt5:Spt4/DSIF (DRB sensitivity-inducing factor) and NELF (negative elongation factor) complexes (Core and Lis 2008; Price 2008). Following recruitment of P-TEFb, many other components of the mature elongation complex are loaded onto the Ser2P CTD, including factors required for splicing, polyadenylation and cleavage, and termination. However, little is known regarding how these different complexes are assembled and arranged relative to each other onto the CTD, nor is it clear how the different cotranscriptional events that target the nascent RNA and the chromatin template are coordinated. We recently discovered that the murine Spt6 elongation factor interacts directly, through its conserved SH2 domain, with the Ser2P RNAPII CTD (Yoh et al. 2007). A point mutation in the SH2 domain (R1358K) eliminates binding of Spt6 to the CTD, but does not impact Spt6-dependent elongation in cell-free assays on nonchromatin templates. However, transcripts formed in vivo by the Spt6-R1358K protein are longer than normal and contain mRNA processing defects, and overexpression of the wild-type Spt6 SH2 domain, but not the R1358K mutant SH2 domain, was found to induce bulk HeLa poly(A)+ mRNAs to accumulate in the nucleus. Similar mRNA processing and export defects were observed in cells depleted of the Spt6 partner protein, Iws1 (“interacts-with-Spt6”) (Krogan et al. 2002; Liu et al. 2007), which strongly suggests that Iws1 functions selectively with the CTD-bound form of Spt6. In Saccharomyces cerevisiae, the yIws1/Spn1 protein is an elongation factor that helps recruit Spt6 to active genes (L. Zhang et al. 2008). In contrast, the mammalian Iws1 protein, although required for cell viability (Liu et al. 2007), does not affect Spt6 occupancy at the genes we examined and, unlike Spt6, fails to stimulate RNAPII transcription elongation in vitro (Yoh et al. 2007).
Interestingly, the human Iws1 protein interacts directly with the REF1/Aly mRNA export factor and stabilizes its binding to the body of the actively transcribed c-Myc gene in 293T cells (Yoh et al. 2007). The yeast ortholog of REF1/Aly, Yra1p, interacts and functions in concert with the Mex67 (Tap-p15 in metazoans) nuclear export receptor (Strasser and Hurt 2000; Zenklusen et al. 2001; Strasser et al. 2002) and is part of the machinery that bridges active genes to the nuclear pore during the process of “gene gating” (Brown and Silver 2007; Luna et al. 2008). REF1/Aly is eventually transferred to the 5′ cap-binding complex to help guide mRNPs through the nuclear pore (Cheng et al. 2006; Nojima et al. 2007). Iws1 knockdown was also correlated with decreased occupancy of Rrp6, a subunit of the nuclear exosome that interacts with REF1/Aly, within the body of active genes in vivo (Yoh et al. 2007). Rrp6 travels with the elongating RNAPII complex for surveillance of nascent transcripts (Andrulis et al. 2002) and is also part of a checkpoint that ensures that transcripts are properly processed prior to release from the site of transcription (Vasudevan and Peltz 2003; Hieronymus et al. 2004). Thus REF1/Aly and Rrp6, and potentially other nuclear export factors, may be carried with the RNAPII elongation complex as components of the Spt6:Iws1:CTD complex.
In addition to its role in transcription, Spt6 functions as a histone H3 chaperone (Bortvin and Winston 1996) in the reassembly of nucleosomes displaced by promoter-bound activators (Adkins and Tyler 2006) and the elongating RNAPII complex (Belotserkovskaya and Reinberg 2004). Nucleosome reassembly during elongation is coordinated with the de novo deposition of several post-translational histone modifications, including trimethylation of histone H3 at Lys-36 (H3K36me3) (Hampsey and Reinberg 2003; Li et al. 2007), which is mediated by the Setd2 (SET domain 2) histone methyltransferase (Strahl et al. 2002; Krogan et al. 2003; Li et al. 2003; Xiao et al. 2003). The Setd2 enzyme directly binds to [Ser2P, Ser5] doubly-phosphorylated CTD repeats, which tether it to the elongation complex (Kizer et al. 2005; Li et al. 2005; Vojnic et al. 2006). In yeast, Setd2 mediates both di- and trimethylation of H3K36, which attracts Rpd3-type histone deacetylases to restore the hypoacetylated state of the reassembled chromatin and block cryptic or intragenic transcription (Carrozza et al. 2005; Keogh et al. 2005; Lee and Shilatifard 2007; Youdell et al. 2008). In contrast, the mammalian HYPB/Setd2 enzyme mediates tri- but not dimethylation of H3K36 and does not affect histone acetylation across coding regions (Edmunds et al. 2008).
Given the prominent role of Spt6 as a histone H3 chaperone that regulates chromatin structure at many genes, it was important to assess whether the CTD-bound Spt6:Iws1 complex might associate with HYPB/Setd2 to link nucleosome reassembly with elongation-coupled H3K36me3 in vivo. We show here that the Spt6 partner protein, Iws1, is required for optimal loading of HYPB/Setd2 and for stable accumulation of H3K36me3 across the coding regions of the c-Myc, PABPC1, and HIV-1 genes in 293T and HeLa cells. Binding of Iws1 to nuclear HYPB/Setd2 is mediated through a unique N-terminal domain that is distinct from the region that binds to Spt6. Unexpectedly, we noted that depletion of either Iws1 or HYPB/Setd2 enhanced H3K27me3 in the promoter-proximal region of the human PABPC1 gene, and that knockdown of Iws1, but not HYPB/Setd2, strongly increased histone acetylation throughout the body of the HIV-1 and PABPC1 genes in vivo. RNA-FISH experiments revealed that depletion of HYPB/Setd2 induced widespread accumulation of HeLa poly(A)+ mRNAs in the nucleus. In vitro, recombinant Spt6 binds specifically to the consensus repeat-rich (R2) region of the Ser2P CTD, and formation of this complex strongly stimulates binding of Iws1. Together, these data define an important role for Iws1 in the function of two distinct CTD-binding proteins, Spt6 and HYPB/Setd2, and integrates H3K36me3 with mRNA biosynthesis during elongation.
Results
The Spt6:CTD-interacting protein, Iws1, is required for elongation-coupled H3K36me3
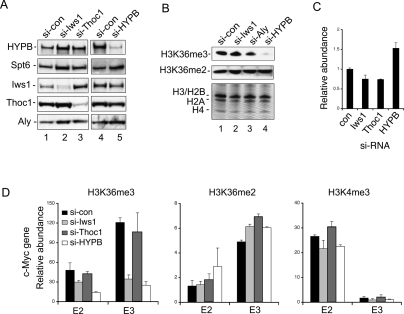
Because Iws1, unlike Spt6, is not essential for the accumulation of steady-state RNA at the genes we examined (c-Myc, HIV-1, and PABPC1), it can be used to probe the possible cotranscriptional activities of the Spt6:CTD complex. We first asked whether depletion of endogenous Iws1 affects elongation-dependent H3K36me3 at the c-Myc gene in 293T cells. Immunoblot analysis of a nuclear extract derived from cells treated with a siRNA directed against Iws1 (si-Iws1) confirmed a sharp decline in steady-state levels of the native Iws1 protein, whereas Spt6, CycT1, and Ser2P RNAPII levels were unaffected (Fig. 1A). As expected, Iws1 knockdown did not significantly alter total levels of c-Myc or β-actin mRNA, as measured by quantitative RT–PCR (qRT–PCR) (Fig. 1B). Quantitative chromatin immunoprecipitation (ChIP) experiments confirmed a sharp decline in Iws1 occupancy in the body of the c-Myc gene in vivo (Fig. 1C). Although transcription was not affected, H3K36me3 levels nevertheless dropped eightfold at exon 3 of the c-Myc gene in the Iws1 knockdown cells (Fig. 1C, E3 primer), as compared with cells treated with a control (scrambled) siRNA (si-con). H3K36me3 levels also declined near the 5′ end of the c-Myc gene (E2), whereas only small or negligible effects were observed at the core promoter (cp) or at a distal downstream region (U3). Knockdown of Iws1 had no effect on binding of Spt6 to the c-Myc gene, nor were total or Ser2P RNAPII or H3K4me3 levels affected, indicating that transcription was not significantly disrupted under the conditions of this experiment (Fig. 1C). We conclude that Iws1 controls H3K36me3 levels without blocking transcription of the c-Myc gene in vivo.
Figure 1.
Knockdown of Iws1 affects H3K36me3 across the coding region of the c-Myc gene in vivo. (A) Immunoblot of HeLa nuclear extracts from cells treated with si-Iws1 or control si-RNA (si-con). Antisera used are indicated to the left of each panel. (B) RT–PCR analysis of c-Myc transcripts in cells treated with si-con (black) or si-Iws1 (gray). (C) ChIP analysis of the c-Myc gene promoter (CP), exon 2 (E2), exon 3 (E3), or 3′ untranslated (U3) regions in cells treated with si-con (black) or si-Iws1 (gray) RNAs. The proteins analyzed in each experiment are listed above each panel.
Iws1, like HYPB/Setd2, regulates tri- but not dimethylation of H3K36
To better evaluate the specific changes in histone methylation at the c-Myc gene, comparative RNAi-ChIP experiments were carried out in 293T cells depleted of Iws1, HYPB/Setd2, or, as a control, the THOC1 subunit of the THO/TREX transcription export complex. Immunoblot analysis of nuclear extracts from these cells revealed that transient siRNA-mediated depletion of Iws1 did not affect steady-state levels of endogenous HYPB/Setd2 (Fig. 2A, cf. lanes 1 and 2), nor vice versa (Fig. 2A, cf. lanes 4 and 5). Interestingly, immunoblot analysis of total acid-extracted histones from these cells revealed that knockdown of HYPB/Setd2, but not Iws1 or REF1/Aly (included as a control), blocks global H3K36me3 in vivo (Fig. 2B). Therefore bulk H3K36me3 in 293T cells depends upon HYPB/Setd2, but not Iws1. RT–PCR analyses further established that steady-state c-Myc mRNA levels were unaffected by knockdown of Iws1 or THOC1 and even increased modestly in HYPB/Setd2-depleted cells, indicating that these factors are not essential for c-Myc gene transcription under the transient knockdown conditions used here (Fig. 2C). Despite the fact that ongoing transcription was not inhibited, ChIP experiments confirmed that reduced levels of either Iws1 or HYPB/Setd2 greatly impaired H3K36me3 at the c-Myc gene (Fig. 2D, left panel), without affecting H3K36me2 (Fig. 2D, center panel) or H3K4me3 (Fig. 2D, right panel). In contrast, H3K36me3 levels at the c-Myc gene were unchanged in cells transfected with control (scrambled) or THOC1-specific siRNAs. We conclude that whereas Iws1 is not universally required for H3K36me3, it is critical for RNAPII elongation-dependent H3K36me3 by HYPB/Setd2 at the c-Myc gene in vivo.
Figure 2.
Iws1, like HYPB/Setd2, selectively regulates trimethylated H3K36 levels in vivo. (A) Western blot analysis of protein levels in cells treated with siRNAs specific to Iws1 (lane 2), THOC1 (lane 3), or HYPB/Setd2 (lane 5) were compared with cells treated with control siRNA (lanes 1,4). (B) Analysis of global H3K36me3 (top), H3K36me2 (middle), and total histones (bottom; Coomassie), acid extracted from cells treated with indicated siRNAs. (C) RT–PCR analysis of c-Myc transcripts in cells treated with a specific siRNA indicated at the bottom. (D) ChIP analysis of histone modifications at the c-Myc gene in control cells or cells treated with the indicated siRNAs. The specific histone modifications examined are listed above each panel.
Iws1 controls HYPB/Setd2 occupancy at the HIV-1 and PABPC1 genes
To address whether Iws1 similarly regulates H3K36me3 at other Spt6 target genes, we tested the Poly(A)-Binding Protein C1 (PABPC1) and Tat-induced HIV-1 genes in HLM107 cells, a HeLa-derived cell line that contains a single integrated, Rev-defective, HIV-1 provirus. As before, Iws1 knockdown did not affect the stability or expression of either HYPB/Setd2 or Spt6 in vivo (Fig. 3A), nor were PABPC1, β-actin, or HIV-1 gene transcript levels significantly affected (Fig. 3B). However, we noticed that Spt6 and RNAPIIo coimmunoprecipitate with HYPB/Setd2 in nuclear extracts from HeLa cells, but not in comparable extracts derived from Iws1-depleted cells (Fig. 3C, cf. lanes 3 and 4). Thus, even though HYPB/Setd2 is a CTD-binding protein, its association with RNAPIIo and Spt6 depends upon Iws1. RNAi-ChIP experiments revealed a sharp decline in H3K36me3 at a distal (23 kb) downstream location in the PABPC1 gene in Iws1-depleted cells, and a modest decrease was also apparent at the 5′ end of the gene (0.5 kb) (Fig. 3D). Moreover, loss of H3K36me3 correlated with reduced binding of HYPB/Setd2 to the PABPC1 gene, whereas Spt6 occupancy was unaffected. Similar to the results obtained at the c-Myc and PABPC1 genes, knockdown of Iws1 also induced a drop in H3K36me3 and impaired recruitment of HYPB/Setd2 to the coding region of the integrated HIV-1 provirus (Fig. 3D). Thus the loss of H3K36me3 in Iws1 knockdown cells can be attributed to reduced HYPB/Setd2 occupancy in gene coding regions, most likely because it is no longer stably associated with the RNAPII elongation complex. Further studies will be needed to assess whether Iws1 might also regulate the histone methyltransferase activity of HYPB/Setd2 on chromatin substrates.
Figure 3.
Iws1 recruits HYPB/Setd2 and is required for H3K36me3 at the PABPC1 gene and integrated HIV-1 provirus in HLM107 cells. (A) Immunoblot analysis of protein expression in si-control or si-Iws1 RNA-treated HLM107 cells, as indicated above each lane. (B) RT–PCR analysis of b-actin and PAPBC1 mRNAs and Northern blot analysis of HIV-1 mRNAs in si-control (black) or si-Iws1-treated (gray) cells. MS, SS, and US denote multiply-spliced, singly-spliced, and unspliced transcripts. (C) Immunoblot analysis of anti-HYPB (lanes 3,4) or normal IgG (lanes 5,6) immunoprecipitates from HeLa nuclear extracts that were prepared from normal cells or cells depleted of endogenous Iws1 proteins. Antisera used are shown to the left of each panel. (D) ChIP analysis at 5′ (0.5 kb) and 3′ (23 kb) sites within the PAPBC1 gene and at the 5′ long terminal repeat (LTR) and coding (5 kb) regions of the HIV-1 genome in HLM107 cells treated with si-con (black) or si-Iws1 (gray) RNAs.
Depletion of Iws1, but not HYPB/Setd2, increases cotranscriptional histone acetylation in vivo
Additional RNAi-ChIP experiments revealed a sharp increase in histone H4 acetylation at the promoter, and more strongly within the transcribed region of the PAPBC1 and HIV-1 genes, in Iws1 knockdown cells (Fig. 3D). In contrast, depletion of HYPB/Setd2 strongly reduced H3K36me3 levels without affecting H4 acetylation at these genes (Supplemental Fig. 1). Thus Iws1 appears to control histone acetylation within the HIV-1 and PABPC1 coding regions independently of HYPB/Setd2 and H3K36me3.
Knockdown of either Iws1 or HYPB/Setd2 increases H3K27me3 at the PABPC1 gene
These findings prompted us to ask whether other transcription-associated histone modifications might be altered in Iws1 knockdown cells. A survey of other histone methylations revealed a significant increase in H3K27me3 levels toward the 5′ end of the PABPC1 gene in the si-Iws1-treated cells (0.5 kb) (Fig. 4A). This effect was selective because mono-methylated H3K27, which peaks near the 3′ end of the PABPC1 gene, was unaffected in the same cells. Moreover, H3K9me3 and H3K79me3, which accumulate at the 5′ end of the PABPC1 gene, were unaffected by the reduction in Iws1 protein levels. Despite these changes at the PABPC1 gene, global H3K27me3 levels were relatively unaffected in extracts from si-Iws1-treated 293T cells, as were levels of H3K9me3 (Fig. 4B, cf. lanes 1 and 2). Most interestingly, an increase in H3K27me3 at the PABPC1 promoter also observed in cells depleted of HYPB/Setd2 (Supplemental Fig. 1), indicating that this effect might be an indirect response to the drop in HYPB/Setd2 or H3K36me3. Indeed, global H3K27me3 levels were reproducibly elevated in cells depleted of HYPB/Setd2, as compared with control or Iws1 knockdown cells (Fig. 4B, cf. lanes 1 and 3).
Figure 4.
Knockdown of Iws1 enhances H3K27me3 at the promoter-proximal end of the PABPC1 gene in vivo. (A) ChIP analysis of histone modifications at the PAPBC1 gene in si-control cells (black) or si-Iws1 (gray) RNA-treated cells. The specific histone modifications examined are listed above each panel. (B) Analysis of global H3K27me3 (top panel), H3K9me3 (second panel) H3K36me3 (third panel), and total histones (bottom panel) acid extracted from cells treated with indicated siRNAs. (C) Pull down of endogenous HYPB/Setd2, Spt6 and RNAPIIo by recombinant wild-type or mutant Flag-Iws1 proteins or control Flag-GFP protein from HeLa nuclear extracts. Proteins analyzed by immunoblot are indicated to the left of each panel, and the Iws1 protein mutants analyzed are listed above each lane. (D) Flag-Iws1proteins coupled to anti-Flag M2 agarose beads were incubated with nuclear extracts prepared from HeLa cells either untreated (−) or treated (+) with 500 nM flavopiridol for 2 h and analyzed for binding to the factors indicated at left by immunoblot.
We next addressed whether Iws1 and Spt6 associate with HYPB/Setd2 in nuclear extracts. Preliminary studies revealed that HYPB/Setd2 coimmunoprecipitates with expressed Myc-tagged Spt6, but not with the Myc-Spt6-R1358K mutant, in HeLa nuclear extracts (data not shown), indicating that Spt6 likely associates with HYPB/Setd2 indirectly, through RNAPIIo. Endogenous HYPB/Setd2 bound avidly to recombinant Flag-Iws1 (amino acids 1–819) coupled beads, but not to control Flag-GFP beads (Fig. 4C, cf. lanes 2 and 3). In contrast, the unrelated NSD1 Setd2 histone methyltransferase did not bind to the Flag-Iws1 beads, indicating that these protein interactions are specific. Moreover, Flag-Iws1 did not associate strongly with two Jumonji C-related H3K27me3 demethylases, UTX and JMJD3 (Fig. 4C). Thus UTX, which travels with the elongating RNAPII complex (Smith et al. 2008), seems unlikely to be recruited directly through Iws1. Unfortunately, we were unable to address this question directly due to the lack of ChIP-grade antisera to UTX and JMJD3. It remains possible that the H3K27me3 demethylases associate with HYPB/Setd2 or H3K36me3-modified chromatin or, alternatively, more indirect mechanisms may be responsible for the observed increase in H3K27me3 at the PABPC1 promoter in Iws1- and HYPB/Setd2 knockdown cells.
In addition to HYPB/Setd2, both Spt6 and RNAPIIo were present in Flag-Iws1 immunoprecipitates from HeLa nuclear extracts, and all three proteins also bound to a truncated Flag-Iws1 protein that lacks the N-terminal domain (amino acids 522–819; Fig. 4C, cf. lanes 3 and 6). Two Flag-Iws1 mutants lacking the conserved CTD (amino acids 1–598 and 281–598) retained binding to HYPB/Setd2, but bound only weakly to RNAPIIo and failed to interact with Spt6 (Fig. 4C, lanes 4,7, respectively). These data suggest that Iws1 can bind HYPB/Setd2 independently of Spt6. As illustrated schematically in Figure 4D, the region of Iws1 that binds to HYPB/Setd2 partially overlaps the Spt6 interaction domain, but also includes residues outside the conserved Iws1-C domain. Interestingly, we also noted in these studies that recombinant Flag-Iws1 protein is unable to bind to either HYPB/Setd2 or Spt6 in nuclear extracts treated with the CDK9 inhibitor, flavopiridol (Fig. 4D, cf. lanes 5 and 6), which suggests that these interactions may require the P-TEFb/Ser2P RNAPII CTD.
Knockdown of HYPB/Setd2 induces nuclear retention of HeLa bulk poly(A)+ mRNA
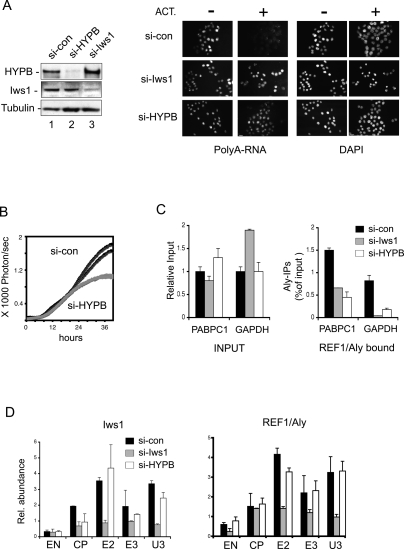
We previously showed that Iws1 recruits REF1/Aly to the c-Myc gene and stimulates the kinetics of mRNA export in vivo (Yoh et al. 2007). Knockdown of Iws1, or over-expression of the Spt6 SH2 domain, led to the nuclear accumulation of total poly(A)+ mRNAs and delayed reporter gene expression. To address whether HYPB/Setd2 similarly affects mRNA exit from the nucleus, cells were treated with si-HYPB/Setd2 RNA and analyzed for a block to mRNA export by RNA fluorescence in situ hybridization (RNA-FISH), using a probe specific for poly(A)+ mRNA (Fig. 5A). In these experiments, the siRNA transfected cells were also treated for 1 h with Actinomycin D to eliminate the background signal arising from on-going nascent transcription. Interestingly, reduction of HYPB/Setd2 protein levels caused bulk poly(A)+ mRNAs to accumulate in the nucleus, as compared with cells treated with the control siRNA (Fig. 5A). This kinetic delay in mRNA release or export reduced the maximal rate of luciferase reporter gene expression (Fig. 5B). In contrast, knockdown of Ash-2, a conserved subunit of the MLL-1/Setd1 H3K4me3 histone methyltransferase, reduced global H3K4me3 levels without inducing a widespread nuclear accumulation of poly(A)+ mRNAs (Supplemental Fig. 2).
Figure 5.
HYPB/Setd2 facilitates the kinetics of mRNA export from the nucleus. (A) Immunoblot of whole-cell lysates prepared from cells treated with control siRNA (si-con) or with siRNAs specific for HYPB or Iws1. The panels on the right show RNA-FISH analysis of bulk poly(A)+ mRNA in the nucleus of wild-type cells or cells depleted of either Iws1 or HYPB, as indicated on the left of each panel. Cells were treated with Actinomycin D for 1 h, as indicated at the top of each panel. (B) Rate of expression of the integrated HIV-1:Luciferase reporter gene from wild-type cells (si-con) or cells depleted of HYPB/Setd2 (si-HYPB). The HIV-1 Tat activator was expressed transiently, and duplicates are shown for each experiment. (C) Analysis of the fraction of PABPC1 or GAPDH mRNAs in the nascent mRNA (nuclear pellet) fraction that associate with REF1/Aly in wild-type cells (si-con; black) or cells depleted of Iws1 (gray) or HYPB/Setd2 (white). Input mRNAs levels are shown in the left panel for comparison. (D) ChIP analysis of Iws1 (left panel) or REF1/Aly (right panel) occupancy at the c-Myc gene in wild-type 293T cells (si-con, black) or cells depleted of Iws1 (gray) or HYPB/Setd2 (white).
Because REF1/Aly function requires that it bind to the 5′-end mRNA cap (Cheng et al. 2006), we asked whether Iws1 or HYPB/Setd2 might be required to transfer REF1/Aly to nascent PABPC1 or GAPDH mRNAs. RT–PCR analysis of nascent poly(A)+ mRNAs in fractionated HeLa nuclear pellets (Fig. 5C) revealed a consistent drop in the fraction of mRNAs that immunoprecipitate with REF1/Aly in either Iws1- or HYPB/Setd2-depleted cells, consistent with the observed block to mRNA export. Control experiments established that REF1/Aly protein levels were comparable in extracts from si-con, si-Iws1, and si-HYPB/Setd2-treated cells (Supplemental Fig. 2D). However, this impaired association of REF1/Aly with nuclear RNA in Iws1 knockdown cells was transient and evident only in nascent mRNA fractions (data not shown). ChIP analyses of the c-Myc gene showed that HYPB/Setd2 knockdown does not influence binding of either Iws1 or REF/Aly to chromatin (Fig. 5D). Thus HYPB/Setd2 acts downstream from Iws1 and appears to play a role in the formation or release of export competent mRNPs.
Spt6 binds selectively to the N-terminal half of the RNAPII CTD
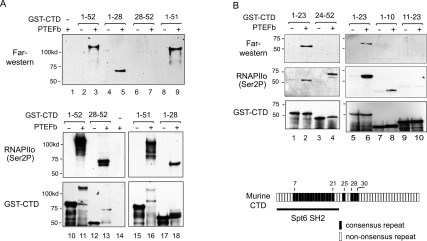
The CTD is composed of distinct functional segments, which are specified by the variations in the repeat sequence and the underlying phosphorylation patterns. To map where Spt6 binds within the CTD, fragments of the murine GST-CTD protein were phosphorylated in vitro with recombinant P-TEFb and analyzed by Far-Western blot for binding to a recombinant GST-Spt6-SH2 protein, which was coupled to glutathione horseradish peroxidase (HRP). As visualized by HRP chemiluminescence, the GST-Spt6 SH2 protein bound avidly to the wild-type 52 repeat GST-CTD (Fig. 6A, lane 3), as well as to a GST-CTD mutant lacking the terminal repeat (repeats 1–51) (Fig. 6A, lane 9). Shorter fragments that span either the N-terminal (repeats 1–28) or C-terminal (28–52) half of the CTD were effective substrates for P-TEFb in vitro, although only the C-terminal fragment displayed the distinctive hyperphosphorylated migration pattern that is characteristic of the full-length CTD. Most interestingly, the GST-Spt6 SH2 protein bound selectively to the N-terminal (Fig. 6A, lane 5), and not the C-terminal (Fig. 6A, lane 7), half of the CTD in vitro. Binding in each case required phosphorylation by P-TEFb (Fig. 6A, cf. lanes 2,4,8 and 3,5,9). As shown in Figure 6B, Spt6 also recognized an even shorter N-terminal CTD fragment containing the first 23 repeats (lane 2), but did not bind the corresponding C-terminal fragment (repeats 24–52) (lane 4), which contains only three consensus heptad repeats (see schematic in Fig. 6B). No binding was observed to shorter CTD fragments (repeats 1–10 and 11– 23) (Fig. 6B, lanes 8,10), which appear to be significantly weaker substrates for P-TEFb under these reaction conditions (Fig. 6B, middle panel, cf. lanes 6 and 8,10). We were unable to detect specific binding of the GST-Spt6 SH2 domain to synthetic Ser2P or doubly Ser2P, Ser5P, tandem repeat CTD phosphopeptides (data not shown), indicating that the Spt6-binding domain may span more than two Ser2P CTD repeats. We conclude that Spt6 binds exclusively to the proximal region of the CTD, which includes a stretch of 15 tandem consensus repeats (repeats 7–21) and corresponds to the region reported previously to be most strongly phosphorylated by P-TEFb in vivo (Ramanathan et al. 2001).
Figure 6.
The Spt6-SH2 domain interacts with the CTD N-terminal consensus-rich repeats in vitro. (A, top, lanes 1–9) Binding of the GST-Spt6 SH2 domain (amino acids 1162–1726) to GST-CTD fragments containing various heptapeptide repeats as indicated above each lane that were either untreated (−) or phosphorylated in vitro with recombinant P-TEFb (+) was analyzed by Far-Western blot. Immunoblots of these same reactions with the H5 (anti-Ser2P) RNAPII antiserum (middle) and anti-GST (bottom) are shown in lanes 10–18. (B) Far-Western analysis similar to A except the following GST-CTD proteins were used: repeats 1–23 (lanes 1,2,5,6), 24–52 (lanes 3,4), 1–10 (lanes 7,8), and 11–23 (lanes 9,10). Immunoblot analysis of Ser2P efficiency and GST-CTD protein levels using anti-Ser2P RNAPII (middle) and anti-GST (bottom) antibodies, respectively. At the bottom is a schematic diagram of murine RNAPII CTD (52 heptad repeats; rectangles), with consensus repeats (YSPTSPS) indicated in black. The region of the CTD targeted by the Spt6 SH2 domain is marked with brackets.
Iws1 binds preferentially to the Spt6:RNAPIIo complex, rather than to free Spt6, in vitro
The observation that Spt6 fails to associate with Iws1 in flavopiridol-treated cell extracts suggests that this interaction might require the P-TEFb/Ser2P CTD. To address this question, GST-Iws1 (amino acids 523–819) beads were preincubated with purified HeLa RNAPII and analyzed for binding to recombinant full-length His-Spt6 protein by immunoblot with anti-His antibodies. Under these conditions, His-Spt6 interacted weakly with GST-Iws1 beads in the absence of purified HeLa RNAPII (Fig. 7A, lane 4), and incubation with recombinant P-TEFb did not affect this interaction (Fig. 7A, lane 3). As expected, both the wild-type Spt6 and the Spt6-R1358K proteins bound equivalently to the GST-Iws1 beads in the absence of RNAPII (Fig. 7A, lane 5). Interestingly, addition of purified HeLa RNAPII to the reaction strongly enhanced binding of wild-type Spt6, but not Spt6-R1358K, to the GST-Iws1 beads (Fig. 7A, cf. lanes 4,6 and 5,7). The Spt6–Iws1 interaction was also enhanced by preincubation of a hypophosphorylated RNAPII fraction with recombinant P-TEFb and ATP (Fig. 7A, cf. lanes 13 and 15), and not if the RNAPII fraction was instead treated with the FCP1a CTD phosphatase (Fig. 7A, cf. lanes 13 and 14).
Figure 7.
Binding of Spt6 to purified HeLa RNAPIIo recruits Iws1 to the complex in vitro. (A) Binding of the recombinant full-length wild-type His-Spt6 (wt) or the His-Spt6 R1358K mutant (mt) to GST-Iws1 (amino acids 523–819) beads was analyzed in the absence (−) or presence (+) of purified HeLa RNAPII. Binding of His-Spt6 was assessed by immunoblot with anti-His antiserum. Total RNAPII and Ser2P RNAPII in each reaction was monitored using N20 anti-RNAPII (middle panel) and H5 anti-Ser2P-RNAPII (bottom panel) antibodies, respectively. Where indicated, RNAPII fractions were preincubated with either P-TEFb or the FCP1a CTD phosphatase, as indicated above each lane. (B) GST-Iws1 (523–819) beads were incubated with either HeLa or J-Lat nuclear extracts and analyzed for associating proteins indicated at the left. (C) Model diagram depicting how Iws1 might connect the Spt6 and HYPB/Setd2 complexes on the RNAPIIo CTD. See the text for details.
Preferential binding of Iws1 to the Spt6:RNAPIIo complex could also be demonstrated in extracts from resting J-LatT cells, which normally express very low levels of CycT1 and Ser2P RNAPII (Fig. 7B). In particular, we noted that GST-Iws1 bound strongly to endogenous Spt6 in HeLa nuclear extracts but was unable to do so in nuclear extracts from J-Lat cells, even though its ability to bind the native REF1/Aly protein was comparable in each extract (Fig. 7B, cf. lanes 3 and 6). These findings indicate that the Ser2P CTD strongly affects the ability of Iws1 to associate with Spt6 but does not affect its binding to REF1/Aly. Collectively, these data implicate a role for Iws1 in the assembly of a CTD complex that controls mRNA export as well as histone methylation by HYPB/Setd2, as illustrated in the hypothetical schematic in Figure 7C and discussed further below.
Discussion
Recruitment of P-TEFb and Ser2 hyperphosphorylation of the Rbp1 CTD is a critical step preceeding the assembly of a mature RNAPII elongation complex at many genes. Although numerous proteins interact directly with RNAPIIo, little is known about when and where the various complexes assemble on the CTD or how they are arranged relative to each other to coordinate the diverse steps in mRNA biosynthesis, elongation-coupled nucleosome reassembly, and cotranscriptional histone modification (Phatnani and Greenleaf 2006; Chapman et al. 2008; Egloff and Murphy 2008). We recently reported that Spt6, a transcription elongation factor and histone H3 chaperone, binds selectively through its SH2 domain to the P-TEFb/Ser2P RNAPII CTD (Yoh et al. 2007). We show here that Spt6 selectively binds in vitro to murine Ser2P-CTD repeats 1–23, which includes a long uninterrupted stretch of tandem consensus heptads (repeats 7–21), and does not recognize the predominantly nonconsensus repeats in the C-terminal half of the CTD. Spt6 also did not bind to shorter N-terminal CTD fragments (repeats 1–10 or 11–23) or synthetic CTD tandem repeat phosphopeptides (data not shown). Future studies are needed to address whether the native Spt6 protein binds exclusively to this region of the CTD in vivo and how the Spt6:Iws1 complex is situated relative to other Ser2P CTD-interacting proteins. Collectively, these data indicate that the Spt6 SH2 domain preferentially targets the unique sequence or conformational features of the highly conserved R2 domain of the CTD.
We previously reported that steady-state c-Myc or HIV-1 mRNA levels decrease dramatically in cells depleted of Spt6, but not Iws1, and that recombinant Spt6, but not Iws1, stimulates RNAPII transcription elongation in vitro (Yoh et al. 2007). These findings indicate that Iws1 does not regulate loading of Spt6 onto responsive promoters and is not essential for Spt6-driven transcription elongation. However, transcripts formed in Iws1-depleted cells contained mRNA processing defects, and global poly(A)+ mRNAs accumulated in the nucleus. Similar mRNA processing and nuclear retention defects were observed with the Spt6-R1358K protein, which otherwise retained the ability to load onto promoters and stimulate elongation in vitro. These findings indicated that the function of Iws1 is linked to that of the CTD-bound form of Spt6, and we subsequently showed that Iws1 facilitates loading of the REF1/Aly mRNA export adaptor and the Rrp6 subunit of the nuclear exosome to the RNAPII elongation complex in vivo. We show here that the purified P-TEFb/Ser2P RNAPII greatly enhances binding of Iws1 to Spt6 in vitro and that Iws1 binds poorly to Spt6-R1358K, even in the presence of RNAPIIo. Thus the observation that Iws1 binds with higher affinity to the Spt6:CTD complex than to free Spt6 may explain why its activity appears to be confined to the CTD-related functions of Spt6 in vivo.
Most interestingly, we find that Iws1 associates strongly with a second CTD-binding protein, HYPB/Setd2. Iws1 is required for stable binding of HYPB/Setd2 to RNAPIIo in nuclear extracts and recruits it to the coding region of active genes for elongation-dependent H3K36me3 in vivo. These results strongly suggest that the Hypb/Setd2 SRI CTD-binding domain is insufficient to direct it to the RNAPII elongation complex in vivo. Moreover, although Iws1 interacts strongly with both Spt6:CTD and HYPB/Setd2, we did not detect a significant interaction between Spt6 and HYPB/Setd2 in extracts (data not shown), indicating that these two CTD-binding proteins may associate indirectly, through Iws1 (Fig. 7C). This model suggests a possible mechanism to couple HYPB/Setd2-mediated H3K36me3 with Spt6-driven nucleosome reassembly, both of which are thought to depend upon the histone H3 chaperone activity of Spt6. Consistent with this model, we find that RNAi-mediated knockdown of Iws1 disrupts binding of HYPB/Setd2 and REF1/Aly, but not Spt6, to actively transcribed genes, whereas depletion of HYPB/Setd2 did not affect binding of any of these factors, indicating that it acts downstream from Spt6 and Iws1.
Although both Spt6 and HYPB/Setd2 are CTD-interacting proteins, only the latter depends upon Iws1 for recruitment to active genes in vivo. In this respect, the human Iws1 protein appears to differ fundamentally from its S. cerevisiae counterpart (yIws1/Spn1), which controls Spt6 occupancy at transcribed genes in vivo (L. Zhang et al. 2008). A recent model of Spt6, based on the structure of the related prokaryotic Tex protein (Johnson et al. 2008), indicates that the N-terminal (Iws1-binding) domain lies on an opposite face of the protein relative to the SH2 (CTD-binding) domain, which may explain how Spt6 can bind simultaneously to Iws1 and the CTD. The observation that Iws1 binds preferentially to the RNAPIIo-bound Spt6, rather than to free Spt6, indicates that the conformation of Spt6 may change upon binding to the CTD to expose the N-terminal Iws1-binding domain. Alternatively, Iws1 may form direct contacts with both Spt6 and the phospho-CTD. Additional studies are needed to test this model by examining, for example, whether the Spt6:CTD complex recruits Iws1 to the elongation complex in vivo, whether Iws1 can bind simultaneously with HYPB/Setd2, Spt6, and the CTD, and whether Iws1 and HYPB/Setd2 have a role in Spt6-mediated nucleosome reassembly during elongation.
The observation that both the Spt6 histone H3 chaperone and HYPB/Setd2 histone H3K36 methyltransferase interact directly with the CTD and are also physically linked through Iws1 strongly supports the long-held idea that elongation-coupled histone modifications are carried out in concert with nucleosome reassembly (Workman and Abmayr 2004). A striking example of the close coupling between nucleosome assembly and histone modification comes from studies showing that FACT/Spt16, a histone H2A:2B chaperone and nucleosome reassembly factor, is required for histone H2B mono-ubiquitination, both on chromatin templates in vitro and in vivo (Pavri et al. 2006) and that H2B ubiquitylation stimulates nucleosome reassembly during transcription (Fleming et al. 2008). In an interesting twist, a recent study showed that REF1/Aly mRNA export adaptor recruits the histone H2A:H2B chaperone Nap-1 to active genes (Del Rosario and Pemberton 2008), which further suggests a link between histone chaperones and mRNA export. Given that Iws1 interacts directly with both Spt6 and REF1/Aly, and recruits the latter to the body of active genes, it will be important to assess whether Iws1 also attracts Nap-1 or other histone chaperones to the elongation complex or whether it may contact the nucleosomal template directly.
The observation that knockdown of Iws1 leads to a sharp drop in H3K36me3 at active genes without affecting global H3K36me3 levels suggests that most HYPB/Setd2-mediated H3K36 methylation is independent of Iws1. The effects of Iws1 on histone methylation appeared to be relatively specific because H3K36me2, H3K4me3, H3K9me3, and H3K79me3 levels remained unchanged in si-Iws1-treated cells. Interestingly, however, RNAi-ChIP experiments revealed that levels of H3K27me3, a modification typically associated with gene silencing, increased at the 5′ end of the PABPC1 gene in si-Iws1-treated cells. A similar increase in PABPC1 H3K27me3 levels was observed in HYPB/Setd2 knockdown cells and therefore may reflect the reduced HYPB/Setd2 levels at the gene. Although H3K27me3 levels were low at the other genes we examined and did not appear to increase significantly in Iws1- or HYPB/Setd2-depleted cells (data not shown), we noted that global H3K27me3 levels were reproducibly higher in HYPB/Setd2 knockdown cells, confirming this inverse relationship on a broader level. A recent study found that H3K27me3 levels are inversely correlated with H3K36me2 among the differentially rearranged proximal and distal genes in the immunoglobulin heavy chain locus (Xu et al. 2008), and it will be interesting to learn whether a similar opposing relationship exists with H3K36me3. Thus disruption of HYPB/Setd2 and H3K36me3, or depletion of Iws1, can result in promoter regions marked with the characteristic “bivalent” epigenetic signature observed at poised genes in embryonic stem cells (namely, H3K4me3 and H3K27me3) (Spivakov and Fisher 2007). The observed increase in H3K27me3 at the PABPC1 gene, coupled with the defects in rapid mRNA exit from the nucleus, suggests that a sustained defect in the Iws1:Spt6:CTD complex might be expected to impede transcription elongation, especially toward the 3′ end of transcribed genes in vivo. Although steady-state mRNA levels are not significantly affected in the Iws1 knockdown cells and we observe no evidence of transcription impairment by ChIP, more sensitive nuclear run-on and high-resolution ChIP studies are warranted to resolve this question in a more definitive manner.
Although the underlying mechanism is unclear, one interesting possibility is that Iws1 or HYPB/Setd2 may recruit an H3K27me3-specific demethylase (Swigut and Wysocka 2007) to chromatin. Indeed, the UTX H3K27me3 demethylase is known to travel with the RNAPIIo elongation complex (Smith et al. 2008). Although we were unable to detect significant binding of Iws1 to nuclear UTX or the related JMJD3 H3K27 demethylase under the stringent binding conditions of our assays, it remains possible that the K27 demethylases are recruited to the RNAPII elongation complex through HYPB/Setd2 or other Iws1-associated factors. Relevant to this question, a recent study from the Tamkun laboratory reported a striking increase in H3K27 methylation in Drosophila mutants lacking the Kismet-L (Kis-L, corresponding to human CHD7) chromatin remodeling enzyme, as visualized by salivary gland polytene chromosome staining and gene-specific ChIP (Srinivasan et al. 2008). Lack of Kis-L blocks transcription elongation downstream from P-TEFb recruitment and leads to a loss in both RNAPII Ser2P and Spt6 recruitment (Srinivasan et al. 2005). Most interestingly, the increase in H3K27 methylation in kis mutants did not correlate with changes in the E(Z) histone methyltransferase and so might involve the K27-specific demethylases, or a failure to exchange histone H3 with the histone H3.3 variant, which generally displays active gene marks and low levels of H3K27 methylation and was correlated with a failure to recruit the ASH1 and TRX trithorax proteins to active genes (Srinivasan et al. 2008).
We also observed an unanticipated increase in histone H4 acetylation in cells treated with si-Iws1 RNA. Unlike the change in H3K27me3, histone H4 acetylation did not change in cells depleted of HYPB/Setd2. Studies in S. cerevisiae have shown that dimethylation of H3K36 attracts Rpd3-related histone deacetylases, which function to block cryptic and intragenic transcription and reset chromatin to its original state before elongation (for review, see Lee and Shilatifard 2007; Li et al. 2007). However, in mammalian cells, HYPB/Setd2 mediates tri-, but not di-, methylation of H3K36me3 and is not associated with changes in histone acetylation (Edmunds et al. 2008). Thus Iws1 may act, directly or indirectly, to recruit a histone deacetylase(s) to restore the original state of the newly transcribed chromatin.
Knockdown of HYPB/Setd2 also induced a strong kinetic delay in global poly(A)+ mRNA release or export, similar to that observed in cells depleted of Iws1 or expressing a dominant-negative Spt6 SH2 domain protein (Yoh et al. 2007). Although both proteins contribute to mRNA biosynthesis, HYPB/Setd2 does not affect the recruitment of Iws1 or REF1/Aly to genes. Bulk poly(A)+ mRNA did not accumulate in the nucleus upon knockdown of Ash2 and loss of global H3K4me3, indicating that the effect on mRNA export is not a general phenomenon associated with loss of any cotranscriptional chromatin modifications, but rather is specific to HYPB/Setd2 or H3K36me3. Further studies are needed to assess whether H3K36me3 chromatin might help stabilize the association of active genes with nuclear pores or whether HYPB/Setd2 might play a role in mRNA biosynthesis.
An intriguing connection between mRNA export and histone modification was observed previously in studies of the Sus1 subunit of the yeast Spt/Ada/Gcn5 acetyltransferase (SAGA) complex (Köhler et al. 2006, 2008; X.Y. Zhang et al. 2008a, b; Zhao et al. 2008). Sus1 forms a submodule of the SAGA complex with Sgf11 and the histone H2B deubiquitinating enzyme, Ubp8, and is required to deubiquitylate histones H2A and H2B during transcription. Importantly, Sus1 is also a resident member of the Sac3:Thp1:Cdc31 mRNA export complex, which functions together with the Mex67 nuclear export receptor to promote interact with and direct nascent transcripts through the nuclear pore (Cabal et al. 2006; Köhler et al. 2006, 2008). Although SAGA plays a critical role at promoters to acetylate histones and remove monoubiquitin from histone H2B, it also found in the transcribed region of genes (Govind et al. 2007), as is ubiquitinated H2B (Minsky et al. 2008). Moreover, it was recently shown that Sus1 is required for gene gating at the nuclear pore, copurifies with Yra1 (yeast homolog of REF1/Aly) and Mex67 receptor, and is recruited to gene coding regions in a Sac3-dependent manner (Pascual-García et al. 2008). It will be particularly interesting to learn whether the Iws1:Spt6:CTD complex recruits the Sus1 complex to active genes or whether these complexes intersect later; for example, upon binding to REF1/Aly, the Mex67 export receptor, or nucleoporins. The observation that both the Sus1:Sgf11:Ubp8 SAGA subcomplex and the Iws1:Spt6:CTD complex have dual roles in histone modification and mRNA trafficking through binding to factors associated with mRNA export underscores the intricate connections between cotranscriptional histone modification and mRNA biosynthesis.
In summary, we find that Iws1 functions as a unique CTD assembly factor in association with Spt6 and HYPB/Setd2 and helps recruit HYPB/Setd2 to the RNAPIIo complex for elongation-coupled H3K36me3. In addition, Iws1 directly or indirectly maintains low levels of at least two other transcription-coupled histone modifications, coding region histone H4 acetylation and promoter-proximal H3K27 methylation. In vitro, the assembly of this complex requires an extended region of the P-TEFb/Ser2P CTD that contains multiple consensus repeats. A more complete inventory of the proteins recruited to the RNAPII elongation complex through the Iws1:Spt6:CTD “megacomplex,” as well as the events that regulate its assembly, will help define how cotranscriptional histone modification is integrated with mRNA export.
Materials and methods
Recombinant proteins, antibodies, and cell lines
The recombinant His-Spt6 (murine), P-TEFb (human), FCP1a (human), GST-CTD (murine), and GST-Iws1 (human) proteins and the purified HeLa RNAPII preparation used in this study were described previously (Yoh et al. 2007). Recombinant Flag-Iws1 protein was affinity purified from recombinant baculovirus-infected Sf9 cell extracts using anti-Flag M2 agarose resin (Sigma). Rabbit polyclonal antisera (Pocano Rabbit Farm and Laboratory, Inc.) was raised against His-Spt6 (amino acids 1508–1726) and His-Iws1 (amino acids 523–819) proteins, as described previously (Yoh et al. 2007). Other antisera used here were obtained commercially: H5/Ser2P RNAPII (Covance); N20/RNAPII, Cyclin T1, NSD1, and His-tag (Santa Cruz Biotechnologies); THOC1 and REF1/Aly (Gene Tex), HYPB/Setd2, H3K36me3 (Abcam), γ-tubulin and Myc-tag (Sigma); H3K4me3 (07,473), H3K36me3 (05,801), H3K36me2 (07,369), acetylated histone H4 (06598; Upstate Biotechnologies/Millipore). Both the HLM107 cell line, which is a HeLa CD4+ cell line containing a single integrated copy of a Rev-defective HIV-1 genome, and the J-Lat cell line, a Jurkat-derived cell line containing an integrated but transcriptionally latent HIV-1 provirus, were purchased from American Type Culture Collection (ATCC).
RNAi, qRT–PCR, and ChIP experiments
siRNAs were transfected into either 293T or HLM107 cells using RNAi MAX (Invitrogen), according to the manufacturer’s protocol. For Iws1 and THOC1 siRNAi, cells were transfected with the specific si-RNA twice at 24-h intervals and harvested 3–4 d after the initial transfection. For knockdown of HYPB/Setd2 and Ash2L, the siRNAs were transfected only once, and cells were harvested 48 h later. The control and human Iws1 siRNA sequences were described previously (Yoh et al. 2007). The human HYPB/Setd2 siRNAs were obtained from Bioneer (sense, 5′-GUGAAGGAGUAUGCACGAAdtdt-3′; antisense, 5′-UUCGUGCAUACUCCUUCACdTdT-3′), and human THOC1 siRNAs were from Ambion (sense, 5′-GCCAUUGAACAGGC AGACCdTdT-3′; anti-sense, 5′- GGUCUGCCUGUUCAAUG GCdTdT-3′). Human Ash2L siRNAs were obtained from Ambion (sense, 5′-GCCUGGUAUUUUGAAAUCAdTdT-3′; anti-sense, 5′-UGAUUUCAAAAUACCAGGCdAdC-3′). Total RNAs were isolated using Trizol (Ambion) and reverse-transcribed using First Strand Synthesis (Invitrogen). For ChIP experiments, H3K36me3 antibody was used from Upstate Biotechnologies (05,801) or, for Figure 2 only, from Abcam (ab9050). Both RT and ChIP samples were analyzed by MX3005P q-PCR machine (Stratagene) using SYBR-green master mix (Applied Biosystems), and mRNA levels were normalized relative to 18S RNA. Chromatin immunoprecipitates were calculated relative to percent input, multiplied by 100, and represented as relative abundance; the data shown were derived from at least three independent experiments.
Protein–protein interaction experiments
The Far-Western procedure was described previously (Yoh et al. 2007). Briefly, the GST-CTD protein was phosphorylated in vitro with recombinant baculovirus-expressed P-TEFb (CycT1: CDK9), separated on a denaturing SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was incubated with GST-Spt6 SH2 (amino acids 1162–1726) labeled with glutathione-coupled HRP (Marligen), and binding was visualized by conventional chemiluminescence. For Flag-Iws1 and GST-Iws1 (amino acids 523–819) pull-down experiments, aliquots containing 5–10 μg of recombinant protein were coupled to anti-Flag M2 or glutathione agarose, respectively, and incubated with 200 μL of HeLa nuclear extract in a final volume of 500 μL of binding buffer (10 mM HEPES-HCl at pH 8.0, 200 mM KCl, 12.5 mM MgCl2, 0.2 mM EDTA, 0.5% Triton, 10% glycerol, 10 mM sodium fluoride, 5 mM activated sodium orthovanadate, 1× protease inhibitor cocktail, 0.2 mM PMSF) for 3 h at 4°C. The beads were washed extensively in washing buffer (binding buffer containing 300 mM KCl and lacking MgCl2) and analyzed by SDS-PAGE and Western blot. For coimmunoprecipitation experiments an aliquot of 500 μg of HeLa nuclear extract, which had been transfected with either control or Iws1 siRNAs, was incubated with 2 μg anti-HYPB/Setd2 antibody and protein A/G agarose beads in binding buffer overnight at 4°C. The beads were rinsed extensively with washing buffer followed by Western analysis.
For the protein interaction experiments in Figure 7A, an aliquot of 2 μg of GST- Iws1 (amino acids 523–819) fusion protein was coupled to Glutathione-S-agarose beads and incubated with 2 μg of wild-type or R1358K mutant His-tagged Spt6 in 500 μL of buffer (10 mM HEPES-HCl at pH 8.0, 300 mM KCl, 0.2 mM EDTA, 10% glycerol, 1 mM sodium fluoride, 1 mM activated sodium orthovanadate, 1× protease inhibitor cocktail, 0.2 mM PMSF, 1 mM DTT), supplemented with 10 mg/mL BSA, for 1 h at 4°C, and was then washed extensively in the absence of BSA in buffer containing 0.05% Triton. Where indicated, 50 ng of purified RNAPII fraction were added, either untreated, phosphorylated in vitro with recombinant P-TEFb, or treated with recombinant FCP1a CTD phosphatase prior to addition to the binding reaction.
RNA-FISH, mRNA export, and analysis of REF1/Aly-associated nascent mRNA
RNA-FISH with Texas red-oligo-dT (18mer) was carried out as described previously (Yoh et al. 2007). To analyze the rate of reporter gene expression, HeLa cells with an integrated HIV-1 LTR:Luciferase reporter gene were transfected with the indicated siRNAs and an HIV-1 Tat expression vector. Four hours after transfection of the Tat vector, the media was replaced with culture medium supplemented with luciferin, and luminescence was measured as counts per second for 30 sec at 10-min intervals over a 36-h period using a Lumicycle (Actimatrics). Excess luciferin in the medium rapidly quenched luciferase activity to ensure that only newly synthesized luciferase proteins were scored. To assay for REF1/Aly association with nascent RNA, HeLa cells expressing si-RNAs were fractionated into cytoplasmic, soluble nuclear extract, and nuclear pellet in the presence of RNase inhibitors. Briefly, 2 × 106 to 3 × 106 cells were incubated in 1 mL of hypotonic buffer (10 mM Tris-HCl at pH 8, 1.5 mM KCl, 2.5 mM MgCl2, 0.004% digitonin, 10 mM sodium fluoride, 5 mM activated sodium orthovanadate, 1× protease inhibitor cocktail, 0.2 mM PMSF, 40 U/mL RNase Out; Invitrogen) for 5 min on ice followed by centrifugation at 1500g for 1 min to remove the cytosolic fraction. The nuclear pellets were resuspended in 1 mL of nuclear extraction buffer (10 mM Tris-HCl at pH 8, 100 mM NaCl, 2.5 mM MgCl2, 0.5% Triton, 10 mM sodium fluoride, 5 mM activated sodium orthovanadate, 1× protease inhibitor cocktail, 0.2 mM PMSF, 40 U/mL RNase Out) and incubated for 30 min at 4°C followed by 10 min centrifugation at 14,000g. The supernatant was saved as soluble nuclear extract, and the remaining pellets were resuspended in 1 mL nuclear extraction buffer and briefly sonicated, followed by 5 min high speed centrifugation to isolate the nuclear pellet. Nuclear pellet fractions were immunoprecipitated with anti-REF1/Aly antibodies, washed extensively with nuclear buffer containing 300 mM KCl, and then subjected to Trizol extraction of the immunoprecipitated RNAs. The extracted RNAs were then analyzed by qRT–PCR, and values were calculated as percentage of input.
Acknowledgments
We thank Jane Mellor (University of Oxford, Oxford, UK) for helpful suggestions regarding this work and for sharing unpublished data. This work was supported by grants to K.J. from the NIH (AI044615) and CHRP (SIBS-164).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1720008.
References
- Adkins M.W., Tyler J.K. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Aguilera A. Cotranscriptional mRNP assembly: From the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Werner J., Nazarian A., Erdjument-Bromage H., Tempst P., Lis J.T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Reinberg D. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 2004;14:139–146. doi: 10.1016/j.gde.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bortvin A., Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Brown C.R., Silver P.A. Transcriptional regulation at the nuclear pore complex. Curr. Opin. Genet. Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chapman R.D., Conrad M., Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol. Cell. Biol. 2005;25:7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.D., Heidemann M., Hintermair C., Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Core L.J., Lis J.T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N., Vivo M., Antoniou M., Carmo-Fonseca M. Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J. Cell Biol. 2007;179:199–207. doi: 10.1083/jcb.200612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida S.F., Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Del Rosario B.C., Pemberton L.F. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Mol. Cell. Biol. 2008;28:2113–2124. doi: 10.1128/MCB.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds J.W., Mahadevan L.C., Clayton A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Fleming A.B., Kao C.F., Hillyer C., Pikaart M., Osley M.A. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Fong N., Bentley D.L. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N., Bird G., Vigneron M., Bentley D.L. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 2003;22:4274–4282. doi: 10.1093/emboj/cdg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C.K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A.G. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Hampsey M., Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Hieronymus H., Yu M.C., Silver P.A. Genome-wide mRNA surveillance is coupled to mRNA export. Genes & Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.J., Close D., Robinson H., Vallet-Gely I., Dove S.L., Hill C.P. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J. Mol. Biol. 2008;377:1460–1473. doi: 10.1016/j.jmb.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Pascual-García P., Llopis A., Zapater M., Posas F., Hurt E., Rodríguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Schneider M., Cabal G.G., Nehrbass U., Hurt E. Yeast Ataxin-7 links histone deubiquitylation with gene gating and mRNA export. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell. Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D.P., Beattie B.K., Emili A., Boone C., et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Li B., Howe L., Anderson S., Yates J.R., Workman J.L. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li M., Phatnani H.P., Guan Z., Sage H., Greenleaf A.L., Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl. Acad. Sci. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu Z., Zhou Z., Chen G., Bao S. A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem. Biophys. Res. Commun. 2007;353:47–53. doi: 10.1016/j.bbrc.2006.11.133. [DOI] [PubMed] [Google Scholar]
- Luna R., Gaillard H., González-Aguilera C., Aguilera A. Biogenesis of mRNPs: Integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P. A structural perspective of CTD function. Genes & Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Nojima T., Hirose T., Kimura H., Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- Pascual-García P., Govind C.K., Queralt E., Cuenca-Bono B., Llopis A., Chavez S., Hinnebusch A.G., Rodríguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes & Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Price D.H. Poised polymerases: On your mark . . . get set . . . go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Ramanathan Y., Rajpara S.M., Reza S.M., Lees E., Shuman S., Mathews M.B., Pe'ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Lee M.G., Winter B., Droz N.M., Eissenberg J.C., Shiekhattar R., Shilatifard A. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol. Cell. Biol. 2008;28:1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M., Fisher A.G. Epigenetic signatures of stem cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Armstrong J.A., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Dorighi K.M., Tamkun J.W. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F., et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with MEx67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R., et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Swigut T., Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Peltz S.W. Nuclear mRNA surveillance. Curr. Opin. Cell Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- Vojnic E., Simon B., Strahl B.D., Sattler M., Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- Workman J.L., Abmayr S.M. Histone H3 variants and modifications on transcribed genes. Proc. Natl. Acad. Sci. 2004;101:1429–1430. doi: 10.1073/pnas.0308506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes & Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.R., Schaffer L., Head S.R., Feeney A.J. Reciprocal patterns of methylation of H3K36 and H3K27 on proximal vs. distal IgVH genes are modulated by IL-7 and Pax5. Proc. Natl. Acad. Sci. 2008;105:8685–8690. doi: 10.1073/pnas.0711758105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh S.M., Cho H., Pickle L., Evans R.M., Jones K.A. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes & Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdell M.L., Kizer K.O., Kisseleva-Romanova E., Fuchs S.M., Duro E., Strahl B.D., Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Strahm Y., Stutz F. The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fletcher A.G., Cheung V., Winston F., Stargell L.A. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol. Cell. Biol. 2008;28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Pfeiffer H.K., Thorne A.W., McMahon S.B. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008a;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., McMahon S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008b;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A., et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]