Abstract
An RNA-based gene silencing pathway that protects bacteria and archaea from viruses and other genome invaders is hypothesized to arise from guide RNAs encoded by CRISPR loci and proteins encoded by the cas genes. CRISPR loci contain multiple short invader-derived sequences separated by short repeats. The presence of virus-specific sequences within CRISPR loci of prokaryotic genomes confers resistance against corresponding viruses. The CRISPR loci are transcribed as long RNAs that must be processed to smaller guide RNAs. Here we identified Pyrococcus furiosus Cas6 as a novel endoribonuclease that cleaves CRISPR RNAs within the repeat sequences to release individual invader targeting RNAs. Cas6 interacts with a specific sequence motif in the 5′ region of the CRISPR repeat element and cleaves at a defined site within the 3′ region of the repeat. The 1.8 angstrom crystal structure of the enzyme reveals two ferredoxin-like folds that are also found in other RNA-binding proteins. The predicted active site of the enzyme is similar to that of tRNA splicing endonucleases, and concordantly, Cas6 activity is metal-independent. cas6 is one of the most widely distributed CRISPR-associated genes. Our findings indicate that Cas6 functions in the generation of CRISPR-derived guide RNAs in numerous bacteria and archaea.
Keywords: CRISPR, Cas, endoribonuclease, RNA processing, Dicer, RNAi
All genomes are potential targets of invasion by molecular parasites such as viruses and transposable elements, and organisms have evolved RNA-directed defense mechanisms to cope with the constant threat of genome invaders (Farazi et al. 2008; Girard and Hannon 2008). The well-known subpathway of RNA silencing referred to as RNAi functions in defense against viruses in eukaryotes (Ding and Voinnet 2007). The RNAi defense response is mediated by short (∼22-nucleotide [nt]) RNAs termed siRNAs. The siRNAs are generated from invading viral RNAs by dsRNA-specific, RNase III-like endonucleases called Dicers (Jaskiewicz and Filipowicz 2008). The mature siRNAs are assembled with host effector proteins and target them to corresponding viral target RNAs to effect viral gene silencing via RNA destruction or other mechanisms (Farazi et al. 2008; Girard and Hannon 2008).
Compelling evidence has recently emerged for the existence of an RNA-mediated genome defense pathway in archaea and numerous bacteria that has been hypothesized to parallel the eukaryotic RNAi pathway (for reviews, see Godde and Bickerton 2006; Lillestol et al. 2006; Makarova et al. 2006; Sorek et al. 2008). Known as the CRISPR-Cas system or prokaryotic RNAi (pRNAi), the pathway is proposed to arise from two evolutionarily and often physically linked gene loci: the CRISPR (clustered regularly interspaced short palindromic repeats) locus, which encodes RNA components of the system, and the cas (CRISPR-associated) locus, which encodes proteins (Jansen et al. 2002; Makarova et al. 2002, 2006; Haft et al. 2005). The individual Cas proteins do not share significant sequence similarity with protein components of the eukaryotic RNAi machinery, but have analogous predicted functions (e.g., RNA binding, nuclease, helicase, etc.) (Makarova et al. 2006).
Unlike the siRNAs of the eukaryotic RNAi system, the effector RNAs of pRNAi are encoded in the host genome. CRISPR loci encode short (typically ∼30- to 35-nt) invader-derived sequences interspersed between short (typically ∼30- to 35-nt) direct repeat sequences (Bolotin et al. 2005; Mojica et al. 2005; Pourcel et al. 2005; Godde and Bickerton 2006; Lillestol et al. 2006; Makarova et al. 2006; Horvath et al. 2008; Sorek et al. 2008). Recent studies have provided clear experimental evidence that correlates the presence of virus-specific CRISPR sequences with viral immunity (Barrangou et al. 2007; Brouns et al. 2008; Deveau et al. 2008). Furthermore, viral infection has been shown to result in the appearance of new corresponding CRISPR elements in surviving strains (Barrangou et al. 2007; Deveau et al. 2008). This rapidly adapting CRISPR-based immunity acts within natural microbial populations to promote host cell fitness and to influence microbial ecology (Andersson and Banfield 2008; Tyson and Banfield 2008).
The primary products of the CRISPR loci appear to be short RNAs that contain the invader targeting sequences, and are termed guide RNAs or prokaryotic silencing RNAs (psiRNAs) based on their hypothesized role in the pathway (Makarova et al. 2006; Hale et al. 2008). RNA analysis indicates that CRISPR locus transcripts are cleaved within the repeat sequences to release ∼60- to 70-nt RNA intermediates that contain individual invader targeting sequences and flanking repeat fragments (Fig. 1A; Tang et al. 2002, 2005; Lillestol et al. 2006; Brouns et al. 2008; Hale et al. 2008). In the archaeon Pyrococcus furiosus, these intermediate RNAs are further processed to abundant, stable ∼35- to 45-nt mature psiRNAs (Hale et al. 2008).
Figure 1.
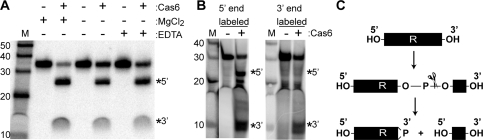
Cas6 is an endoribonuclease that cleaves CRISPR RNAs within repeat sequences. (A) psiRNA biogenesis pathway model. The primary CRISPR transcript contains unique invader targeting or guide sequences (colored blocks) flanked by direct repeat sequences (R). Cas6 catalyzes site-specific cleavage within each repeat, releasing individual invader targeting units. The Cas6 cleavage products undergo further processing to generate smaller mature psiRNA species. (B) Purified recombinant PfCas6 expressed in E. coli. The sizes (in kilodaltons) of protein markers (M) are indicated. (C) Radiolabeled RNAs (repeat–guide–repeat [R–g–R] or repeat alone [R], as diagrammed) were either uniformly or 5′-end-labeled and incubated in the absence (−) or presence (+) of PfCas6 protein (500 nM). Products were resolved by denaturing gel electrophoresis and visualized using a phosphorimager. The main cleavage products are indicated by a star or asterisk on the gel and in the diagram.
The primary goal of this study was to begin to understand the biogenesis of psiRNAs through identification and characterization of the enzyme that cleaves within the repeat sequences of CRISPR RNA transcripts to liberate the many individual psiRNA species that function in defense against molecular invaders. Our results indicate that Cas6, one of the six highly conserved or “core” Cas proteins (Haft et al. 2005), functions as a CRISPR repeat RNA-specific endoribonuclease in P. furiosus and likely numerous other archaea and bacteria.
Results
The psiRNAs, which are thought to be primary agents in prokaryotic genome defense, are derived from CRISPR RNA transcripts that consist of a series of individual invader targeting sequences separated by a common repeat sequence (Fig. 1A). To identify the enzyme required for dicing CRISPR RNA transcripts and releasing the individual embedded psiRNAs, we screened a number of recombinant P. furiosus Cas proteins for the ability to cleave CRISPR repeat sequences. We identified a single protein, Cas6 (PF1131), that cleaves specifically within the repeat sequence of radiolabeled substrate RNAs consisting of either a guide (invader targeting or “spacer”) sequence flanked by two repeat sequences or the repeat sequence alone (Fig. 1B,C). Examination of the cleavage products generated from uniformly labeled and 5′-end-labeled RNA substrates indicates that cleavage occurs ∼20–25 nt from the 5′ end of the repeat. Cleavage also occurs within each repeat of an extended substrate RNA containing two guide sequences and flanking repeats (Fig. 2).
Figure 2.
PfCas6 cleavage of a CRISPR RNA containing two repeat-guide RNA units. A uniformly radiolabeled substrate RNA containing two guide (invader targeting) sequences (yellow and green), two repeats (R) and a short (natural) 5′ leader (L) sequence was incubated with 1 μM PfCas6 protein and samples were analyzed by denaturing gel electrophoresis at the indicated times. The expected sizes and compositions of the RNA products (based on site-specific cleavage within each repeat) are indicated, as are the sizes of the marker RNAs (M).
More than 40 CRISPR-associated genes have been identified; however, only a subset of the cas genes is found in any given genome, and no cas gene appears to be present in all organisms that possess the CRISPR-Cas system (Haft et al. 2005; Makarova et al. 2006). Cas6 is among the most widely distributed Cas proteins and is found in both bacteria and archaea (Haft et al. 2005). A distinct protein with similar activity was very recently reported in Escherichia coli (Brouns et al. 2008). This protein, Cse3 (CRISPR-Cas system subtype E. coli, also referred to as CasE), is found in some bacteria that lack Cas6 (Haft et al. 2005). Both Cas6 and Cse3 are members of the RAMP (repeat-associated mysterious protein) superfamily, as are a large number of the Cas proteins (Makarova et al. 2002, 2006). RAMP proteins contain G-rich loops and are predicted to be RNA-binding proteins (Makarova et al. 2002, 2006). Cas6 is distinguished from the many other RAMP family members by a conserved sequence motif within the predicted C-terminal G-rich loop (consensus GhGxxxxxGhG, where h is hydrophobic and xxxxx has at least one lysine or arginine) (Makarova et al. 2002; Haft et al. 2005). Nuclease activity was not predicted for Cas6 based on sequence analysis.
To determine the precise PfCas6 cleavage site within the CRISPR repeat sequence, 5′-end-labeled repeat RNA was incubated with the purified enzyme and the 5′ cleavage product was mapped relative to RNase T1 (cuts after guanosines) and alkaline hydrolysis (cuts after each nucleotide) cleavage products (Fig. 3A). A 22-nt 5′ cleavage product was identified indicating that cleavage occurs between adenosine 22 and adenosine 23 of the 30-nt repeat sequence (Fig. 3A,B). The resulting 5′ end generated by PfCas6 is the same as that observed in mature psiRNA species isolated from P. furiosus cells (C. Hale, R. Terns, and M. Terns, unpubl.). Mutation of the 2 nt spanning the cleavage site (AA to GG) drastically reduced the cleavage activity of PfCas6 (Fig. 3C) without preventing binding of the enzyme to the RNA (assayed by RNA gel mobility shift; Fig. 3D). The site of cleavage is at a junction within a potential stem–loop structure that may form by base-pairing between weakly palindromic sequences commonly found at the 5′ and 3′ termini of CRISPR repeat sequences (Fig. 3B; Godde and Bickerton 2006; Kunin et al. 2007).
Figure 3.
Identification of the site of PfCas6 cleavage within the CRISPR repeat RNA. (A) The site of PfCas6 cleavage within the CRISPR repeat RNA was mapped by incubating 5′ end labeled repeat RNA with PfCas6 nuclease and comparing the size of the 5′ RNA cleavage product (arrow) with RNAse T1 (T1) and alkaline hydrolysis (OH) sequence ladders. (B) Potential secondary structure of P. furiosus repeat RNA with cleavage site indicated. (C) Analysis of cleavage of wild-type and cleavage site mutant (AA to GG) repeat RNAs with increasing concentrations (0, 1, 50, 200, and 500 nM) of PfCas6. (D) Native gel mobility shift analysis of wild-type and mutant repeat RNAs with increasing concentrations of PfCas6. The positions of the free (RNA) and protein-bound (RNP) RNAs are indicated. 5′ and 3′ cleavage products are indicated in both C and D. The sizes of RNA markers (M) are indicated in A and C.
We next investigated the RNA sequence requirements of Cas6 binding and endonucleolytic cleavage. To identify the RNA-binding determinants, we performed gel mobility shift assays with a series of RNAs (Fig. 4A). The results indicate that sequences in the 5′ region of the CRISPR repeat are important for PfCas6 binding. Under normal assay conditions, rapid cleavage prevents unambiguous observation of PfCas6 binding to the intact repeat (Fig. 3C,D), although binding can be observed with the cleavage site mutant (Fig. 3D) and at reduced temperatures where PfCas6 cleavage activity is inhibited (Supplemental Fig. S1). However, incubation of PfCas6 with the repeat RNA (Fig. 3D) or with a guide sequence flanked by two repeat sequences (Fig. 4A, panel a) under conditions compatible with cleavage reveals interaction of the protein with the 5′ cleavage product generated during incubation. PfCas6 also interacts with the gel-purified 5′ cleavage product, but not with the 3′ cleavage product (Fig. 4B). Furthermore, we found that PfCas6 binds each tested RNA that contains the repeat sequences found upstream of the cleavage site (i.e., the first 22 nt of the repeat) (Fig. 4A, panels c,f,g), but not an RNA that contains only the downstream region (last 8 nt) of the repeat (Fig. 4A, panel b).
Figure 4.
CRISPR repeat sequence requirements for PfCas6 binding. (A) Detailed analysis of binding with a series of CRISPR-derived RNAs and mutants. The left panel illustrates the RNAs tested, with repeat (R) and invader targeting (yellow blocks) sequences, and PfCas6 cleavage site (dashed lines) indicated. Blue block denotes an insertion, dashed block denotes an internal deletion, and red blocks denote substitutions (with complementary sequence). DNA indicates a DNA repeat sequence substrate. PfCas6 binding is summarized relative to binding to the 5′ cleavage product (++++). Corresponding RNA diagrams and data panels are designated with lowercase letters. The right panels show gel mobility shift analysis of the indicated RNAs with increasing concentrations (0, 1, 50, 200, and 500 nM) of PfCas6. Substrates are uniformly radiolabeled except for those shown in panels a, b, c, and l, which are 5′-end-labeled. Data for the intact repeat (*) and cleavage site mutant (**) are shown in Figure 3D. (B) PfCas6 interacts with the gel-purified 5′ cleavage product. The left panel shows the products of incubation of uniformly radiolabeled repeat RNA with (+) or without (−) PfCas6 (1 μM). The positions of the 5′ and 3′ cleavage products are indicated. The right panel shows native gel mobility shift analysis of the gel-purified 5′ and 3′ PfCas6 cleavage products (from the left panel) with increasing concentrations (0, 1, 50, 200, and 500 nM) of PfCas6. The positions of free (RNA) and protein-bound RNA (RNP) are indicated. (C) Model summarizing the minimal PfCas6-binding site within the CRISPR repeat RNA relative to the cleavage site.
Further analysis indicates that the first 12 nt of the 5′ region of the CRISPR repeat play a critical role in Cas6 binding. PfCas6 binds to an RNA comprised of the first 12 nt of the repeat with similar affinity as the 5′ cleavage product (Fig. 4A, panel h). Furthermore, protein binding is abolished by substitution or deletion of the first 8 nt of the repeat (Fig. 4A, panels d,e). In addition, substitution, insertion or deletion in the region of nucleotides 9–12 appears to have slightly reduced interaction (Fig. 4A, panels i,j,k). No binding was observed with a DNA repeat sequence (Fig. 4A, panel l). Taken together, the results indicate that PfCas6 requires sequence and/or structure information present within the first 12 nt of the CRISPR repeat RNA for stable interaction (Fig. 4C).
While nucleotides at the 5′ end of the CRISPR repeat are sufficient for robust PfCas6 binding, cleavage appears to involve additional elements. As expected, mutations that disrupt protein binding also eliminate cleavage activity (Fig. 5, panels d,e). However, other mutations dramatically reduced cleavage efficiency without disrupting PfCas6 binding. As indicated above, substitution of the two adenosines at the cleavage site disrupts cleavage but not binding (Fig. 3C,D). In addition, substitution of the last 8 nt of the repeat specifically disrupted cleavage (Fig. 5, panel f). PfCas6 cleavage activity was also significantly reduced by small (4-nt) insertions or deletions between the PfCas6-binding site and cleavage site (Fig. 5, panels i,j). Substitution of 6 nt between the binding and cleavage sites also disrupted cleavage (Fig. 5, panel k). No cleavage activity was observed with a DNA repeat sequence (Fig. 5, panel l). These results suggest that cleavage depends upon sequence elements along the length of the repeat and perhaps upon the distance between the binding and cleavage sites, and are consistent with a requirement for a specific RNA fold such as the predicted hairpin structure (Fig. 3B; Godde and Bickerton 2006; Kunin et al. 2007).
Figure 5.
CRISPR repeat sequence requirements for PfCas6 cleavage. Detailed analysis of cleavage with a series of CRISPR-derived RNAs and mutants. The left panel illustrates the RNAs tested as in Figure 4. PfCas6 cleavage is summarized relative to cleavage of the intact repeat RNA (++++). PfCas6 binding is summarized from Figure 4. Corresponding RNA diagrams and data panels are designated with lowercase letters. The right panels show cleavage assays using uniformly radiolabeled repeat RNA with (+) or without (−) PfCas6 (500 nM). Data for the intact repeat (*) is shown on right and data for the cleavage site mutant (**) is shown in Figure 3C.
P. furiosus has seven CRISPR loci with five slightly varied repeat sequences, and the elements that we identified as most important for Cas6 recognition and cleavage map to the regions of greatest sequence conservation. Variation is observed at only one position within each the first 12 and last 11 nt of the P. furiosus repeat sequences, consistent with the importance of these two regions in Cas6 binding and cleavage. On the other hand, variation occurs at three positions between the binding and cleavage sites (positions 14, 16, and 19), suggesting that nucleotide identities are less important in this region.
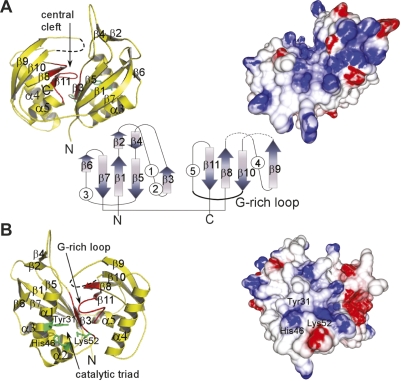
To gain a more detailed understanding of PfCas6, we obtained a crystal structure of the protein at 1.8 Å resolution (Fig. 5; see Supplemental Table S3 for structure determination details). PfCas6 contains a duplicated ferredoxin-like fold linked by an extended peptide (residues 118–123). The close arrangement of the β-sheets of the two ferredoxin-like folds creates a well-formed central cleft (Fig. 6A). The ferredoxin fold is a common protein fold also found in the structures of other RNA-binding proteins including the well-characterized RNA recognition motif (RRM), which primarily functions in ssRNA binding (Maris et al. 2005). However, PfCas6 appears to exploit a distinct mechanism of base-specific ssRNA recognition. Most notably, PfCas6 lacks the prevalent aromatic and positive residues that characterize the β-sheets of RRMs (Maris et al. 2005). The central regions of both the front and back surfaces of PfCas6 display positive potential that coincides with regions of conserved amino acids (Fig. 6) suggesting that the composite surfaces formed by the tandem ferredoxin-like folds correspond to RNA-binding sites.
Figure 6.
Structural features of PfCas6. Front (A) and back (B) views of the structure of PfCas6 represented in ribbon diagrams (left) and colored electrostatic surface potential (right). In the center, the fold topology is illustrated with arrows (β-strands) and circles (α-helices). In the ribbon diagrams, the G-rich loop characteristic of RAMP proteins is designated in red and the predicted catalytic triad residues are indicated in green. The electrostatic potential was computed using the GRASP2 program (Petrey and Honig 2003) and is colored red and blue, for negative and positive potentials, respectively.
The structure of PfCas6 allows us to predict the site of catalysis and catalytic mechanism of the enzyme. Several candidate catalytic residues are evident as strictly conserved residues in aligned Cas6 sequences (Supplemental Fig. S2). These include Tyr31, His46, and Lys52, which cluster within 6 Å of each other and are found in close proximity to the G-rich loop that contains the Cas6 signature motif (Fig. 6B). We suggest that these three residues form a catalytic triad for RNA cleavage similar to that of the tRNA intron splicing endonuclease (Calvin and Li 2008). The G-rich loop is located immediately above the putative catalytic triad and may facilitate the placement of CRISPR repeat RNA substrates. Consistent with the corresponding predicted general acid-base catalytic mechanism (proposed for the splicing endonuclease) (Calvin and Li 2008), PfCas6 does not require divalent metals and like other metal-independent nucleases cleaves on the 5′ side of the phosphodiester bond, likely generating 5′ hydroxyl (OH) and 2′, 3′ cyclic phosphate RNA end groups (Fig. 7). Finally, while binding of the enzyme occurs over a wide temperature range, PfCas6 cleavage activity is sharply temperature-dependent with significantly more activity at 70°C than 37°C (Supplemental Fig. S1).
Figure 7.
Catalytic features of PfCas6 cleavage activity. (A) Cleavage activity is not dependent on divalent metal ions. Uniformly radiolabeled repeat RNA was incubated with 1 μM PfCas6 in the absence (−) or presence (+) of 1.5 mM MgCl2 or 20 mM metal chelator EDTA as indicated. (B) Analysis of the termini of PfCas6 cleavage products. The products of cleavage reactions performed with unlabeled repeat RNA substrates (initially containing hydroxyl groups at both the 5′ and 3′ termini) were radiolabeled at either their 5′ ends (using 32P-ATP and polynuclotide kinase) or 3′ ends (using 32pCp and RNA ligase). The positions of the 5′ and 3′ cleavage products are indicated in A and B. (C) The pattern of radiolabeling of the RNA cleavage products (B) indicates that PfCas6 cleaves on the 5′ side of the phosphodiester bond, as is the case for other metal-independent ribonucleases. Cleavage likely generates 5′ hydroxyl (OH) and 2′, 3′ cyclic phosphate (>P) RNA termini.
Discussion
The results presented here indicate that Cas6 plays a central role in the production of the psiRNAs in the emerging prokaryotic RNAi pathway. Cas6 is a novel riboendonuclease. Through direct binding and cleavage of CRISPR repeat sequences, Cas6 is capable of dicing long, single-stranded CRISPR primary transcripts into units that consist of an individual guide sequence flanked by a short (8-nt) repeat sequence at the 5′ end and by the remaining repeat sequence at the 3′ end of the RNA (Fig. 1A). Mature psiRNAs retain the short repeat-derived sequence established by Cas6 at their 5′ ends in P. furiosus (C. Hale, R. Terns, and M. Terns, unpubl.), which we speculate functions as a psiRNA identity tag that allows recognition of the guide RNAs by components of the pRNAi machinery. A repeat sequence of the same length was observed on the 5′ ends of RNAs associated with E. coli Cse3, indicating that this may indeed be a generally conserved feature (Brouns et al. 2008). The 3′ ends of Cas6 cleavage products appear to be further processed since mature psiRNAs lack repeat sequences at their 3′ termini in P. furiosus (C. Hale, R. Terns, and M. Terns, unpubl.). Because Cas6 remains bound to the CRISPR repeat sequences at the 3′ end of the cleavage product (Figs. 3, 4B), Cas6 could influence the subsequent 3′ end processing of the RNA. Additional studies may reveal if Cas6 is also an important component of pRNAi effector complexes (serving to couple biogenesis and function), as is the case for eukaryotic Dicer enzymes (Jaskiewicz and Filipowicz 2008).
Cas6 is evolutionarily, structurally, and catalytically distinct from the Dicer proteins that function in the release of individual RNAs that mediate gene silencing in eukaryotes (Hammond 2005; Jaskiewicz and Filipowicz 2008). However, Cas6 is one of three different ferredoxin fold Cas proteins recently found to possess nuclease activity. Cas2, another protein found in many of the prokaryotes that possess the CRISPR-Cas system, cleaves U-rich ssRNA (Beloglazova et al. 2008). The mechanism of action of Cas6 seems to be distinct from that of Cas2, which appears to be a metal-dependent, hydrolytic enzyme (Beloglazova et al. 2008). The role of Cas2 in the pRNAi pathway is currently unknown. The E. coli Cse3 protein functions like Cas6 as a CRISPR repeat cleaving enzyme (Brouns et al. 2008). Cse3 also cleaves RNA in a divalent metal-independent manner (Brouns et al. 2008). The substrate RNA recognition requirements and the precise cleavage site have not yet been defined for Cse3. Interestingly, despite the lack of significant sequence homology, the Cas6 and Cse3 proteins appear to adopt similar structures to perform a common function in psiRNA biogenesis. Moreover, some bacteria with the CRISPR-Cas system do not appear to contain either a cas6 or a cse3 gene, suggesting that there is another Cas6 functional homolog among the Cas proteins, and illustrating the diversity of the CRISPR-Cas systems present in prokaryotes.
Materials and methods
Purification of PF1131 protein for cleavage and RNA-binding assays
N-terminal, 6x-histidine-tagged PF1131 protein (PfCas6 from P. furiosus DSM 3638 strain) was expressed in Escherichia coli BL21 codon + (DE3, Invitrogen) cells harboring a pET24d plasmid containing the appropriate gene insert (gift of Michael Adams, University of Georgia). Protein expression was induced by growing the cells to an OD600 of 0.6 and adding isopropylthio-β-D-galactoside (IPTG) to a final concentration of 1 mM. The cells were disrupted by sonication (Misonix Sonicator 3000) in buffer A (20 mM sodium phosphate [pH 7.0], 500 mM NaCl and 0.1 mM phenylmethylsulfonyl fluoride). The lysate was then cleared by centrifugation and the supernatant was incubated for 20 min at 70°C. This sample was centrifuged and the supernatant was applied to a Ni-NTA agarose column (Qiagen) that had been equilibrated with Buffer A. The protein was eluted from the column with Buffer A containing 350 mM imidazole. The purity of the protein was evaluated by SDS-PAGE and staining with coomassie blue. Buffer exchange into 40 mM HEPES-KOH (pH 7.0), 500 mM KCL was carried out using Microcon PL-10 filter columns (Millipore). The protein concentration was determined by the BCA assay (Pierce).
Generation of RNA substrates
Synthetic RNAs (listed in Supplemental Table S1) and the RNA size standards (Decade Markers) were purchased from Integrated DNA Technologies (IDT) and Ambion, respectively. These RNAs were 5′-end-labeled with T4 Polynucleotide kinase (Ambion) in a 20-μL reaction containing 20 pmol of RNA, 500 μCi of [γ32P] ATP (3000 Ci/mmol; MP Biomedicals), and 20 U of T4 kinase. The RNAs were separated by electrophoresis on denaturing (7 M urea) 15% polyacrylamide gels, and the appropriate RNA species were excised from the gel with a sterile razor blade guided by a brief autoradiographic exposure. The RNAs were eluted from the gel slices by end-over-end rotation in 400 μL of RNA elution buffer (500 mM NH4OAc, 0.1% SDS, 0.5 mM EDTA) for 12–14 h at 4°C. The RNA was then extracted with phenol/chloroform/isoamyl alcohol (PCI, 25:24:1 at pH 5.2), and precipitated with 2.5 vol of 100% ethanol in the presence of 0.3 M sodium acetate and 20 μg of glycogen after incubation for 1 h at −20°C.
All other RNAs were generated by in vitro transcription using T7 RNA polymerase (Ambion) and uniformly labeled with [α-32P] UTP (700 Ci/mmol; MP Biomedicals) as described (Baker et al. 2005). The templates used were either annealed DNA oligonucleotides or PCR products (see Supplemental Tables S1, S2), both containing the T7 promoter sequence. A typical reaction contained 200 ng of PCR product or annealed deoxyoligonucleotides, 1 mM DTT, 10 U SUPERase-IN RNase inihibitor (Ambion), 500 μM ATP, CTP, and GTP, 50 μM UTP, 30 μCi [α-32P] UTP, 1× transcription buffer (Ambion), and 40 U T7 RNA polymerase in a total volume of 20 μL.
RNA-binding and cleavage reactions
Typically, identical reaction conditions were used to assay the ability of PfCas6 protein to bind to and to cleave substrate RNAs. These reactions were initiated by incubating 0.05 pmol of 32P-radiolabed RNAs (either uniformly or 5′-end-labeled) with up to 1 μM (as indicated in the figure legends) of PfCas6 protein in 20 mM HEPES-KOH (pH 7.0), 250 mM KCl, 0.75 mM DTT, 1.5 mM MgCl2, 5 μg of E. coli tRNA, and 10% glycerol in a 20-μL reaction volume for 30 min at 70°C. Half of the reactions were directly run on native 8% polyacrylamide gels to assay RNA binding by gel mobility shift essentially as described (Baker et al. 2005). RNA cleavage was assayed using the remaining half of the reaction by deproteinizing (PCI extraction and ethanol precipitation) the RNAs and separating them by electrophoresis on denaturing (7 M urea), 12%–15% polyacrylamide gels. Gels were dried and the radiolabeled RNAs visualized by phosporimaging.
Cleavage site mapping
In order to map the site of RNA cleavage by Cas6, a standard cleavage reaction was set up using 5′ end labeled repeat RNA as described above. Alkaline hydrolysis and RNase T1 (0.1 U) ladders were generated as described previously (Youssef et al. 2007). Following the reactions, the RNAs were extracted with PCI, ethanol precipitated, and separated by electrophoresis on large, denaturing (7 M urea), 15% polyacrylamide (19:1 acrylamide:bis) gels. The gels were dried and the RNAs visualized by phosphorimaging.
Purification of PfCas6 for structure determination
N-terminal polyhistidine-tagged wild-type and selenomethionine-labeled PF1131 protein was expressed in E. coli and purified from cell extract by heat-denaturation and two chromatography steps. The cells were disrupted by sonication in a buffer containing 25 mM sodium phosphate (pH 7.5), 5% (v/v) glycerol, 1 M NaCl, 5 mM β-mercaptoethanol (βME), and 0.2 mM phenylmethylsulfonyl fluoride. The cell lysate was heated for 15 min to 70°C before being pelleted. The supernatant was then directly loaded at room temperature onto a Ni-NTA (Qiagen) column equilibrated with 25 mM sodium phosphate (pH 7.5), 5% (v/v) glycerol, 1 M NaCl, and 5 mM imidazole. The column was washed with the loading buffer containing 25 mM imidazole and then the bound protein was eluted using the loading buffer containing 350 mM imidazole. Fractions containing PF1131 were pooled and loaded onto a Superdex 200 (Hiload 26/60, Pharmacia) size-exclusion column equilibrated with 20 mM Tris-HCl (pH 7.4), 500 mM KCl, 5% glycerol, 0.5 mM ethylenediaminetetraacetic acid (EDTA), and 5 mM βME. The fractions corresponding to PF1131 were pooled and concentrated to 100 mg/mL for crystallization.
Crystallization of PF1131 and selenomethionine-labeled PF1131
Both the wild-type and selenomethionine-labeled PF1131 protein were crystallized using vapor diffusion in a hanging drop at 30°C. The droplets of PF1131 at 40 mg/mL were combined in equal volume with a well solution that contained 50 mM MES (pH 6.0), 30 mM MgCl2, and 15% (v/v) isopropanol. The crystals formed in 1–5 d with a cubic shape and to a size of ∼0.4 mm × 0.4 mm × 0.4 mm.
Data collection and structure determination
Crystals were soaked briefly in a cryo-protecting solution containing the mother liquor plus 20% (w/v) polyethylene glycol 4000 before being flash frozen in a nitrogen stream at 100 Kelvin. The crystals of the native and selenomethionine-labeled PF1131 diffracted to dmin = 1.8–2.2 Å at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22ID. The space group of the crystals was determined to be P3221 and the cell dimensions are listed in Supplemental Table S3. A single wavelength data set was collected at the anomalous peak of selenine from a selenomethionine-labeled crystal. The solvent content was calculated to be 54.9% if the crystal was assumed to contain one PF1131 in one asymmetric unit. The structure of PF1131 was solved by a SAD phasing method using the automated crystallographic structure solution program SOLVE (Terwilliger and Berendzen 1999). The initial model traced by SOLVE was further improved by the program COOT (Emsley and Cowtan 2004), followed by refinement using CNS (Brunger et al. 1998) and REFMAC5 (Murshudov et al. 1997) to Rwork/Rfree of 23.6/27.3. The quality of the structure model was checked by PROCHECK (Laskowski et al. 1993) and was found to be of satisfactory stereochemical properties.
Acknowledgments
We thank Caryn Hale (Terns laboratory) for contributions to the early stages of this project, Caryn Hale and Claiborne Glover (University of Georgia) for critical review of the manuscript, and Michael Adams (University of Georgia) for providing a PF1131 expression construct. This work was supported by NIH grant R01 GM54682 to M.T. and R.T., and NIH grant R01 GM66958 to H.L. X-ray diffraction data were collected from the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions for APS beamlines may be found at http://necat.chem.cornell.edu and http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1742908.
References
- Andersson A.F., Banfield J.F. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Baker D.L., Youssef O.A., Chastkofsky M.I., Dy D.A., Terns R.M., Terns M.P. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes & Dev. 2005;19:1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beloglazova N., Brown G., Zimmerman M.D., Proudfoot M., Makarova K.S., Kudritska M., Kochinyan S., Wang S., Chruszcz M., Minor W., et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Calvin K., Li H. RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H., Barrangou R., Garneau J.E., Labonte J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W., Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farazi T.A., Juranek S.A., Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Girard A., Hannon G.J. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde J.S., Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: Evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C., Kleppe K., Terns R.M., Terns M.P. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008 doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Horvath P., Romero D.A., Coute-Monvoisin A.C., Richards M., Deveau H., Moineau S., Boyaval P., Fremaux C., Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L., Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- Kunin V., Sorek R., Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lillestol R.K., Redder P., Garrett R.A., Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Grishin N.V., Rogozin I.B., Koonin E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Petrey D., Honig B. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Sorek R., Kunin V., Hugenholtz P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Tang T.H., Bachellerie J.P., Rozhdestvensky T., Bortolin M.L., Huber H., Drungowski M., Elge T., Brosius J., Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.H., Polacek N., Zywicki M., Huber H., Brugger K., Garrett R., Bachellerie J.P., Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G.W., Banfield J.F. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- Youssef O.A., Terns R.M., Terns M.P. Dynamic interactions within sub-complexes of the H/ACA pseudouridylation guide RNP. Nucleic Acids Res. 2007;35:6196–6206. doi: 10.1093/nar/gkm673. [DOI] [PMC free article] [PubMed] [Google Scholar]