Abstract
Covalent modifications of histones integrate intracellular and extracellular cues to regulate the genome. H3 Lys 9 methylation (H3K9me) can direct heterochromatin formation and DNA methylation, while phosphorylation of H3 Ser 10 (H3S10p) drives gene activation and chromosome condensation. To examine the relationship between H3S10p, H3K9me, and DNA methylation in Neurospora crassa, we built and tested mutants of the putative H3S10 phosphatase, PP1. A PP1-impaired mutant showed increased H3S10p and selective reduction of methylation of H3K9 and DNA. Similarly, amino acid substitutions of H3S10 abolished methylation of H3K9 and DNA. Thus, H3S10 dephosphorylation by PP1 is required for DNA methylation of some loci.
Keywords: Neurospora crassa, DNA methylation, protein phosphatase PP1, Histone H3 Ser 10 phosphorylation, histone H3 mutation
A key feature of eukaryotic genomes is their segregation into active and inactive components, generally referred to as euchromatin and heterochromatin, respectively. The state of chromatin influences genetic processes including replication, chromosome segregation, transcription, repair, and recombination. In the last decade there has been an explosion of evidence suggesting that histone modifications play important roles in functioning of the associated DNA. Heterochromatin and euchromatin show different histone modifications (Strahl and Allis 2000; Jenuwein and Allis 2001). For example, methylation of Lys 9 on H3 (H3K9me) is involved in the establishment of heterochromatin, DNA methylation, and gene silencing. In contrast, the neighboring residue, Ser 10 (H3S10) is subject to phosphorylation by a number of kinases, either in response to environmental signals to activate genes, or in response to internal cues in the cell to drive the events associated with chromosome condensation and segregation during mitosis and meiosis (Prigent and Dimitrov 2003; Nowak and Corces 2004; Johansen and Johansen 2006).
Effects of H3S10p may be mediated by its influence on methylation of H3K9, which in turn can control other processes, such as DNA methylation in Neurospora (Tamaru and Selker 2001). Structural studies showed that the Neurospora H3K9 methyltransferase DIM-5 directly interacts with H3S10, suggesting that S10 phosphorylation may influence methylation of K9 (Zhang et al. 2003), and mutation of a DIM-5 residue that interacts with S10 (D209) abolishes its methyltransferase activity (Rathert et al. 2008). Moreover, phosphorylation of Ser 10 in an H3 peptide substrate prevents the activity of H3K9 methyltransferases that have been tested in vitro, SUV39H1, Clr4, and DIM-5 (Rea et al. 2000; Nakayama et al. 2001; our unpublished results). H3S10 phosphorylation is also known to influence the binding of effector proteins such as HP1, which recognizes methylation of H3K9 to direct heterochromatin formation (Fischle et al. 2005; Hirota et al. 2005). This forms the basis of a “methyl-phospho switch” in which H3S10 phosphorylation by Aurora B at the onset of mitosis causes the dissociation of heterochromatin protein HP1 from pericentric heterochromatin (Fischle et al. 2005; Hirota et al. 2005). Similarly, in fission yeast, H3S10 phosphorylation by the Aurora kinase Ark1 causes a dissociation of the HP1 homolog, Swi6, from heterochromatin (Chen et al. 2008; Kloc et al. 2008). Reassociation of HP1 with chromatin can occur after dephosphorylation of H3S10p (Fischle et al. 2005; Hirota et al. 2005; Chen et al. 2008). Protein phosphatase PP1 is believed to be the H3S10 phosphatase in Saccharromyces cerevisiae and Caenorhabditis elegans (Hsu et al. 2000; Murnion et al. 2001) but no studies have addressed its possible role in heterochromatin formation. Intriguingly, a point mutation in PP1 (Gly200Ser/Asp) results in suppression of position effect variegation (PEV) in Drosophila suppressor Su-var(3)-6 (Dombradi and Cohen 1992).
Because methylation of H3K9 is critical for DNA methylation in N. crassa and some other systems, we sought to investigate the possible role of H3S10 phosphorylation in H3K9 and DNA methylation in Neurospora. DNA methylation in this filamentous fungus is an outcome of a genome defense mechanism, repeat-induced point mutation (RIP) (Selker 1990; Galagan and Selker 2004). During premeiosis, the RIP machinery recognizes duplications and peppers them with C:G to T:A mutations. In general, the remaining cytosines of mutated sequences are subject to methylation by the DNA methyltransferase (DNMTase) DIM-2 (Kouzminova and Selker 2001). Indeed, most DNA methylation in Neurospora is found in relics of RIP (Galagan et al. 2003; Selker et al. 2003). Cytosine DNA methylation depends on H3K9 methylation by DIM-5 (Tamaru and Selker 2001; Tamaru et al. 2003). The resulting trimethyl mark (H3K9me3) is read by the heterochromatin protein HP1, which directly interacts with DIM-2 (Freitag et al. 2004a; Honda and Selker 2008). Establishment and maintenance of DNA methylation does not require the RNAi machinery (Freitag et al. 2004b). Here we show that dephosphorylation of H3S10 is a prerequisite for establishment of H3K9 methylation. We created a partial loss-of-function mutant in the ppp-1 gene (ppp-1RIP2) and generated substitutions of the target of PP1, H3S10 (hH3S10A/G/E). Our in vivo results demonstrate that dephosphorylation of H3S10 by PP1 is required for normal H3K9 methylation and DNA methylation in Neurospora.
Results and Discussion
PP1 activity is essential for growth and development of Neurospora
As a first step to explore the effect of H3S10 phoshorylation on DNA methylation we generated and examined mutants in the putative H3S10 phosphatase, PP1. Studies with a partial loss-of-function mutant obtained previously (ppp-1RIP1) suggested that PP1 may be essential in Neurospora (Yang et al. 2004). We confirmed this (see below) but managed to use RIP to make a strain (his-3+∷Pccg-1-ppp-1+-sgfp+; ppp-1RIP2) with a greater deficiency for PP1. In this mutant strain the endogenous ppp-1 allele is expected to be a null due to the presence of two nonsense and multiple missense mutations (Supplemental Material). This strain also has a GFP tagged copy of ppp-1 gene at an ectopic location (his-3) driven by the ccg-1 promoter. The presence of the bulky GFP tag and expression under a heterologous promoter caused incomplete complementation of the null mutant, leading to a partial loss-of-function phenotype. Hereafter, we will refer to this mutant strain as ppp-1RIP2. Both the mutants grew slower than wild type, had reduced aerial hyphae, and produced fewer conidia (vegetative spores) (Fig. 1A,C). They were also defective in sexual development (Fig. 1B). These defects were rescued upon complementation of the mutant with the wild-type allele (Supplemental Fig. 1; data not shown). We were unable segregate this partially complementing wild-type copy from the RIP generated null allele (ppp-1RIP2) in crosses, indicating that PP1 is essential in Neurospora (Supplemental Material; data not shown). This conclusion was confirmed by finding that it was impossible to isolate viable progeny bearing a deletion of this gene from a sheltered heterokaryotic knockout strain generated by the Neurospora Knockout Project (Colot et al. 2006).
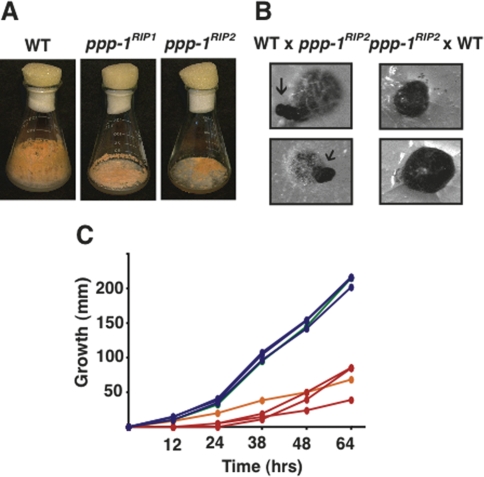
Figure 1.
PP1 is required for normal vegetative and sexual development. (A) ppp-1 mutants have reduced aerial hyphae and conidia. Wild type (N150), ppp-1RIP1 (N3482), and ppp-1RIP2 (N3468) after 7 d of growth at 32°C on Vogel’s N medium containing 1.5% sucrose. (B) Reciprocal crosses of wild type (N150) with ppp-1RIP2 (N3468) on synthetic crossing medium containing 0.1% sucrose. The female parent was inoculated 4 d before fertilization. Photographs were taken after 20 d at 25°C. When wild type was used as the female, development was normal and perithecia formed beaks (arrows). When ppp-1RIP2 was used as the female, beaks were not formed and perithecia burst to release ascospores on the agar surface. (C) Reduced growth rates of the ppp-1 mutants. Linear growth rates were measured in “race tubes” for wild type (N150, green), ppp-1RIP1 (N3482, orange), and siblings from a cross of wild type with ppp-1RIP2 (three wild-type progeny N3463, N3464, and N3465 [blue] and three mutant progeny N3466, N3467, and N3468 [red]). Points represent averaged measurements from three tubes.
PP1 is responsible for the dephosphorylation of H3S10 and influences methylation of H3K9 and DNA
To determine if PP1 is responsible for the dephosphorylation of H3S10 in Neurospora, we performed Western analyses on nuclear extracts using antibodies that recognize specific histone H3 modifications. Very low levels of H3S10 phosphorylation were observed in the wild-type strain, but substantial phosphorylation was found in the mutants, indicating that PP-1 is indeed important in the control of S10 phosphorylation (Fig. 2A). Despite ppp-1RIP2 being maintained in a PP1-GFP background that provided some PP1 activity, the ppp-1RIP2 strain showed greater phosphorylation than the leaky ppp-1RIP1 mutant. Western analysis did not show substantial changes in global levels of methylation at H3K4 or K9 (Fig. 2A) but chromatin immunoprecipitation (ChIP) revealed reduction of H3K9me3 in regions of DNA methylation (Fig. 2B,C; Supplemental Fig. 2).
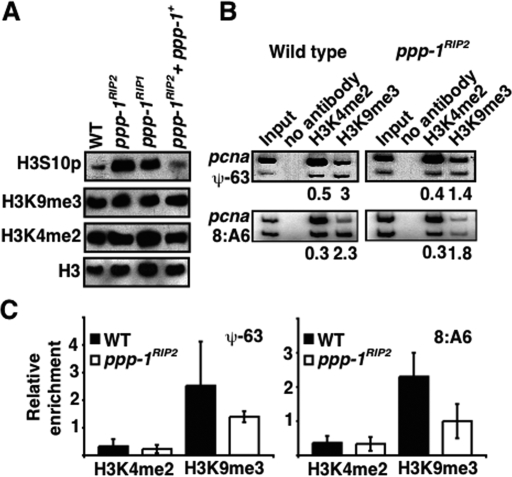
Figure 2.
Increased H3S10 phosphorylation leads to a decrease in H3K9 methylation in genomic regions that loose DNA methylation in ppp-1 mutants. (A) Increased levels of H3S10 phosphorylation in ppp-1 mutants. Nuclear extracts from wild type (N150), ppp-1RIP2 (N3468), ppp-1RIP1 (N3482), and complemented strain (ppp-1RIP2 + ppp-1+; N3470) were analyzed for H3K4me2, H3K9me3, and H3S10p by Western blotting. Loading of proteins was monitored by a Western blot for histone H3. (B) Reduction in H3K9me3 at genomic loci that loose DNA methylation in ppp-1RIP2. ChIP was performed with wild type (N150, left panel) and ppp-1RIP2 (N3468, right panel) using antibodies to immunoprecipitate either H3K4me2 or H3K9me3. Primer pairs were used in duplex PCRs to amplify an unmethylated region (pcna) and methylated regions (ψ-63 or 8:A6) (Tamaru et al. 2003). The relative enrichment of the modification (H3K4me2 or H3K9me3) at methylated regions compared with that of an unmethylated region (ψ-63/pcna or 8:A6/pcna) is indicated. (C) Quantitation of reduced H3K9me3 at genomic loci that lost DNA methylation. Data were obtained from duplex PCRs to amplify an unmethylated region (pcna) with two methylated regions (ψ-63 or 8:A6) (Supplemental Fig. 2).
Because trimethylation of H3K9 is read by HP1 to direct DNA methylation (Freitag et al. 2004a), we expected that reduction in H3K9 methylation would cause a corresponding reduction of DNA methylation. To test this possibility, we analyzed DNA methylation in the ppp-1 mutants by Southern hybridization. Consistent with the regional reduction of H3K9me3, we observed a loss of DNA methylation in the PP1 mutants at some chromosomal regions (Fig. 3). Interestingly, unlike the case in some mutants, such as dim-2 (which encodes the DNMTase), loss of DNA methylation was not complete in the ppp-1 mutants (Fig. 3); we observed a partial or complete loss of methylation in eight of 10 regions tested, suggesting that some methylated regions of the genome require PP1 while others do not. Alternatively, it is possible that the residual level of PP1 required to sustain viability is sufficient to support the low levels of DNA methylation. Not surprisingly, the loss of DNA methylation was more pronounced in the ppp-1RIP2 strain than in ppp-1RIP1 strain (Fig. 2). Introduction of a wild-type copy of ppp-1 at an ectopic location (his-3) resulted in complementation of the mutant phenotypes (Fig. 2; Supplemental Fig. 1). Normal levels of H3K9me3 were found in the regions that retain DNA methylation in the ppp-1RIP2 strain (Supplemental Fig. 3).
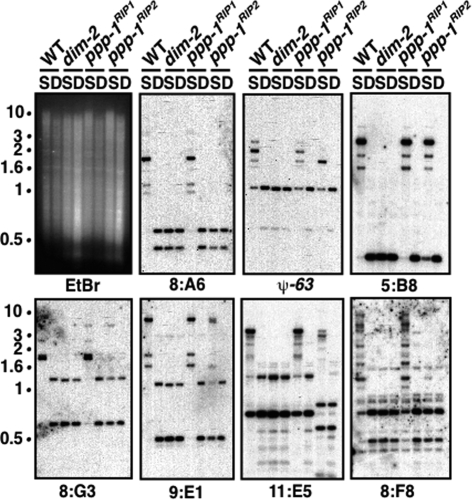
Figure 3.
PP1 is required for normal DNA methylation. Loss of DNA methylation in the ppp-1 mutants. DNA samples from wild type (N150), dim-2 (N1860), ppp-1RIP1 (N3482), and ppp-1RIP2 (N3468) were digested with methylation-sensitive and methylation-insensitive isoschizomers, Sau3AI (S) or DpnII (D), respectively, and used in Southern hybridizations with probes for methylated regions (8:A6, ψ-63, 5:B8, 8:G3, 9:E1, 11:E5, and 8:F8) (Selker et al. 2003). Blots were stripped and reprobed for an unmethylated region (set-1) (K. Adhvaryu and E. Selker, unpubl.) to confirm complete digestion of DNA (data not shown). The ethidium bromide (EtBr)-stained gel is shown revealing equivalent patterns with Sau3AI- and DpnII-digested dim-2 DNA because this strain is completely defective in DNA methylation (Kouzminova and Selker 2001). Position of the size standards (in kilobases) are shown.
Mutants with substitutions of H3 S10 are viable but show developmental defects and are defective in DNA methylation
In principle, the effect of PP1 on methylation of H3K9 and DNA could be indirect. To determine if residue 10 of H3 plays a role in histone and DNA methylation we tested the effect of amino acid substitutions at this position. Neurospora harbors just one histone H3 gene (Hays et al. 2002), facilitating tests of mutations in this gene. Using RIP, we generated a nonfunctional allele of H3 that has two nonsense and multiple missense mutations (hH3RIP1) (Supplemental Material). An additional copy of H3 with a serine to alanine substitution at residue 10 (hH3S10A) was introduced at an ectopic position (his-3), which successfully sheltered this null mutation (his-3+∷hH3S10A; hH3RIP1). We also successfully introduced alleles in which S10 was replaced by glycine or glutamic acid (hH3S10G, hH3S10E). In these strains all histone H3 had Ser 10 mutated (Fig. 4A). These histone mutants grew slower than wild type, produced conidia sparsely, and were female sterile (Supplemental Fig. 4; data not shown). A loss of H3K9me3 was observed in these mutants using two independent preparations of anti-H3K9me3 antibodies (Fig. 4B; Supplemental Fig. 5; data not shown), suggesting that S10 is indeed important for K9 methylation. Fluorescence microscopy showed that, as expected, HP1-GFP localization was also perturbed by mutation of S10, as by mutation of DIM-5 itself (Fig. 4C). Most importantly, the loss of H3K9me3 and mislocalization of HP1 was associated with a complete loss of DNA methylation in these strains (Fig. 4D). The fact that this effect is even more striking than that observed in the ppp-1 mutants (Figs. 3, 4D) may reflect the importance of Ser 10 in recognition of H3 by DIM-5 (K.K. Adhvaryu, E. Berge, H. Tamaru, M. Freitag, and E.U. Selker, in prep.). It is interesting that, unlike PP1, S10 does not appear to be essential since S10 substitution strains are viable. Similar substitutions were found to cause defects in chromosome condensation and segregation in Tetrahymena, S. cerevisiae, and Schizosaccharomyces pombe (Wei et al. 1998; Hsu et al. 2000; Mellone et al. 2003).
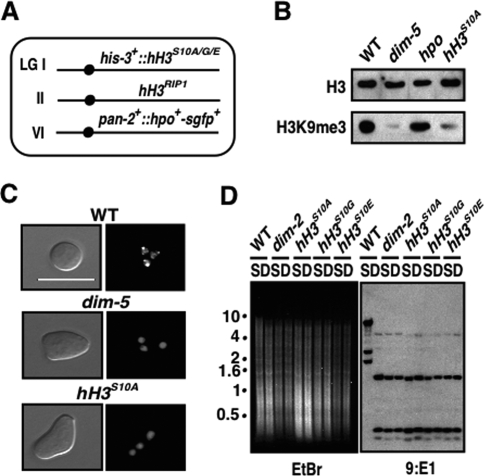
Figure 4.
Mutation of H3S10 phenocopies ppp-1. (A) Schematic of histone H3 mutant genotypes. A null allele of hH3 at the endogenous locus (hH3RIP1) (Supplemental Material) was combined with the desired substitutions (hH3S10A/G/E) at his-3. These strains also have HP1-GFP (Freitag et al. 2004a). (B) Global loss of H3K9me3 was observed in the hH3S10A mutant. Western blots were performed with ∼100 μg of nuclear proteins from wild-type (WT; N150), dim-5 (N2264), hpo (N2532), or hH3S10A (N3474) strains using antibodies against H3K9me3. The hpo mutant showed an apparently normal level of H3K9me3. The faint bands in the dim-5 and hH3S10A mutants presumably reflect background signals due to cross-reactivity of the antibody (Tamaru et al. 2003; Collins et al. 2005). Levels of H3 were analyzed to evaluate loading. (C) HP1 localization to heterochromatin is lost in the hH3S10A mutant. Visualization of GFP-tagged HP1 in nuclei of germinating conidia is shown for wild-type (N2534), dim-5 (N2542), and hH3S10A (N3481) strains. Bar, 10 μm. (D) Loss of DNA methylation in hH3 mutants [hH3S10A (N3474), hH3S10G (N3477), and hH3S10E (N3478)]. Southern blots of DNA (of hH3 mutants and wild type [N150] and dim-2 [N1860] controls) digested with Sau3AI (S) or DpnII (D) and probed for 9:E1and other methylated regions (not shown). The ethidium bromide (EtBr)-stained gel shows equivalent patterns with Sau3AI-and DpnII-digested hH3 mutant DNA, as with dim-2, indicative of loss of DNA methylation.
Accurate partitioning of the genome into domains of euchromatin and heterochromatin depends on the regulation of H3S10 phosphorylation. For example, in Drosophila the activity of the H3S10 kinase JIL-1 helps maintain the distinction between these euchromatic and heterochromatic domains by preventing ectopic H3K9 methylation and HP1 binding (Zhang et al. 2006; Bao et al. 2007). H3S10 phosphorylation is also required for the faithful propagation of heterochromatin during the cell cycle. In the fission yeast, increased H3S10 phosphorylation caused by Aurora kinase Ark1 during S phase drives the transcription of repeats in these regions. This results in the generation of small RNAs that nucleate the formation of heterochromatin by recruiting the RNAi machinery and the ClrC complex (Chen et al. 2008; Kloc et al. 2008; Zhang et al. 2008). Decreased H3S10 phosphorylation allows reassociation of HP1 with chromatin (Chen et al. 2008). A failure to remove the phosphate group from S10 would prevent this reassociation of HP1 with chromatin. Thus, regulated dephosphorylation of H3S10 appears to be important for maintenance and propagation of heterochromatin.
Although H3S10 phosphatase activity has been attributed to PP1 (Hsu et al. 2000; Murnion et al. 2001), its role in formation of heterochromatin was not previously established. Using a partial loss-of-function mutant, we demonstrate that PP1-dependent dephosphorylation of H3S10 is required prior to methylation of H3K9 by DIM-5, binding of HP1 and DNA methylation. The elevated global level of H3S10 phosphorylation in this mutant suggests that PP1 is the major H3S10 phosphatase in Neurospora. Increased H3S10 phosphorylation could cause disassociation of HP1 from heterochromatin leading to a loss of DNA methylation, but considering that a S10A replacement or phosphorylation of S10 results in a loss of DIM-5 activity in vitro (K.K. Adhvaryu, E. Berge, H. Tamaru, M. Freitag, and E.U. Selker, in prep.) it seems likely that H3S10 phosphorylation interferes with establishment of H3K9 methylation leading to a loss of DNA methylation. As noted above, other H3K9 methyltransferases including SUV39H1 and Clr4 are similarly sensitive to H3S10 phosphorylation in vitro (Rea et al. 2000; Nakayama et al. 2001).
Based on our observations, we propose a model for the role of PP1 in the establishment of DNA methylation in Neurospora (Fig. 5). PP1 is recruited to chromatin by an unidentified regulatory protein, causing dephosphorylation of H3S10, and this prepares the site for DIM-5 to methylate the neighboring K9. The trimethyl mark laid down by the DIM-5 is read by HP1 to directly recruit the DNMTase DIM-2 leading to DNA methylation (Honda and Selker 2008). It will be interesting to learn what targets various chromosomal regions for phosphorylation and dephosphorylation. Regulators of PP1 such as the “14–3–3” group of proteins or Repo-Man may be involved (Trinkle-Mulcahy et al. 2006; Winter et al. 2008). Future work will clarify the emerging connections between cell cycle-dependent phosphorylation of H3S10, methylation of H3K9, and DNA methylation.
Figure 5.
Model for role of protein phosphatase PP1 in establishment of DNA methylation. Unknown regulatory factor(s) recruit the catalytic subunit PP1 to the chromatin causing dephosphorylation of H3S10. This permits methylation of H3K9 by DIM-5. HP1 recognizes and binds this trimethyl mark, setting the stage for subsequent events that lead to methylation of DNA by DIM-2. Open ovals indicate putative unknown factors in various complexes.
Materials and methods
Neurospora strains and methods
Strains used in this study are listed in Supplemental Table 1 and were grown, maintained and crossed according to standard procedures (Davis 2000). Generation of ppp-1RIP2 and histone H3 Ser 10 substitutions are described in the Supplemental Material. For isolation of genomic DNA, strains were grown with shaking in Vogel’s minimal medium N with 1.5% sucrose and appropriate supplements for 3 d at 32°C, and DNA was isolated essentially as described previously (Irelan et al. 1993).
Genomic Southern analyses
Approximately 1 μg DNA was digested overnight with 3–5 U of restriction enzyme, fractionated in 0.8%–1.0% agarose gels, and transferred to nylon membranes and hybridized with probes prepared by random oligomer priming (Feinberg and Vogelstein 1983) as described previously (Selker and Stevens 1987).
Western blotting
Nuclei were isolated by an adaptation of the method of Baum and Giles (1986). The following reagents were added to all the buffers: 1 mM sodium butyrate, 1 μM Trichostatin A, 1 μM PMSF, 3 mM DTT, 10 mM sodium fluoride, 1 mM sodium vanadate, 0.1% phosphatase inhibitor cocktail (Sigma, P2850), and 1 μM each of leupeptin (Roche, 11034626001), pepstatin (Roche, 11524488001), and E-64 (Roche, 11585681001). Western blotting was performed with ∼100 μg of nuclear protein per sample as described previously (Tamaru et al. 2003). The following antibodies were used: α-H3 (Abcam, #1791), α-H3K4me2 (Upstate Biotechnologies, 07-030), α-H3K9me3 (Cowell et al. 2002), and α-H3S10p (Mitosis marker; Upstate Biotechnologies, 06-570). All antibodies were used at a dilution of 1:5000. HRP-conjugated goat antibody against rabbit IgG was used to detect antibody–peptide complexes by chemiluminescene (Pierce).
ChIP
ChIP experiments were performed as described previously (Adhvaryu et al. 2005) with the following antibodies: α-H3K4me2 (Upstate Biotechnologies, 07-030) and α-H3K9me3 (Cowell et al. 2002). To calculate relative enrichment band intensities were quantified using phosphor imager and the ImageQuant software (Molecular Dynamics). The values for methylated regions were divided by the values for the unmethylated region and normalized to signals of total input.
Microscopy
Suspensions of conidia were germinated on Vogel’s N agar medium containing 1.5% sucrose and the required supplements in Petri plates for 2 h at 30°C. Pieces of agar with germinating conidia were cut from plates and placed on glass slides, flooded with a few drops of liquid medium, and then overlayed with coverslips. Bright-field and fluorescence images were collected using a Zeiss Axioplan 2 Imaging system with a 10× objective and a 100× oil immersion lens. Images were processed using the Axiovision (version 4.6.3) and Adobe Photoshop CS (version 8) software. About 100 cells were examined for each sample.
Acknowledgments
We thank Dr. Yi Liu (University of Texas Southwest Medical Center) for the ppp-1RIP1 strain. We also thank Zachary Lewis, Shinji Honda, and Tamir Khalfallah for helpful suggestions on the manuscript. This work was supported by NIH grant GM025690-22 to E.U.S. K.K.A was supported partly by a fellowship from the American Heart Association (02275370Z).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1738008.
References
- Adhvaryu K.K., Morris S.A., Strahl B.D., Selker E.U. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell. 2005;4:1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Deng H., Johansen J., Girton J., Johansen K.M. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in Drosophila heterochromatin. Genetics. 2007;176:1355–1358. doi: 10.1534/genetics.107.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J.A., Giles N.H. DNase I hypersensitive sites within the inducible qa gene cluster of Neurospora crassa. Proc. Natl. Acad. Sci. 1986;83:6533–6537. doi: 10.1073/pnas.83.17.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Zhang K., Nicolas E., Cam H.P., Zofall M., Grewal S.I. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Collins R.E., Tachibana M., Tamaru H., Smith K.M., Jia D., Zhang X., Selker E.U., Shinkai Y., Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell I.G., Aucott R., Mahadevaiah S.K., Burgoyne P.S., Huskisson N., Bongiorni S., Prantera G., Fanti L., Pimpinelli S., Wu R., et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- Davis R.H.2000. Neurospora: Contributions of a model organism Oxford University Press; New York [Google Scholar]
- Dombradi V., Cohen P.T. Protein phosphorylation is involved in the regulation of chromatin condensation during interphase. FEBS Lett. 1992;312:21–26. doi: 10.1016/0014-5793(92)81402-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity (I) Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fischle W., Tseng B.S., Dormann H.L., Ueberheide B.M., Garcia B.A., Shabanowitz J., Hunt D.F., Funabiki H., Allis C.D. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Freitag M., Hickey P.C., Khlafallah T.K., Read N.D., Selker E.U. HP1 is essential for DNA methylation in Neurospora. Mol. Cell. 2004a;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- Freitag M., Lee D.W., Kothe G.O., Pratt R.J., Aramayo R., Selker E.U. DNA methylation is independent of RNA interference in Neurospora. Science. 2004b;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- Galagan J.E., Selker E.U. RIP: The evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Galagan J.E., Calvo S.E., Borkovich K.A., Selker E.U., Read N.D., Jaffe D., FitzHugh W., Ma L.J., Smirnov S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Hays S.M., Swanson J., Selker E.U. Identification and characterization of the genes encoding the core histones and histone variants of Neurospora crassa. Genetics. 2002;160:961–973. doi: 10.1093/genetics/160.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Lipp J.J., Toh B.H., Peters J.M. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Honda S., Selker E.U. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora. Mol. Cell. Biol. 2008;28:6044–6055. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Irelan J., Miao V., Selker E.U. Small scale DNA preps for Neurospora crassa. Fungal Genet. Newsl. 1993;40:24. [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johansen K.M., Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 2006;14:393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- Kloc A., Zaratiegui M., Nora E., Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzminova E.A., Selker E.U. Dim-2 encodes a DNA-methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B.G., Ball L., Suka N., Grunstein M.R., Partridge J.F., Allshire R.C. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 2003;13:1748–1757. doi: 10.1016/j.cub.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Murnion M.E., Adams R.R., Callister D.M., Allis C.D., Earnshaw W.C., Swedlow J.R. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nowak S.J., Corces V.G. Phosphorylation of histone H3: A balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Prigent C., Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- Rathert P., Zhang X., Freund C., Cheng X., Jeltsch A. Analysis of the substrate specificity of the Dim-5 histone lysine methyltransferase using peptide arrays. Chem. Biol. 2008;15:5–11. doi: 10.1016/j.chembiol.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Selker E.U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Selker E.U., Stevens J.N. Signal for DNA methylation associated with tandem duplication in Neurospora crassa. Mol. Cell. Biol. 1987;7:1032–1038. doi: 10.1128/mcb.7.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E.U., Tountas N.A., Cross S.H., Margolin B.S., Murphy J.G., Bird A.P., Freitag M. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tamaru H., Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamaru H., Zhang X., McMillen D., Singh P.B., Nakayama J., Grewal S.I., Allis C.D., Cheng X., Selker E.U. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andersen J., Lam Y.W., Moorhead G., Mann M., Lamond A.I. Repo-Man recruits PP1 γ to chromatin and is essential for cell viability. J. Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Mizzen C.A., Cook R.G., Gorovsky M.A., Allis C.D. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S., Simboeck E., Fischle W., Zupkovitz G., Dohnal I., Mechtler K., Ammerer G., Seiser C. 14–3–3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., He Q., Cheng P., Wrage P., Yarden O., Liu Y. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes & Dev. 2004;18:255–260. doi: 10.1101/gad.1152604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yang Z., Khan S.I., Horton J.R., Tamaru H., Selker E.U., Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Deng H., Bao X., Lerach S., Girton J., Johansen J., Johansen K.M. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development. 2006;133:229–235. doi: 10.1242/dev.02199. [DOI] [PubMed] [Google Scholar]
- Zhang K., Mosch K., Fischle W., Grewal S.I. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]