Abstract
Fermentation of glucose to d-lactic acid under aerobic growth conditions by an evolved Escherichia coli mutant deficient in three terminal oxidases is reported in this work. Cytochrome oxidases (cydAB, cyoABCD, and cbdAB) were removed from the E. coli K12 MG1655 genome, resulting in the ECOM3 (E. coli cytochrome oxidase mutant) strain. Removal of cytochrome oxidases reduced the oxygen uptake rate of the knockout strain by nearly 85%. Moreover, the knockout strain was initially incapable of growing on M9 minimal medium. After the ECOM3 strain was subjected to adaptive evolution on glucose M9 medium for 60 days, a growth rate equivalent to that of anaerobic wild-type E. coli was achieved. Our findings demonstrate that three independently adaptively evolved ECOM3 populations acquired different phenotypes: one produced lactate as a sole fermentation product, while the other two strains exhibited a mixed-acid fermentation under oxic growth conditions with lactate remaining as the major product. The homofermenting strain showed a d-lactate yield of 0.8 g/g from glucose. Gene expression and in silico model-based analyses were employed to identify perturbed pathways and explain phenotypic behavior. Significant upregulation of ygiN and sodAB explains the remaining oxygen uptake that was observed in evolved ECOM3 strains. E. coli strains produced in this study showed the ability to produce lactate as a fermentation product from glucose and to undergo mixed-acid fermentation during aerobic growth.
Escherichia coli is one of the most commonly used host organisms for metabolic engineering and overproduction of metabolites due to its fast growth rate, amenability to genetic manipulation, and ability to produce a wide variety of anaerobic fermentation products, such as organic acids. E. coli has also been extensively characterized with respect to its metabolic physiology (22), enabling the utilization of rational model-based engineering strategies (14, 16, 20, 21). Rational model-based approaches to engineering of E. coli that aim to couple specific metabolite overproduction to growth combined with adaptive evolution have shown promise for strain optimization (16, 17). For engineered strains that couple desirable by-product secretion to growth, adaptation to higher growth rates has been shown to lead to the increased production of the product (16).
The anaerobic growth of E. coli is characterized by the formation of a number of reduced by-products as a result of mixed-acid fermentation, with the majority of the metabolic engineering designs relying on anoxic growth conditions. Maintenance of a strict anoxic condition is a challenging task and complicates the procedure of experimental adaptation.
The goal of this study was to develop an E. coli strain that could show similar phenotypic behaviors under oxic and anoxic growth conditions. The resulting strain could be used as a platform strain in evolutionary engineering, where long-term laboratory evolution under aerobic conditions is used to optimize desirable phenotypic traits (16).
It has been reported that simultaneous deletion of the cyd and cyo genes has resulted in a significant decrease, but not elimination, of oxygen uptake (5). However, no secretion analysis or other physiological characterization of this mutant strain has been described (5, 28). Based on these results, we hypothesized that removal of all of three cytochrome oxidases would result in anaerobic growth characteristics even under oxic conditions. Moreover, we expected activation of anaerobic pathways responsible for mixed-acid fermentation as a means of NADH recycling, leading to the production of fermentation products aerobically.
A wild-type E. coli strain (MG1655) was subjected to genetic manipulation, and all active cytochrome oxidases were removed from its genome. The resulting ECOM3 (E. coli cytochrome oxidase mutant) strain was subjected to adaptive evolution and phenotypic characterization through out the course of evolution. The experimental setup for the adaptive evolution of ECOM3 and the nomenclature used in this study are presented in Fig. 1. The three evolved populations were extensively characterized, and results are presented.
FIG. 1.
Overall experimental design and nomenclature. The wild-type E. coli K12 MG1655 strain was converted to the parental ECOM3 strain through a series of genetic manipulations. A single clone (colony) of ECOM3 was isolated from the solid medium and subjected to adaptive evolution (ECOM31, ECOM32, and ECOM33), resulting in three end-point populations (eECOM31, eECOM32, and eECOM33; “e” indicates the evolved strain). Following the phenotypic characterization, the strain with the highest lactate yield was identified (eECOM31LA).
MATERIALS AND METHODS
Strains and media.
E. coli K12 MG1655 (ATCC 700926), obtained from the American Type Culture Collection (Manassas, VA), was used as a parent strain for all gene deletions in this study. During the gene deletion process, the strains were cultured on Luria-Bertani medium supplemented with 50 μg/ml kanamycin and 100 μg/ml ampicillin when necessary. Evolution and phenotype assessments of the mutant strain were carried out using M9 minimal medium (26) with glucose (2 g/liter) as the carbon source containing Na2HPO4·7H2O (6.8 g), KH2PO4 (3 g), NaCl (0.5 g), NH4Cl (1 g), MgSO4 (2 mM), and CaCl2 (0.1 mM) and trace elements (15). During the early stage of adaptive evolution, minimal medium was additionally supplemented with EZ supplements (Technova) containing a mixture of l-amino acids at the following concentrations (numbers indicate milimolarity): Ala, 0.8; Arg, 5.2; Asn, 0.4; Asp, 0.4; Cys, 0.1; Glu, 0.6; Gln, 0.6; Gly, 0.8; His, 0.2; Ile, 0.4; Leu, 0.8; Lys, 0.4; Met, 0.2; Phe, 0.4; Pro, 0.4; Ser, 10.0; Thr,0.4; Trp, 0.1; Tyr, 0.2; Val, 0.6; adenine, guanine, cytosine, and uracil, 0.2 each; and thiamine, calcium pantothanate, p-aminobenzoic acid, p-hydroxybenzoic acid, and 2,3-dihydroxybenzoic acid, 0.01 each (23).
Generation of mutant strains.
All strains and plasmids used in this study are listed in Table 1. The Escherichia coli K-12 MG1655 (ECOM3 or E. coli cytochrome oxidase mutant: ΔcydAB cyoABCD cbdAB) strain lacking three known cytochrome oxidases was generated by homologous recombination using the lambda Red recombinase system (6, 9) with primers listed in Table S1 in the supplemental material. In short, the gene to be deleted was replaced by a kanamycin gene flanked by FRT sites, and the insert was removed with an FLP recombinase. The cydAB operon was removed first, followed by the cyoABCD operon and then cbdAB. For the cbdAB operon deletion, the resistance cassette was not removed with an FLP recombinase. In order to verify the genotypes of all evolved mutants, colonies were isolated from solid medium and tested with PCR. Primers used for deletion verification are presented in Table S1 in the supplemental material. Wild-type E. coli colonies were tested in parallel as a negative control.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Escherichia coli (wild type) | ATCC (cat. no. 47076) |
| ECOM3 | MG1655, Δ(cydAB appBC cyoABCD)::FRT-kan-FRT | This study |
| ECOM3ygiN | MG1655, Δ(cydAB appBC cyoABCD ygiN)::FRT-kan-FRT | This study |
| eECOM31 | Evolved ECOM3 strain 1 (60 days) | This study |
| eECOM32 | Evolved ECOM3 strain 2 (60 days) | This study |
| eECOM33 | Evolved ECOM3 strain 3 (60 days) | This study |
| eECOM31LA | Best lactate producer isolated from eECOM31 culture | This study |
| Plasmids | ||
| pKD46 | bla γβ exo (Red recombinase), temp-conditional pSC101 operon | 9 |
| pKD13 | Template plasmid with FRT-kan-FRT (kanamycin cassette) | 9 |
| pCP20 | FLP+ λ cI857+ λ pr RepTS, Apr Cmr | 6 |
temp, temperature; cat., catalog.
Adaptive evolution.
The mutant strain was adapted though continuous passage in M9 minimal medium supplemented with 2 g/liter d-glucose and trace elements as reported earlier (15, 17). To initiate evolutions, an ECOM3 mutant was plated on the solid M9 minimal medium containing 2 g/liter glucose, trace elements, EZ supplements, and kanamycin antibiotic and incubated overnight at 37°C. A single colony was selected from the ECOM3 plate, resuspended in 10 μl of sterile water, and inoculated into three 500-ml Erlenmeyer flasks containing 250 ml of M9 minimal medium supplemented with 20 ml of 5× supplement EZ (Teknova). Flasks were incubated at 37°C using a stir bar for mixing and aeration (∼1,000 rpm). Every day, optical density (OD) measurements were taken at 600 nm and cells were passed into a fresh medium. The volume of the inoculum for each passage was adjusted to account for changes in the growth rate and to ensure that cultures would not enter the stationary phase before the next passage. The amount of EZ supplements added to the medium was reduced exponentially within the first 2 weeks of evolution. The evolutions were propagated under oxic condition for 60 days (∼700 generations) until a stable growth rate was reached. Cultures were screened every other day for contamination using PCR. The evolutions were also supplemented with 50 μg/ml kanamycin once a week in order to prevent contamination. Each evolved cell population was sampled to investigate the effects of adaptive evolution on cellular metabolism at day 1, day 30 (∼268 doublings), and day 60 (∼700 doublings). Samples were also frozen on day 1 and every 2 days throughout the evolution.
Phenotype assessment.
To measure growth rates and by-product secretion, each population was grown in batch culture at 37°C under oxic and anoxic conditions. Aerobic cultivation was conducted in 500-ml Erlenmeyer flasks containing 250 ml of M9 minimal medium with trace elements and 2 g/liter glucose as a sole carbon source. The temperature was controlled at 37°C by a circulating water bath; mixing and aeration were controlled with a stir bar at ∼1,000 rpm. Anaerobic cultivation was conducted in 250-ml Erlenmeyer flasks with 200 ml of medium, sealed with rubber stoppers containing necessary inlet tubing. Anoxic conditions were achieved by continuous flashing of cultures with a 95% N2-5% CO2 gas mixture at flow rate of 1 ml/min. The temperature was controlled by using a circulating water bath; the mixing was controlled with a stir speed of ∼200 rpm. Samples were taken from the batch cultures regularly (every 30 min), filtered through a 0.2-μm filter, and stored at −20°C for by-product secretion analysis. The glucose concentration in the medium was assessed using an enzymatic assay kit (R-Biopharm), while d-lactate secretion was measured using refractive index detection by high-performance liquid chromatography (HPLC) (Waters, Milford, MA) with a Bio-Rad Aminex HPX87-H ion exclusion column (injection volume, 50 μl) and 5 mM H2SO4 as the mobile phase (0.6 ml/min, 65°C). The identities of metabolites and organic acids in the fermentation broth were further verified using enzymatic kits (R-Biopharm). The oxygen uptake rate of each aerobic culture was determined by measuring the rate of dissolved oxygen depletion in an enclosed respirometer chamber using a polarographic dissolved oxygen probe (Cole-Parmer Instruments, Vernon Hills, IL).
qPCR.
RNA samples were taken from exponentially growing cells and added to two volumes of RNA Protect (Qiagen, Valencia, CA). Total RNA was isolated using an RNeasy minikit (Qiagen, Valencia, CA). Reverse transcription was performed on 10 μg of total RNA. The reverse transcription mixture (60 μl) contained 10 μg total RNA, 75 μg random primers, 1× 1st Strand buffer, 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphates, 30 U of Superase, and 1,500 U of Superscript II. The mixture was incubated in a thermocycler (Bio-Rad, Hercules, CA) at 25°C for 10 min, 37°C for 1 h, and then 42°C for 1 h. The reaction was followed by an incubation at 70°C for 10 min to inactivate the superscript. The RNA was then degraded by adding 20 μl of 1 N NaOH and incubation at 65°C for 30 min. After the incubation, 20 μl of 1 N HCl was added to neutralize the solution. QIAquick PCR purification kits were used to clean up the cDNA synthesis product. Following the purification, the cDNA was quantified and then directly used in quantitative PCR (qPCR) reactions. The 50 μl of qPCR reaction volume contained 25 μl of Sybr green tag master mix (Qiagen), 0.2 μM forward primer, 0.2 μM reverse primer, and cDNA as a template. Each qPCR reaction was run in triplicate in the Bio-Rad thermocycler (Bio-Rad, Hercules, CA) with the following settings: 95°C for 15 min, 94°C for 15 s, 52°C for 30 s, and 72°C for 30 s; the denaturation, annealing, and extension steps were repeated for 40 cycles. Gene expression of evolved ECOM3 strains was analyzed under oxic and anoxic growth conditions and compared to that of the wild-type strain under similar growth conditions. In order to determine the binding affinity of each primer set, a standard curve was calculated for each primer and reaction efficiency obtained from it. Using the standard curve, the relative cDNA quantity was obtained for each gene by normalizing it to the quantity of acpP (acyl carrier protein) cDNA in the same sample. acpP was chosen as the internal control gene since it is constitutively expressed in the wild type and mutants under both aerobic and anaerobic conditions (7).
Clonal analysis.
Evolved ECOM3 populations (day 60) were cultured overnight on solid M9 medium with 2 g/liter glucose and 50 μg/ml kanamycin. Ten random individual colonies were selected from each plate and grown overnight in M9 minimal medium with 2 g/liter glucose. Cells were harvested at 5,000 rpm in the centrifuge (Thermo CR3i), washed three times with M9 minimal medium without a carbon source, and loaded on a Bioscreen C machine (Growth Curves). Cultures were inoculated into 300-μl wells containing M9 minimal medium with 2 g/liter glucose and trace elements, and the initial OD of each well was kept bellow the 0.05. Cells were grown for 8 h at 37°C with continuous shaking to ensure good mixing and aeration, and OD measurements were taken at 600 nm every 15 min. Once the cells reached stationary phase, the assay was stopped and the final d-lactate concentration (g/liter) was assessed by HPLC. Strains with the highest production yield were identified and subjected to aerobic batch cultivation. Strains were grown in 500-ml Erlenmeyer flasks with 250 ml of M9 minimal medium for 8 h at 37°C with continuous agitation as described above. Samples were taken every 30 min, filtered, and analyzed using HPLC (Waters, Milford, MA). The growth rate (1/h), oxygen uptake rate (mmol/g [dry weight {DW}]/h), sugar uptake rate (mmol/g [DW]/h), and product secretion rates (mmol/g [DW]/h) were measured as described above.
Computational analysis.
The computational analyses were done using the iAF1260 genome-scale metabolic model of E. coli K-12 MG1655 (13). The simulations were performed using the Simpheny software platform (Genomatica, San Diego, CA) and the Matlab COBRA Toolbox (2) using established methods for gene deletion (11) and robustness analysis (12). The metabolic reconstruction has been examined with flux balance analysis. Flux balance analysis provides a solution space that contains all of the possible steady-state flux distributions satisfying given constrains (10, 27). Phase plane analysis (4, 12) was used to calculate the range of characteristic phenotypes that a network can display as a function of variations in the activities of two reactions, such as lactate dehydrogenase (LDH) and the biomass function (growth) (see the supplemental material). The model was constrained by experimental data by setting lower/upper bounds of uptake/secretion fluxes to the experimentally measured values. In order to allow for experimental uncertainty, the bounds were set to within one experimental standard deviation of the experimentally measured mean value.
RESULTS
Strain construction and growth adaptation.
In an effort to develop an E. coli strain that would exhibit similar phenotypic behaviors under oxic and anoxic culture conditions, we constructed a triple mutant of the wild-type E. coli K-12 MG1655 strain (ECOM3) that had the genes coding for the cytochrome oxidase bd (cydAB; b0733-b0734), cytochrome oxidase bo (cyoABCD; b0432 to b0429), and putative cytochrome oxidase (cbdAB; b0979-b0978) completely removed from the genome. The cbd locus is annotated as a putative cytochrome oxidase, and studies indicate that its gene products do not form a fully functional terminal oxidase (5, 29). Deletions of terminal cytochrome oxidases, encoded by the cydAB (cytochrome bd complex), cyoABCD (cytochrome bo complex), and cbdAB (also known as appBC) operons, have been previously found in E. coli (8, 19, 24, 25, 29).
Initial phenotypic characterization and adaptive evolution.
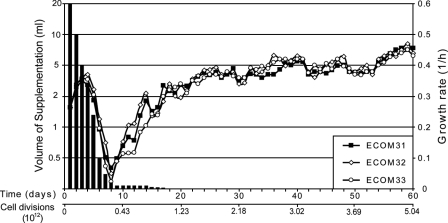
The resulting ECOM3 strain was initially incapable of growing on M9 minimal medium supplemented with 2 g/liter glucose and also demonstrated slow growth on LB medium (0.034 ± 0.002 1/h). Significant growth (0.25 ± 0.02 1/h) was observed only on rich defined medium (Technova) and on M9 minimal medium supplemented with a full mixture of amino acids (supplement EZ; Technova). Three parallel adaptive evolutions (denoted by ECOM31, ECOM32, and ECOM33) were conducted to adapt the initial ECOM3 strain to growth on M9 minimal medium with glucose as the sole carbon source (Fig. 1). Initially, a rapid decrease in the growth rate was observed upon reduction of the amino acid supplements (Fig. 2), but the growth rate increased once the supplement volume was reduced to 60 μl. The cells were then allowed to adapt to the new environment with the amount of supplement remaining unchanged for an additional 6 days. At day 14 of adaptive evolution, amino acid supplements were further reduced to 30 μl, resulting in a significant change in the growth rate. A further reduction in amino acid supplements had no effect on the cell growth rate, and starting on day 17, amino acid supplements were no longer added to the medium (see Fig. S1 in the supplemental material). A rapid increase in the growth rate followed complete removal of supplements, and the growth rate reached a maximum of 0.44 ± 0.01 1/h on day 56 of evolution. The evolutions were continued for an additional 4 days with no further observed growth rate increases (Fig. 2). The number of cell divisions that occurred during the term of evolution was estimated based on the amount of cells passed each day and the doubling time. The total number of cell divisions assuming a small death rate was 5.00 × 1012 ± 0.5 × 1012 (average is reported) (Fig. 2).
FIG. 2.
Evolutionary trajectories of ECOM3 populations. Growth rate measurements for three evolved ECOM3 strains and cell divisions are shown as a function of time of evolution. The final average growth rate was 0.42 ± 0.02 1/h. Anaerobic growth rate of wild-type E. coli is 0.45 ± 0.02 1/h. The EZ amino acid supplement amount (in ml) is shown with black bars. The total number of cell divisions for the entire period of adaptation is presented on a secondary abscissa.
The three independently evolved end-point populations (eECOM31, eECOM32, and eECOM33) showed similar growth rate gains and acquired the ability to grow on glucose minimal medium without amino acid supplementation. Evolutions were stopped once the observed growth rates for the three end-point populations (average reported, 0.42 ± 0.02 1/h) became equivalent to the growth rate of wild-type E. coli cultivated under anoxic conditions (0.45 ± 0.02 1/h), indicating a similarity of the evolved ECOM3 populations to the anaerobic phenotype of the wild-type strain. In order to further probe the metabolic phenotypes of the populations during and after evolutions, growth rates, oxygen uptake rates, sugar uptake rates, and product secretion rates were measured for each of the three on day 1, day 30, and day 60.
Phenotypic characterization of the evolved populations.
Phenotypic characterization of the evolved populations revealed that the three evolutionary endpoints had slightly different metabolic phenotypes (Table 2). As a common feature, a twofold increase in the substrate uptake rate was observed for all three populations from 9.98 ± 2.0 mmol/g (DW)/h to 20.80 ± 0.7 mmol/g (DW)/h on average within the first 30 days of evolution. Similarly, the d-lactate secretion rate increased more than twofold by day 30 of adaptive evolution (from 17.00 ± 2.9 mmol/g [DW]/h to 35.44 ± 5.5 mmol/g [DW]/h on average), but the end-point populations showed a higher variance in lactate secretion than in glucose uptake.
TABLE 2.
Phenotypic characteristics of ECOM3 populations during adaptive evolutiona
| Strain | Day | SUR (mmol/g [DW]/h) | OUR (mmol/g [DW]/h) | LactSR (mmol/g [DW]/h) | AcSR (mmol/g [DW]/h) |
|---|---|---|---|---|---|
| MG1655 | 9.02 ± 0.23 | 14.92 ± 0.21 | 0.40 ± 0.01 | 3.4 ± 0.02 | |
| ECOM3 | 11.88 ± 2.01 | 6.9 ± 1.52 | 17.60 ± 0.65 | 3.84 ± 0.45 | |
| ECOM31 | 1 | 7.67 ± 0.60 | 7.90 ± 1.00 | 16.63 ± 0.70 | 3.75 ± 0.49 |
| 30 | 20.88 ± 0.89 | 3.81 ± 0.45 | 35.09 ± 0.36 | 0.33 ± 0.05 | |
| 60 | 19.89 ± 0.94 | 5.61 ± 0.30 | 36.36 ± 0.25 | 0.50 ± 0.02 | |
| ECOM32 | 1 | 11.07 ± 0.78 | 7.75 ± 0.42 | 15.94 ± 0.98 | 4.32 ± 0.58 |
| 30 | 20.53 ± 1.01 | 5.26 ± 0.24 | 35.15 ± 2.64 | 2.73 ± 0.47 | |
| 60 | 21.15 ± 1.50 | 3.69 ± 0.60 | 35.63 ± 1.56 | 2.31 ± 0.31 | |
| ECOM33 | 1 | 11.23 ± 1.20 | 5.04 ± 0.50 | 17.24 ± 0.98 | 3.43 ± 0.68 |
| 30 | 16.90 ± 2.16 | 4.29 ± 0.21 | 25.84 ± 2.19 | 3.48 ± 0.22 | |
| 60 | 21.28 ± 0.97 | 2.22 ± 0.38 | 29.98 ± 2.81 | 8.17 ± 0.91 |
SUR, substrate uptake rate; OUR, oxygen uptake rate; LactSR, lactate secretion rate; AcSR, acetate secretion rate.
Acetate, one of the major fermentation products of wild-type E. coli, was not a major growth by-product of the ECOM3 parental strain prior to evolution. However, the three evolutionary end-point populations had significantly different acetate secretion rates. Acetate secretion was strongly reduced for eECOM31, while only a moderate reduction was seen for eECOM32. In contrast, the acetate secretion rate of eECOM33 increased more than twofold by day 60 (Table 2).
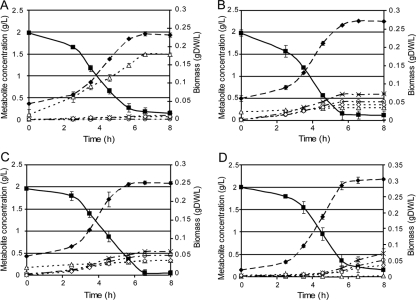
Detailed secretion analysis of the ECOM3 end-point populations under oxic conditions (Fig. 3) indicated that the strains acquired the ability to secrete d-lactate as a dominant fermentation product at yields of 0.76, 0.73, and 0.65 g lactate/g glucose for eECOM31, eECOM32, and eECOM33, respectively. Compared to those of the wild-type strain, the lactate secretion rates increased by 94.5-, 92.3- and 77.2-fold, while the substrate uptake rates increased by 2.3-, 2.2-, and 2.3-fold, respectively, for the three evolved ECOM3 populations. These results are consistent with gene expression analysis using reverse transcription-PCR, which showed that the ldhA gene, encoding the LDH protein, was upregulated by 21-, 8-, and 23-fold, respectively, in the three evolved strains compared with the level in the wild-type strain (see Fig. 5). The increase in glucose uptake and lactate secretion, without the production of any other major fermentation products, results in a significantly elevated flux through the glycolytic pathway. Consistent with the physiological characteristics of the end-point strains, we did not observe a significant increase in the expression of the pyruvate dehydrogenase gene (aceF) or pyruvate formate lyase (pflA) except in the case of the eECOM32 population (Fig. 4).
FIG. 3.
Aerobic growth and secretion profiles of three end-point populations. eECOM31 (A), eECOM32 (B), eECOM33 (C), and wild-type (D) strains are analyzed. Data were taken on day 60 of evolutions; all measurements were done in triplicate. The solid black line (▪) indicates the concentration of glucose remaining in the culture; dashed black lines with symbols are as follows: ▵, amount of d-lactate produced; ○, concentration of acetate; ⧫, cell density (g [DW]/liter).
FIG. 5.
Anaerobic growth and secretion profile of three end-point populations. eECOM31 (A), eECOM32 (B), eECOM33 (C), and wild-type (D) strains are analyzed. The solid black line (▪) indicates the decrease in concentration of glucose remaining in the culture; dashed black lines are as follows: ▵, amount of d-lactate produced; ○, acetate concentration; ×, formate concentration; ⋄, ethanol concentration; ⧫, cell density (g [DW]/liter).
FIG. 4.
Mechanism of d-lactate production and associated gene expression analysis. Pathways of conversion of pyruvate to common organic acids are presented with corresponding enzyme names. Expression of the ldhA, aceF, and pflA genes was measured and is presented by the bar diagrams. Gene expression was measured under oxic (dark gray bars) or anoxic (light gray bars) conditions. ldhA showed a significant upregulation, while no upregulation was observed for the aceF and pflA genes.
Clonal analysis.
In order to characterize and study heterogeneity in the evolved populations, we used clonal analysis. Evolved populations were plated on solid medium at day 60 of evolution, and 10 random colonies from each population were selected for the analysis. We observed a higher level of heterogeneity within the eECOM33 population. The clone with the highest lactate yield (eECOM31LA) was identified within the eECOM31 population based on the highest final lactate concentration (see Fig. S2 in the supplemental material). Phenotypic assessment of eECOM31LA showed growth rates compatible to those of the eECOM31 population under oxic conditions, with slightly higher glucose uptake and lactate secretion rates (glucose uptake rate, 21.61 ± 0.16 mmol/g [DW]/h; lactate secretion rate, 42.32 ± 2.52 mmol/g [DW]/h). The final lactate concentration for the eECOM31LA strain was 1.58 ± 0.1 g/liter, which is equivalent to an 80% conversion of glucose to lactate (0.8 g lactate/g glucose). The growth rate of eECOM31LA (0.39 ± 0.01 1/h) was slightly lower than that of the eECOM31 population. Moreover, we observed that the eECOM31LA mutant had a significantly lower oxygen uptake rate (2.44 mmol O2/g [DW]/h) than the eECOM31 population, demonstrating a wide range of oxygen requirements established during adaptive evolution.
Oxygen dependencies of ECOM strains.
In order to determine whether the presence or absence of oxygen affects the phenotype of the ECOM3 strain, we subjected three evolved populations to anaerobic growth on M9-glucose minimal medium. We observed a slight decrease in the growth rate, 12%, 5%, and 7% for the eECOM31, eECOM32, and eECOM33 populations, respectively, as well as radically different secretion profiles from the aerobic phenotype (Table 3). Secretion analysis demonstrated that the eECOM31 population preserved its aerobic phenotype and produced d-lactate as the sole by-product at a concentration similar to the one observed under oxic growth conditions. In contrast, the eECOM32 and eECOM33 populations lost their ability to secrete d-lactate as a sole by-product and presented a phenotype similar to that of wild-type E. coli under anoxic growth conditions with formate, acetate, and ethanol as by-products (Fig. 5). Gene expression analysis of the ldhA, pflA, and aceF genes under anoxic growth conditions failed to reveal a clear mechanistic basis for the observed physiological differences between the three strains.
TABLE 3.
Phenotypic comparison of ECOM3 populations between oxic and anoxic growth environmentsa
| Strain | Aerobic GR (1/h) | Anaerobic GR (1/h) | Aerobic lactate titer (g/liter) | Anaerobic lactate titer (g/liter) |
|---|---|---|---|---|
| Unevolved wild-type | 0.71 ± 0.01 | 0.45 ± 0.02 | 0 | 0.04 ± 0.01 |
| eECOM31 | 0.42 ± 0.01 | 0.37 ± 0.01 | 1.51 ± 0.01 | 1.47 ± 0.01 |
| eECOM32 | 0.40 ± 0.02 | 0.38 ± 0.02 | 1.45 ± 0.04 | 0.29 ± 0.02 |
| eECOM33 | 0.44 ± 0.02 | 0.42 ± 0.01 | 1.30 ± 0.06 | 0.31 ± 0.03 |
GR, growth rate.
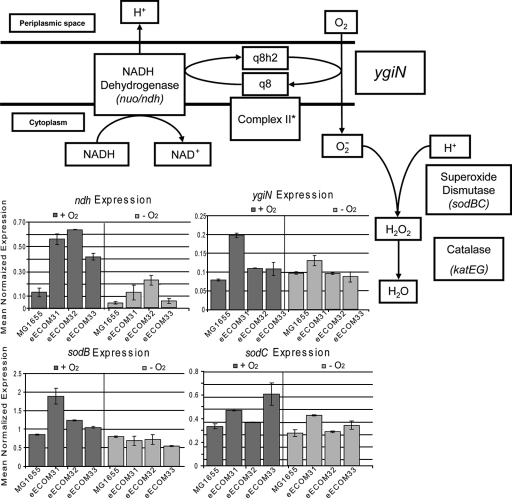
We originally hypothesized that deletion of cytochrome oxidases would completely eliminate oxygen consumption by the ECOM3 strain. However, while oxygen consumption was significantly reduced in the evolved strains, some residual oxygen uptake remained. Prior to evolution (day 0), the oxygen uptake rate of the ECOM3 strain was 6.89 ± 1.61 mmol O2/g (DW)/h, which is almost three times lower than the oxygen uptake rate of the wild-type strains. The oxygen consumption was further reduced to 3.84 ± 1.7 mmol O2/g (DW)/h (average reported for all evolved populations) by the end of the adaptive evolution (Table 2). Evolved populations showed significantly different oxygen uptake rates: 5.62 ± 0.34, 3.69 ± 0.66, and 2.21 ± 0.38 mmol O2/g (DW)/h for the eECOM31, eECOM32, and eECOM33 populations, respectively. Gene expression analysis together with in silico phenotypic modeling (see Discussion) revealed that oxygen uptake levels were consistent with observed levels of expression of the ygiN gene (annotated as quinol monooxygenase [1]; however, since the proposed biochemical reaction (1) does not involve incorporation of oxygen into an electron donor, the term monooxygenase should not be used). The eECOM31 population showed the highest oxygen uptake rate, consistent with the highest ygiN expression. In order to determine if YgiN is involved in oxygen uptake, we conducted an additional gene deletion and removed ygiN from the original unevolved ECOM3 strain. The removal of ygiN almost completely eliminated oxygen uptake (see Discussion).
DISCUSSION
To our knowledge, we describe here the first E. coli strain that is able to homoferment glucose to lactic acid under aerobic growth conditions. This strain (ECOM3) was engineered by removing all active cytochrome oxidases. Genes were removed using homologous recombination techniques, and the resulting strain was evolved to achieve growth on M9 minimal medium with trace elements and 2 g/liter glucose. The observed growth rate after 60 days of evolution was equivalent to the growth rate of wild-type E. coli under anoxic conditions. Adaptive evolution produced three end-point populations that exhibited similar behavior aerobically and had radically different phenotypic characteristics anaerobically. Lactic acid was identified as a major product of aerobic fermentation for all three end-point populations. The best representative of the eECOM31 population exhibited the highest lactate secretion and glucose uptake rate. The yield of lactate from glucose was close to 80%. We used gene expression analysis to investigate genetic perturbations that underlay secretion of lactic acid and the remaining oxygen uptake rate. We also utilized a genome-scale metabolic model of E. coli (iAF1260) to understand the mechanism of oxygen utilization in the ECOM3 phenotype.
In silico analysis using a genome-scale model.
In order to identify potential metabolic fates of oxygen in the ECOM3 strain, we employed a genome-scale metabolic model of E. coli (iAF1260) (13). The in vivo genotype of the ECOM3 strain was implemented computationally though the removal of reactions catalyzed by the deleted genes. The in silico model was further constrained using experimental data to set the glucose and oxygen uptake rates and acetate secretion rates (with experimental error accounted for by allowing a range of uptake/secretion rates). Analysis of the computationally predicted flux distributions utilizing these constraints provided insights into the observed residual oxygen utilization. The model predicted that the mechanism that could account for residual oxygen uptake at the observed level would be through the activity of the ygiN gene. The simulation of the ECOM3 phenotype with a computational model and scientific evidence showed flux coupling between the NADH dehydrogenase (nuo/ndh operons) and the reaction catalyzed by the ygiN gene, forming a ubiquinone cycle. It has been shown that ubiquinone is the electron acceptor for the NADH dehydrogenase (ndh/nuo) (18) and a preferred electron carrier for E. coli during aerobic growth (3). Furthermore, Adams and Jia (1) indicate that ygiN can potentially react with the ubiquinol molecule and oxidize it to the ubiquinone form through coupling of this oxidation reaction with reduction of the molecular oxygen. Based on these findings, we proposed the mechanism for oxygen utilization (Fig. 6).
FIG. 6.
Proposed mechanism of oxygen utilization by ECOM3 strains and corresponding gene expression. A possible mechanism of oxygen utilization by ECOM3 strains is presented. This mechanism was elucidated based on gene expression analysis (bar diagrams) and scientific evidence (1, 3, 18). Gene expression was measured under oxic (dark gray bars) and anoxic (light gray bars) conditions. Expression for the catalase is not shown. *, complex II was not included in this figure due to a lack of evidence of its involvement.
The removal of the ygiN gene in silico predicted elimination the oxygen uptake. In order to determine if ygiN accounts for the oxygen uptake, we removed it from the parental ECOM3 strain and observed a nearly complete elimination of oxygen uptake: 0.03 ± 0.04 mmol O2/g (DW)/h. This experimental evidence demonstrates that the observed oxygen uptake can be attributed to the activity of YgiN. Consistent with the role of this pathway in residual oxygen utilization, we found that levels of aerobic expression of all the genes in this pathway were increased in the ECOM3 populations over those of the wild-type strain (Fig. 6). In particular, the expression of the ygiN gene was increased nearly three times in the eECOM31 population. Additional computational analysis of the observed phenotype is presented in the supplemental material.
In summary, we have engineered an E. coli strain (ECOM3) capable of homofermenting glucose to lactate under both aerobic and anaerobic conditions by deleting all cytochrome oxidase genes and adapting the strain to grow on minimal medium without amino acid supplementation. Clonal analysis allowed identification of the best lactic acid producer from the eECOM31 population, with lactate yields close to 80% from glucose. Interestingly, cell populations derived from the adaptive evolution showed significant residual oxygen uptake. We identified the mechanisms accounting for the observed residual oxygen uptake using a combination of a genome-scale metabolic model of Escherichia coli and gene expression analysis of specific pathways. The resulting ECOM3 populations have been shown to be amenable to genetic manipulation (results not shown) and thus can be used as a platform strain for further metabolic engineering that redirects lactate flux into other desirable by-products.
Supplementary Material
Acknowledgments
We thank Eric M. Knight and Larisa Kagan for help with generation of the cydAB E. coli mutant strain, Ines Thiele, Karsten Zengler, and Byung Kwan Cho for the critical review of the manuscript, and Dae-Hee Lee for helpful discussions. We also thank anonymous reviewers for their insightful comments and suggestions that helped us improve the manuscript.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, M. A., and Z. Jia. 2005. Structural and biochemical evidence for an enzymatic quinone redox cycle in Escherichia coli: identification of a novel quinol monooxygenase. J. Biol. Chem. 280:8358-8363. [DOI] [PubMed] [Google Scholar]
- 2.Becker, S. A., A. M. Feist, M. L. Mo, G. Hannum, B. O. Palsson, and M. J. Herrgard. 2007. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 3.Bekker, M., G. Kramer, A. F. Hartog, M. J. Wagner, C. G. de Koster, K. J. Hellingwerf, and M. J. de Mattos. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153:1974-1980. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. L., and B. O. Palsson. 2004. Phenotype phase plane analysis using interior point methods. Comput. Chem. Eng. 29:481-486. [Google Scholar]
- 5.Calhoun, M. W., K. L. Oden, R. B. Gennis, M. J. de Mattos, and O. M. Neijssel. 1993. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J. Bacteriol. 175:3020-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Covert, M. W., E. M. Knight, J. L. Reed, M. J. Herrgard, and B. O. Palsson. 2004. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429:92-96. [DOI] [PubMed] [Google Scholar]
- 8.Dassa, J., H. Fsihi, C. Marck, M. Dion, M. Kieffer-Bontemps, and P. L. Boquet. 1991. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol. Gen. Genet. 229:341-352. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19:125-130. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, J. S., and B. O. Palsson. 2000. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, J. S., and B. O. Palsson. 2000. Robustness analysis of the Escherichia coli metabolic network. Biotechnol. Prog. 16:927-939. [DOI] [PubMed] [Google Scholar]
- 13.Feist, A. M., C. S. Henry, J. L. Reed, M. Krummenacker, A. R. Joyce, P. D. Karp, L. J. Broadbelt, V. Hatzimanikatis, and B. O. Palsson. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feist, A. M., and B. O. Palsson. 2008. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 26:659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270:880-891. [DOI] [PubMed] [Google Scholar]
- 16.Fong, S. S., A. P. Burgard, C. D. Herring, E. M. Knight, F. R. Blattner, C. D. Maranas, and B. O. Palsson. 2005. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 91:643-648. [DOI] [PubMed] [Google Scholar]
- 17.Fong, S. S., A. Nanchen, B. O. Palsson, and U. Sauer. 2006. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J. Biol. Chem. 281:8024-8033. [DOI] [PubMed] [Google Scholar]
- 18.Jaworowski, A., H. D. Campbell, M. I. Poulis, and I. G. Young. 1981. Genetic identification and purification of the respiratory NADH dehydrogenase of Escherichia coli. Biochemistry 20:2041-2047. [DOI] [PubMed] [Google Scholar]
- 19.Kita, K., K. Konishi, and Y. Anraku. 1984. Terminal oxidases of Escherichia coli aerobic respiratory chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 259:3375-3381. [PubMed] [Google Scholar]
- 20.Lee, K. H., J. H. Park, T. Y. Kim, H. U. Kim, and S. Y. Lee. 2007. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol. Syst. Biol. 3:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S. J., D. Y. Lee, T. Y. Kim, B. H. Kim, J. Lee, and S. Y. Lee. 2005. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl. Environ. Microbiol. 71:7880-7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neidhardt, F. C., et al. (ed.). 1996. Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, DC.
- 23.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton, G., and R. B. Gennis. 1991. In vivo assembly of the cytochrome d terminal oxidase complex of Escherichia coli from genes encoding the two subunits expressed on separate plasmids. Biochim. Biophys. Acta 1089:8-12. [DOI] [PubMed] [Google Scholar]
- 25.Puustinen, A., M. Finel, T. Haltia, R. B. Gennis, and M. Wikstrom. 1991. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30:3936-3942. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. A2.2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schilling, C. H., J. S. Edwards, D. Letscher, and B. O. Palsson. 2000. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol. Bioeng. 71:286-306. [PubMed] [Google Scholar]
- 28.Shioi, J., R. C. Tribhuwan, S. T. Berg, and B. L. Taylor. 1988. Signal transduction in chemotaxis to oxygen in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 170:5507-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturr, M. G., T. A. Krulwich, and D. B. Hicks. 1996. Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a Δcyo Δcyd strain complemented by genes from Bacillus firmus OF4. J. Bacteriol. 178:1742-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.