Abstract
Anaplasma species are tick-transmitted pathogens that impact veterinary and human health. Sicily is one of the locations where these pathogens are endemic. Sicily represents a typical Mediterranean ecosystem to study Anaplasma infection and tick habitat suitability. The aims of this study were (i) to characterize by 16S rRNA and species-specific msp4 gene PCR the prevalence and genotypes of A. marginale, A. phagocytophilum, and A. ovis in the most abundant host species in Sicilian provinces and (ii) to correlate differences between hosts and between western and eastern Sicily with the habitat suitability for ticks in these regions. Differences were found in the prevalence of Anaplasma spp. between different hosts and between western and eastern provinces. The differences in Anaplasma prevalence between different hosts may be explained by pathogen host tropism. The differences between western and eastern provinces correlated with the tick habitat suitability in these regions. The analysis of Anaplasma genotypes suggested a higher host and regional specificity for A. phagocytophilum than for A. marginale and A. ovis strains, a finding probably associated with the broader host range of A. phagocytophilum. The presence of identical A. marginale genotypes in the two regions may reflect cattle movement. The results for A. ovis suggested the possibility of some genotypes being host specific. These results provide information potentially useful for the management of tick-borne diseases caused by Anaplasma spp. in Sicily and other Mediterranean regions and may contribute to the development of models to predict the risks for these tick-borne pathogens.
The genus Anaplasma (Rickettsiales: Anaplasmataceae) includes tick-transmitted pathogens that impact veterinary and human health (12, 17). Anaplasma spp. include ruminant pathogens such as A. marginale, which causes bovine anaplasmosis, and A. ovis, which infects sheep, goat, and deer species (10, 17). This genus also includes A. phagocytophilum (previously recognized as Ehrlichia equi, Ehrlichia phagocytophila, and the human granulocytic ehrlichiosis agent), which infects a wide range of hosts including humans and wild and domesticated animals and causes human, canine, and equine granulocytic anaplasmosis and tick fever of ruminants (12).
Anaplasma spp. are endemic in Sicily, where they constitute a veterinary and human health problem (2, 5-7, 9, 24, 25). Information is available on ticks and the prevalence and genetic diversity of Anaplasma spp. in Sicily (4-7, 9, 23, 25). However, a study covering all Sicilian provinces has not been previously conducted.
The aim of this study was to characterize the prevalence and genotypes of Anaplasma spp. in the most abundant host species in Sicilian provinces and to correlate differences between hosts and between western and eastern Sicily with the habitat suitability for ticks in these regions. Since ticks are the biological vectors of Anaplasma spp., it is interesting to correlate the presence of these pathogens with tick distribution patterns on the island. Because of the lack of long-standing and extensive tick surveys, we used habitat suitability models describing the expected distribution area for the most abundant tick species in the Mediterranean region (14). This is an increasingly popular method aimed at understanding the climate niche of an organism and using these findings to predict its potential distribution.
Sicily represents a typical Mediterranean ecosystem and offers a good milieu to study the prevalence of Anaplasma spp. in relation to the habitat suitability for ticks. These results provide information potentially useful for the management of tick-borne diseases caused by Anaplasma spp. in Sicily and other Mediterranean regions and may contribute to the development of models to predict the risks for these tick-borne pathogens.
MATERIALS AND METHODS
Study site.
Sicily is an autonomous region of Italy and is the largest island in the Mediterranean Sea at 25,708 km2. The island has a typical Mediterranean ecosystem. The climate is characterized by hot and dry summers and mild winters with light rainfall. Sicilian vegetation is remarkably diverse with a great variety of agricultural crops (e.g., citrus fruits, grapes, olives, artichokes, pistachios, and mulberries), flowering shrubs and grasses, cactus, palm, fir trees, and stone (umbrella) pines. Wild fauna is not abundant in the island, but sheep, cattle, goats, pigs, and horses are economically important. Administratively, Sicily is divided into nine provinces: Agrigento, Caltanissetta, Catania, Enna, Messina, Palermo, Ragusa, Syracuse, and Trapani. Each province is divided into communes that vary in number between 12 and 108 per province (average ± standard deviation, 43 ± 33). For this study, Sicily was divided into western (Agrigento, Caltanissetta, Palermo, and Trapani) and eastern (Enna, Catania, Messina, Ragusa, and Syracuse) regions (Fig. 1).
FIG. 1.
Map of Sicily with the administrative (province) divisions and separation of eastern (white) and western (gray) regions of the island as considered in the current study.
Animals and sample collection.
Cattle (n = 374 from 29 herds in 10 communes), goats (n = 134 from five herds in four communes), sheep (n = 286 from 12 herds in six communes), horses (n = 134 from seven herds in two communes), domestic dogs (n = 46, all from different households in three communes), and mice (Mus domesticus and Apodemus sylvaticus) (n = 69 captures in two communes) were sampled in western and eastern Sicilian provinces. One porcupine (Hystrix cristata) and one domestic cat were sampled in western Sicily. Blood was collected during 2006 and 2007 from veins of live animals into sterile tubes with anticoagulant (EDTA) and stored at −20°C until used for DNA extraction. Animal capture and sampling protocols complied with Intituto Zooprofilattico Sperimentale della Sicilia institutional animal welfare regulations.
DNA tests for Anaplasma spp. and sequence analysis.
DNA was extracted from blood samples using the GenElute mammalian genomic DNA Miniprep kit (Sigma, St. Louis, MO). The DNA was resuspended in sterile distilled water and stored at −20°C until used. PCR and sequence analyses of Anaplasma 16S rRNA (8), A. marginale major surface protein 4 (msp4) (3), A. ovis msp4 (10), and A. phagocytophilum msp4 (4) genes were conducted as previously described (3, 4, 8, 10). PCRs were performed with 1 μl (0.1 to 10 ng) DNA using 10 pmol of each primer and the Ready-To-Go PCR beads (Amersham, Piscataway, NJ) or the Access reverse transcription-PCR system (Promega, Madison, WI). Reactions were performed in an automated DNA thermal cycler (Techne model TC-512; Cambridge, England, United Kingdom) for 35 cycles. PCR products were electrophoresed on 1% agarose gels to check the size of amplified fragments by comparison to a DNA molecular size marker (1-kb DNA ladder; Promega). Control reactions were done without the addition of DNA to the reaction mixture to rule out contaminations during PCR.
A 16S rRNA gene fragment (468 bp) from all A. phagocytophilum msp4-positive samples and all (if the number of positive samples was ≤16) or randomly selected (if the number of positive samples was >16) A. marginale and A. ovis msp4 genes (complete coding sequence) was amplified, and the products were resin purified (Wizard; Promega) and cloned into the pGEM-T vector (Promega) for sequencing of both strands by double-stranded dye termination cycle sequencing (Secugen SL, Madrid, Spain). Three randomly selected 16S rRNA amplicons from each cattle, goat, sheep, and horse sample negative in the msp4 PCR were also cloned and sequenced. At least two independent clones were sequenced for each PCR. Multiple sequence alignment was performed using the program AlignX (Vector NTI suite V 5.5; InforMax, North Bethesda, MD) with an engine based on the Clustal W algorithm (22). BLAST (1) was used to search the NCBI databases to identify previously reported sequences with identity to those obtained in the study described herein.
Modeling of habitat suitability for ticks.
The MaxEnt method was used to estimate habitat suitability in the multivariate space of environmental variables, using presence-only data. MaxEnt is a machine learning method that is based on finding the probability distribution of maximum entropy (i.e., closest to uniform) to estimate a target probability distribution. MaxEnt offers other advantages, including its ability to use both continuous and categorical data and an efficient algorithm with a concise mathematical description (20). Models for Boophilus annulatus, Dermacentor marginatus, Rhipicephalus bursa, Rhipicephalus turanicus, Hyalomma marginatum marginatum, and Ixodes ricinus were trained with already-compiled records for these ticks in the Mediterranean region (16). According to previous surveys, these are the most common tick species infesting livestock in Sicily (23). Climate variables were obtained and used as previously described at a resolution of 4 km (14). Because of the low number of accurately referenced records of these ticks in the island, it was decided to train the model with all data from the Mediterranean region and apply the output of the model to Sicily. As reported previously (14), these models provide an accuracy value of approximately 0.9 for the tick species included in the study. The resolution of the models was further improved by including a qualitative layer of vegetation categories. This layer was obtained from the CORINE European seamless vector database, freely available at http://dataservice.eea.europa.eu. A habitat suitability index (HS) was produced for each cell at the given resolution, covering the whole island. Model development, evaluation sets, and model performance tests have been previously described (14). HS was averaged by province in order to have a coarse estimation of the different distribution patterns of ticks over the island and to provide a framework to correlate with rates of pathogen prevalence.
Statistical analysis.
The confidence intervals and standard errors at 95% confidence levels of the prevalence of Anaplasma infections in cattle were calculated using Wilson's continuity correction. Two-by-two χ2 tests were performed to compare infection prevalence between hosts, Anaplasma spp., and western and eastern provinces of Sicily. Since many of the marginal tables were very uneven, we used Fisher's exact tests for small values (less than 10). The differences were considered statistically significant at P ≤ 0.05. The SPSS 11.0 statistical program (SPSS Inc., Chicago, IL) was used.
Nucleotide sequence accession numbers.
The GenBank accession numbers for A. phagocytophilum 16S rRNA and for A. marginale and A. ovis msp4 gene sequences are EU436153 to EU436161.
RESULTS
Prevalence of Anaplasma spp.
The infection prevalence was determined as the percentage of animals positive for the pathogen DNA detected by PCR (Tables 1 and 2). The prevalence of Anaplasma spp. was determined by 16S rRNA gene PCR. The prevalence of A. marginale and A. ovis was determined by species-specific msp4 PCR. For A. phagocytophilum, infection prevalence was determined by species-specific msp4 PCR, after sequencing 16S rRNA genes in all positive samples because in most cases A. phagocytophilum msp4 amplicons were difficult to detect and could not be cloned and sequenced. The msp4 gene could not be amplified from all 16S rRNA gene-positive samples (Table 1), but the sequence of randomly selected 16S rRNA gene amplicons from msp4 PCR-negative samples did not result in sequences with homology to Anaplasma spp.
TABLE 1.
Prevalence of Anaplasma spp. in western and eastern provinces of Sicilya
| Region and animal type sampled (n) | Prevalence of infection (95% CI)
|
P (Fisher test)
|
|||||
|---|---|---|---|---|---|---|---|
| Anaplasma spp. | A. marginale | A. ovis | A. phagocytophilum | A. marginale vs A. ovis | A. marginale vs A. phagocytophilum | A. ovis vs A. phagocytophilum | |
| Western region | |||||||
| Cattle (222) | 52.7-65.9 | 38.9-52.3 | 0-2.1 | 0.6-4.8 | *** | *** | NS |
| Goats (75) | 17.2-13.8 | 0-6.1 | 17.2-13.8 | 0-6.1 | NS | NS | |
| Sheep (193) | 45.1-59.5 | 0-2.4 | 14.9-26.7 | 0.4-4.8 | *** | NS | *** |
| Horses (25) | 0-16.6 | 0-16.6 | 0-16.6 | 0-16.6 | |||
| Dogs (29) | 0-14.6 | 0-14.6 | 0-14.6 | 0-14.6 | |||
| Mice (37) | 0-4.6 | 0-4.6 | 0-4.6 | 0-4.6 | |||
| Eastern region | |||||||
| Cattle (152) | 15.1-28.6 | 6.3-16.8 | 0-3.1 | 0-3.1 | *** | *** | |
| Goats (59) | 40.1-67.1 | 0-7.6 | 21.0-45.7 | 0-7.6 | *** | *** | |
| Sheep (93) | 32.9-53.7 | 0-4.94 | 5.6-19.3 | 0-6.7 | *** | NS | ** |
| Horses (109) | 2.2-12.1 | 0-4.2 | 0-4.2 | 0-5.8 | NS | NS | |
| Dogs (17) | 0-22.9 | 0-22.9 | 0-22.9 | 7.8-50.2 | NS | NS | |
| Mice (32) | 1.1-22.2 | 0-13.3 | 0-13.3 | 1.1-22.2 | NS | NS | |
| Total | |||||||
| Cattle (374) | 38.9-49.1 | 26.7-36.3 | 0-12.7 | 0-2.9 | *** | *** | NS |
| Goats (134) | 19.8-35.3 | 0-3.5 | 11.4-24.8 | 0-3.5 | *** | *** | |
| Sheep (286) | 4.4-55.2 | 0-1.7 | 13.1-22.1 | 0-3.8 | *** | NS | *** |
| Horses (134) | 1.8-9.9 | 0-3.5 | 0-3.5 | 0-4.7 | NS | NS | |
| Dogs (46) | 2.83-21.7 | 0-9.6 | 0-9.6 | 2.8-21.7 | NS | NS | |
| Mice (69) | 0-11.0 | 0-6.6 | 0-6.6 | 0-11.0 | NS | NS | |
Blood samples were collected during 2006 and 2007. The prevalence of Anaplasma spp. was determined by 16S rRNA gene PCR. The prevalence of Anaplasma infections is expressed as the lower and upper limits of the 95% confidence interval (CI) for using the Wilson procedure with a correction for continuity. Significant differences are indicated (Fisher's exact test; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; NS, not significant). Differences in pathogen prevalence between western and eastern regions are shown in bold.
TABLE 2.
Comparison of Anaplasma prevalences between different hostsa
| Comparison | Significance by region and species
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Western region
|
Eastern region
|
Total
|
||||||||||
| Anaplasma spp. | A. marginale | A. ovis | A. phagocytophilum | Anaplasma spp. | A. marginale | A. ovis | A. phagocytophilum | Anaplasma spp. | A. marginale | A. ovis | A. phagocytophilum | |
| Cattle vs | ||||||||||||
| Goats | *** | *** | ** | NS | 33.44*** | ** | *** | 11.92*** | *** | *** | NS | |
| Sheep | NS | *** | *** | NS | 13.41*** | *** | *** | NS | *** | *** | NS | |
| Horses | 31.93*** | *** | NS | *** | *** | NS | NS | *** | *** | NS | ||
| Dogs | *** | *** | NS | * | NS | NS | *** | *** | *** | ** | ||
| Mice | *** | *** | NS | * | * | NS | * | *** | *** | NS | ||
| Goats vs | ||||||||||||
| Sheep | *** | ** | NS | 6.38** | *** | NS | 18.84*** | NS | NS | |||
| Horses | NS | *** | *** | NS | *** | *** | NS | |||||
| Dogs | NS | *** | ** | ** | ** | *** | ** | |||||
| Mice | * | * | *** | *** | NS | *** | *** | NS | ||||
| Sheep vs | ||||||||||||
| Horses | *** | ** | NS | *** | *** | NS | *** | *** | NS | |||
| Dogs | *** | ** | NS | *** | NS | ** | *** | *** | * | |||
| Mice | *** | *** | NS | *** | * | NS | *** | *** | NS | |||
| Horses vs | ||||||||||||
| Dogs | NS | ** | NS | * | ||||||||
| Mice | NS | NS | NS | NS | ||||||||
| Dogs vs mice | NS | NS | NS | NS | ||||||||
The infection prevalence between host species for Anaplasma spp., A. marginale, A. ovis, and A. phagocytophilum was analyzed as described for Table 1 (the χ2 value is included when the χ2 test was used and the value was significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; NS, not significant).
The prevalences of Anaplasma spp., A. marginale, A. ovis, and A. phagocytophilum were analyzed and compared between different hosts and regions (Table 1). A. marginale and A. phagocytophilum were detected in cattle, but the prevalence of A. marginale was higher than that for A. phagocytophilum. Only A. ovis was detected in goats. A. ovis and A. phagocytophilum were detected in sheep, but the prevalence of A. ovis was higher. Only A. phagocytophilum was detected in horses, dogs, mice, one porcupine, and one cat.
Statistically significant differences were observed in the prevalence of Anaplasma spp. between western and eastern provinces (Table 2). These differences were different for different hosts and pathogens. For Anaplasma spp., the prevalence was higher for cattle in the western region (χ2, 54.05, 1 df, P < 0.001), but it was higher for goats (Fisher's exact test, P < 0.001) in the eastern region. The prevalence of A. marginale in cattle was higher in the western than in the eastern region (χ2, 51.32, 1 df, P < 0.001). The prevalence of A. ovis was different between sheep and goats, with higher prevalences for goats in the eastern region and for sheep in the western region (Table 2). The prevalence of A. phagocytophilum was higher in the eastern region for both dogs and mice (Table 2).
Analysis of Anaplasma genotypes.
16S rRNA and msp4 gene amplicons were cloned and sequenced to corroborate the results of the PCR and to compare the Anaplasma genotypes identified in different hosts and regions. The 16S rRNA gene was used for genotype characterization in A. phagocytophilum, and msp4 sequences were used for analysis of A. marginale and A. ovis strains.
Five A. phagocytophilum genotypes were identified infecting cattle, sheep, mice, a porcupine, and a cat (Table 3). The genotypes API and APII were found in different hosts in the western region only. The genotype APIII, with 98.9% identity to a sequence of an isolate from a human case (DQ029028 [7]), was identified in cattle in the eastern region only. The genotypes APIV and APV were found in the eastern region in sheep and mice, respectively.
TABLE 3.
Characterization of Anaplasma genotypes in different hosts and regionsa
| Species and host | Region | No. (%)b | Genotype | Sequence identity (%)c | GenBank accession no. |
|---|---|---|---|---|---|
| A. phagocytophilum | |||||
| Cattle | Western | 2 (50) | API | 95 | EU436153 |
| 1 (25) | APII | 95 | EU436154 | ||
| 1 (25) | APIII | 99 | EU436155 | ||
| Sheep | Eastern | 1 (100) | APIV | 94 | EU436156 |
| Western | 3 (100) | APII | 95 | EU436154 | |
| Mice | Eastern | 2 (100) | APV | 95 | EU436157 |
| Porcupine | Western | 1 (100) | API | 95 | EU436153 |
| Cat | Western | 1 (100) | API | 95 | EU436153 |
| A. marginale | |||||
| Cattle | Eastern | 10 (50) | AMI | 100 | EU436158 |
| 10 (50) | AMII | 99.8 | EU436159 | ||
| Western | 16 (100) | AMII | 99.8 | EU436159 | |
| A. ovis | |||||
| Sheep | Eastern | 10 (100) | AOI | 100 | EU436160 |
| Western | 10 (100) | AOI | 100 | EU436160 | |
| Goats | Eastern | 10 (100) | AOII | 99.9 | EU436161 |
| Western | 4 (100) | AOI | 100 | EU436160 |
The 16S rRNA and msp4 gene amplicons were cloned and sequenced for the characterization of Anaplasma genotypes. All A. phagocytophilum 16S rRNA gene-positive fragments were used for genotype characterization in A. phagocytophilum, and randomly selected msp4 sequences were used for analysis of A. marginale and A. ovis strains.
Number of strains with this genotype (percentage of total samples analyzed).
Percent sequence identity at the nucleotide level with respect to the reference sequence (DQ029028 for A. phagocytophilum 16S rRNA genes and AY702920 and AY702923 for A. marginale and A. ovis msp4, respectively).
For A. marginale, two genotypes were identified in cattle (Table 3). The genotype AMI was found in the eastern region only and was identical to a sequence previously identified in cattle in the western region (AY702920 [5]). The most abundant genotype, AMII, was found in both eastern and western regions. The genotype AMII differed at three nucleotide positions with respect to AMI (A versus G, A versus G, and G versus A at positions 270, 312, and 324, respectively).
Two genotypes were identified for A. ovis (Table 3). The most abundant genotype, AOI, was found in sheep and goats in both eastern and western regions and was identical to a sequence previously identified in sheep in the western region (AY702923 [5]). The genotype AOII was found in goats in the eastern region only. AOI and AOII differed at a single nucleotide (C versus T at position 366 in AOII).
Habitat suitability for ticks.
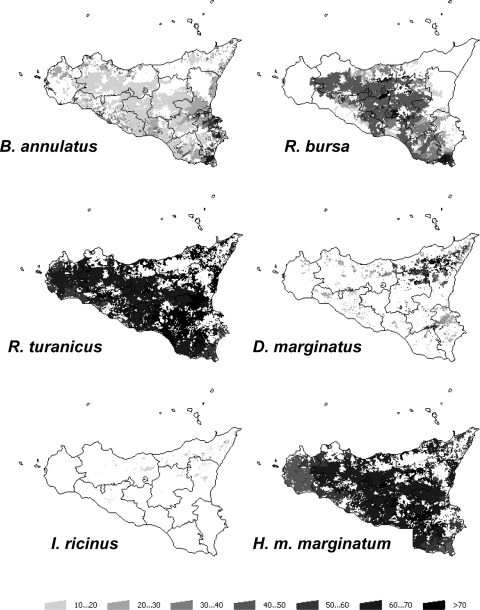
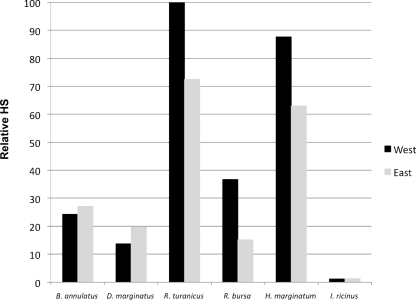
The analysis of tick habitat suitability was done for the most common hard tick species (Acari: Ixodidae) infesting livestock in the island (Fig. 2 and 3). The results showed that the habitat suitability for B. annulatus and D. marginatus was higher in the eastern region than in the western region. While the HS for I. ricinus was relatively low in the whole island, foci of adequate habitat were found in Messina in the eastern region (more than 40% of total suitable area). The main foci of positive HS for D. marginatus were also concentrated in the provinces of Messina and Catania in the eastern region. The habitat suitabilities for R. bursa, R. turanicus, and H. marginatum marginatum were higher in the western region. Remarkably, the western region of the island concentrated around 70% of the area of suitable habitat for R. bursa.
FIG. 2.
Predicted HSs for the six tick species modeled in the current study using climate and vegetation (qualitative) features. Levels of gray represent the HSs for the different tick species in the range 0 to 100.
FIG. 3.
Relative HSs for tick species in western and eastern regions of Sicily. Each histogram represents the average HS for a given tick species in the region of reference. The tick species with maximum suitability in the island was ascribed a value of 100, and the others were referred to that value. For this analysis, provinces were divided into eastern and western regions according to the map in Fig. 1.
DISCUSSION
Previous studies have documented the presence and genetic diversity of Anaplasma spp. in a limited geographic area of Sicily (2, 5-7, 9, 24, 25). Sicily represents a typical Mediterranean ecosystem for the study of Anaplasma prevalence and habitat suitability for ticks. The prevalence of Anaplasma infection as determined by 16S rRNA gene PCR resulted in values higher than those determined by msp4 PCR analysis. The sequence of randomly selected 16S rRNA gene amplicons from msp4 PCR-negative samples did not result in sequences with homology to Anaplasma spp., suggesting the presence of false positives in the 16S rRNA gene PCR. Therefore, the infection prevalence determined for A. marginale, A. ovis, and A. phagocytophilum was considered more reliable than that determined for Anaplasma spp. Nevertheless, the results for Anaplasma prevalence were used in the analysis because of the possibility that in some of the 16S rRNA gene PCR-positive hosts the infection levels were below the sensitivity of the msp4 PCR (four to five copies of msp4/ng DNA) or, in the case of A. phagocytophilum, differences in the sequence of the msp4 locus in these strains may have prevented amplification of the gene (4, 8).
In the study reported herein, differences were found in the prevalence of Anaplasma infection between different hosts and between western and eastern provinces. The differences in Anaplasma infection prevalence between different hosts may be explained by pathogen host tropism. For example, A. marginale and A. ovis are primarily pathogens of cattle and sheep/goats, respectively (10, 17). A. phagocytophilum has a broad host range, infecting all studied host species (4, 12). Although A. platys has been detected with low prevalence in dogs in the province of Palermo in Sicily (9), it was not found in this study. Tick-host specificity may also affect the prevalence of Anaplasma spp. (8).
The differences in Anaplasma infection prevalence between western and eastern Sicilian provinces may result from differences in husbandry practices, wildlife reservoir hosts, and/or habitat suitability for ticks. Although husbandry practices may affect Anaplasma prevalence (6), they are similar in both western and eastern regions (2). The presence of wildlife reservoir hosts may also affect the prevalence of Anaplasma spp., a fact that is particularly important for A. phagocytophilum (8). However, except for wild rodents (mice and one porcupine) that were found infected with A. phagocytophilum, wildlife was not analyzed in this study. Therefore, to explain the differences in Anaplasma infection prevalence between western and eastern provinces, these data were correlated with the habitat suitability for ticks in the two regions. The performance of the models used to evaluate habitat suitability for ticks has been reported elsewhere (13, 14) and will not be discussed here. These models provided a performance index of 0.92 to 0.95 (in the range of 0 to 1) in the whole Mediterranean region (13, 14). Although adaptations to local prevailing conditions may obviously occur in the tick populations found in Sicily, HS estimation is an adequate guide to outline the areas of suitable habitat for ticks. In the absence of extensive tick surveys in Sicily, this seems to be the candidate methodology to compare pathogen prevalence against defined patterns of tick habitat suitability across a wide geographical region.
The prevalence of A. marginale and A. ovis for cattle and sheep was higher in the western region. This finding correlated with higher habitat suitability for R. bursa in this region of Sicily. R. bursa commonly feeds on cattle and sheep but is rare on goats (6, 15, 23) and has been proposed as vector of A. marginale and A. ovis in Mediterranean ecosystems (3, 8, 15). The fact that the higher HS for B. annulatus was found in the eastern region whereas the prevalence of A. marginale was higher in the western region suggested that this tick species does not play an important role in the transmission of A. marginale in Sicily and other Mediterranean ecosystems. The prevalence of A. ovis in goats was higher in the eastern region, a finding that correlated with higher habitat suitability for D. marginatus. This tick species feeds on goats and may be a vector of A. ovis (15). The lack of information about goat density and management practices in both regions of the island precluded further discussion about differences in the prevalence of A. ovis between sheep and goats.
The main vectors of A. phagocytophilum are ticks of the I. ricinus complex, and infections are maintained in nature, at least in part, in rodents and other small and medium-sized mammals (18, 19, 21). In Sicily, the habitat suitability for I. ricinus was relatively low in the whole island but foci of adequate habitat were found in Messina province in the eastern region of the island. These foci may serve as a source of infected ticks for transmission of A. phagocytophilum to dogs and mice in this region. The finding of a porcupine infected with A. phagocytophilum suggested that porcupines need to be studied as a possible pathogen wildlife reservoir. Furthermore, the finding of cattle, a porcupine, and a cat infected with A. phagocytophilum in the western region may reflect the presence of I. ricinus concentrated in small patches where climate and vegetation provide adequate protection for this tick species (5) but also reinforced the suggestion that other tick species may subsequently prove to be vectors of A. phagocytophilum in Mediterranean ecosystems (3, 8).
The other tick species analyzed, R. turanicus and H. marginatum marginatum, may also be involved in the transmission of Anaplasma spp. (3, 5, 8), and their higher habitat suitability in the western region may also explain differences in pathogen prevalence between western and eastern regions. Other tick species that are found in Sicily for which habitat suitability was not predicted due to lack of enough information to use the models (e.g., Hyalomma marginatum lusitanicum and Haemaphysalis punctata) may also act as vectors of Anaplasma spp. (5). Finally, mechanical transmission of Anaplasma spp. by blood-contaminated mouth parts of biting flies or fomites may be important, at least for A. marginale, for pathogen transmission in Sicily and other Mediterranean regions (8, 17).
The analysis of Anaplasma genotypes suggested the existence of A. phagocytophilum genotypes in particular hosts (APIII to APV) and regions (API to APV). In agreement with previous reports (5), A. marginale genotypes AMI and AMII were found in cattle in both eastern and western regions. The A. ovis genotype AOI was found in sheep and goats in both eastern and western regions, but the genotype AOII may be associated with goats in the eastern region only. Collectively, these results suggested a higher host and regional specificity for A. phagocytophilum than for A. marginale and A. ovis strains, a finding probably associated with the broader host range of A. phagocytophilum (4). The presence of identical A. marginale genotypes in both regions may reflect cattle movement, a fact that is common in all regions of endemicity in the world (11). For A. ovis the results of this and previous (10) works suggested the possibility of some genotypes being host specific, but more information is necessary before conclusions are reached.
The results reported herein have important implications for the control of Anaplasma spp. in Sicily. The information about pathogen prevalence and genotypes in different hosts and regions helps the establishment of surveillance and control programs for these pathogens. The correlation between Anaplasma prevalence and habitat suitability for ticks supports the need for studies to characterize the prevalence of Anaplasma spp. in field-collected ticks. These results may be used to construct models to predict the risks for tick-borne pathogens in Mediterranean ecosystems.
Acknowledgments
We thank Franco Ferraro and Nicola Galati for animal sampling.
This work was supported by project IZS SI 05/04 of the Ministry of Health, Italy. Partial support was also provided by the Instituto de Ciencias de la Salud, Spain (ICS-JCCM) (project 06036-00).
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Caracappa, S. 1999. Livestock production and animal health in Sicily, Italy. Parassitologia 41(Suppl. 1):17-23. [PubMed] [Google Scholar]
- 3.de la Fuente, J., V. Naranjo, F. Ruiz-Fons, J. Vicente, A. Estrada-Peña, C. Almazán, K. M. Kocan, M. P. Martín, and C. Gortázar. 2004. Prevalence of tick-borne pathogens in ixodid ticks (Acari: Ixodidae) collected from European wild boar (Sus scrofa) and Iberian red deer (Cervus elaphus hispanicus) in central Spain. Eur. J. Wildl. Res. 50:187-196. [Google Scholar]
- 4.de la Fuente, J., R. F. Massung, S. J. Wong, F. K. Chu, H. Lutz, M. Meli, F. D. von Loewenich, A. Grzeszczuk, A. Torina, S. Caracappa, A. J. Mangold, V. Naranjo, S. Stuen, and K. M. Kocan. 2005. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 43:1309-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Fuente, J., A. Torina, S. Caracappa, G. Tumino, R. Furlá, C. Almazán, and K. M. Kocan. 2005. Serologic and molecular characterization of Anaplasma species infection in farm animals and ticks from Sicily. Vet. Parasitol. 133:357-362. [DOI] [PubMed] [Google Scholar]
- 6.de la Fuente, J., A. Torina, V. Naranjo, S. Caracappa, J. Vicente, A. J. Mangold, D. Vicari, A. Alongi, S. Scimeca, and K. M. Kocan. 2005. Genetic diversity of Anaplasma marginale strains from cattle farms with different husbandry systems in the Province of Palermo, Sicily. J. Vet. Med. B 52:226-229. [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente, J., A. Torina, V. Naranjo, S. Caracappa, V. Di Marco, A. Alongi, M. Russo, A. R. Maggio, and K. M. Kocan. 2005. Infection with Anaplasma phagocytophilum in a seronegative patient in Sicily, Italy: case report. Ann. Clin. Microbiol. Antimicrob. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Fuente, J., V. Naranjo, F. Ruiz-Fons, U. Höfle, I. G. Fernández de Mera, D. Villanúa, C. Almazán, A. Torina, S. Caracappa, K. M. Kocan, and C. Gortázar. 2005. Potential vertebrate reservoir hosts and invertebrate vectors of Anaplasma marginale and A. phagocytophilum in central Spain. Vector Borne Zoonotic Dis. 5:390-401. [DOI] [PubMed] [Google Scholar]
- 9.de la Fuente, J., A. Torina, V. Naranjo, S. Nicosia, A. Alongi, F. La Mantia, and K. M. Kocan. 2006. Molecular characterization of Anaplasma platys strains from dogs in Sicily, Italy. BMC Vet. Res. 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Fuente, J., M. W. Atkinson, V. Naranjo, I. G. Fernández de Mera, A. J. Mangold, K. A. Keating, and K. M. Kocan. 2007. Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet. Microbiol. 119:375-381. [DOI] [PubMed] [Google Scholar]
- 11.de la Fuente, J., P. Ruybal, M. S. Mtshali, V. Naranjo, I. Shuqing, A. J. Mangold, S. D. Rodríguez, R. Jiménez, J. Vicente, R. Moretta, A. Torina, C. Almazán, P. M. Mbati, S. Torioni de Echaide, M. Farber, R. Rosario-Cruz, C. Gortazar, and K. M. Kocan. 2007. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet. Microbiol. 119:382-390. [DOI] [PubMed] [Google Scholar]
- 12.Dumler, J. S., A. C. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of the genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 13.Estrada-Peña, A., J. Quílez, E. del Cacho, and C. Sánchez Acedo. 2006. A method to map abundance of tick metapopulations. Ecol. Model. 193:663-674. [Google Scholar]
- 14.Estrada-Peña, A., and J. M. Venzal. 2007. Climate niches of tick species in the Mediterranean region: modeling of occurrence data, distributional constraints, and impact of climate change. J. Med. Entomol. 44:1130-1138. [DOI] [PubMed] [Google Scholar]
- 15.Friedhoff, K. T. 1997. Tick-borne diseases of sheep and goats caused by Babesia, Theileria or Anaplasma spp. Parassitologia 39:99-109. [PubMed] [Google Scholar]
- 16.Integrated Consortium on Ticks and Tick-Borne Diseases. 2004. Ticks of veterinary and medical importance: the Mediterranean basin. CD-ROM published by contract ICA4T-CT-2000-30006 of the European Commission.
- 17.Kocan, K. M., J. de la Fuente, E. F. Blouin, and J. C. Garcia-Garcia. 2004. Anaplasma marginale (Rickettsiales: Anaplasmataceae): recent advances in defining host-pathogen adaptations of a tick-borne rickettsia. Parasitology 129:S285-S300. [DOI] [PubMed] [Google Scholar]
- 18.Levin, M. L., W. L. Nicholson, R. F. Massung, J. W. Sumner, and D. Fish. 2002. Comparison of the reservoir competence of medium-sized mammals and Peromyscus leucopus for Anaplasma phagocytophilum in Connecticut. Vector Borne Zoonotic Dis. 2:125-136. [DOI] [PubMed] [Google Scholar]
- 19.Petrovec, M., A. Bidovec, J. W. Sumner, W. L. Nicholson, J. E. Childs, and T. Avsic Zupanc. 2002. Infection with Anaplasma phagocytophila in cervids from Slovenia: evidence of two genotypic lineages. Wien. Klin. Wochenschr. 114:641-647. [PubMed] [Google Scholar]
- 20.Phillips, S. J., R. P. Anderson, and R. E. Schapire. 2006. Maximum entropy modelling of species geographic distributions. Ecol. Model. 190:231-259. [Google Scholar]
- 21.Telford, S. R., III, J. E. Dawson, P. Katavolos, C. K. Warner, C. P. Kolbert, and D. H. Persing. 1996. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc. Natl. Acad. Sci. USA 93:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torina, A., C. Khoury, S. Caracappa, and M. Maroli. 2006. Ticks infesting livestock on farms in western Sicily, Italy. Exp. Appl. Acarol. 38:75-86. [DOI] [PubMed] [Google Scholar]
- 24.Torina, A., and S. Caracappa. 2007. Anaplasmosis in cattle in Italy. Vet. Res. Commun. 31(Suppl. 1):73-78. [DOI] [PubMed] [Google Scholar]
- 25.Torina, A., J. Vicente, A. Alongi, S. Scimeca, R. Turlá, S. Nicosia, V. Di Marco, S. Caracappa, and J. de la Fuente. 2007. Observed prevalence of tick-borne pathogens in domestic animals in Sicily, Italy during 2003-2005. Zoonoses Public Health 54:8-15. [DOI] [PubMed] [Google Scholar]