Abstract
We demonstrate herein the ability of Kluyveromyces marxianus to be an efficient ethanol producer and host for expressing heterologous proteins as an alternative to Saccharomyces cerevisiae. Growth and ethanol production by strains of K. marxianus and S. cerevisiae were compared under the same conditions. K. marxianus DMKU3-1042 was found to be the most suitable strain for high-temperature growth and ethanol production at 45°C. This strain, but not S. cerevisiae, utilized cellobiose, xylose, xylitol, arabinose, glycerol, and lactose. To develop a K. marxianus DMKU3-1042 derivative strain suitable for genetic engineering, a uracil auxotroph was isolated and transformed with a linear DNA of the S. cerevisiae ScURA3 gene. Surprisingly, Ura+ transformants were easily obtained. By Southern blot hybridization, the linear ScURA3 DNA was found to have inserted randomly into the K. marxianus genome. Sequencing of one Lys− transformant confirmed the disruption of the KmLYS1 gene by the ScURA3 insertion. A PCR-amplified linear DNA lacking K. marxianus sequences but containing an Aspergillus α-amylase gene under the control of the ScTDH3 promoter together with an ScURA3 marker was subsequently used to transform K. marxianus DMKU3-1042 in order to obtain transformants expressing Aspergillus α-amylase. Our results demonstrate that K. marxianus DMKU3-1042 can be an alternative cost-effective bioethanol producer and a host for transformation with linear DNA by use of S. cerevisiae-based molecular genetic tools.
Ethanol production at high temperature has received much attention because fermentation processes conducted at elevated temperatures will significantly reduce cooling costs (14). Other advantages of elevated temperatures include more-efficient simultaneous saccharification and fermentation, a continuous shift from fermentation to distillation, reduced risk of contamination, and suitability for use in tropical countries (3, 5, 27). However, the temperatures suitable for conventional strains of Saccharomyces cerevisiae are relatively low (25 to 30°C). While screens for S. cerevisiae mutants able to produce ethanol efficiently at high temperature have been performed, only a modest increase in temperature has been obtained, 40°C maximum (35, 40). Alternatively, attention has also focused on thermotolerant yeast species capable of producing ethanol at elevated temperatures. Isolates of Kluyveromyces marxianus appear to be particularly promising (3, 5, 20, 27). This species has been reported to grow at 47°C (3), 49°C (20), and even 52°C (6) and to produce ethanol at temperatures above 40°C (14). Moreover, K. marxianus offers additional benefits (36) including a high growth rate (34) and the ability to utilize a wide variety of industrially relevant substrates such as sugar cane, corn silage juice, molasses, and whey powder (17, 27, 32, 42, 49). Because of these advantages, K. marxianus is currently being promoted as a viable alternative to S. cerevisiae as an ethanol producer. However, systematic comparison of the ethanol productivity of K. marxianus with that of S. cerevisiae, or of those among K. marxianus strains isolated from different areas under the same conditions, has not been reported, making it difficult to determine which yeast strain is the best for specified conditions. Therefore, the present study was undertaken to assess growth and ethanol productivity at high temperature and the carbon source utilization of K. marxianus strains (11, 25-27, 45) and two S. cerevisiae strains, of which NCYC3233 is an isolate from a Brazilian ethanol plant. Furthermore, because K. marxianus is relatively undeveloped as a host for heterologous gene expression, in contrast to S. cerevisiae and K. lactis (15, 17), we demonstrate that K. marxianus DMKU3-1042 can be efficiently transformed with linear DNA to yield random insertions, which promises to facilitate both strain improvements and efforts to learn more about the biology of this important thermotolerant species.
MATERIALS AND METHODS
Strains and culture media.
Yeast strains used are listed in Table 1. YPD medium (1% yeast extract, 2% peptone, and 2% glucose) and minimal medium (MM; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 2% glucose) were prepared as described previously (23). YP medium (1% yeast extract, 2% peptone) or MM without glucose was supplemented with 5% glucose or with 2% glucose, galactose, fructose, mannose, sucrose, raffinose, inulin, arabinose, cellobiose, glycerol, lactose, xylose, or xylitol. Starch medium (YPD supplemented with 1% soluble starch) and MM supplemented individually with uracil, adenine, lysine, histidine, methionine, tryptophan, or leucine were also used. 5-Fluoroorotic acid (5-FOA) and uracil dropout media were prepared as described previously (1, 2). For the preparation of solid plates, 2% agar was added. Anaerobic cultures were grown in an AnaeroPouch (Mitsubishi Gas Chemical, Tokyo, Japan).
TABLE 1.
Yeast strains used in this study
| Straina | Description | Reference(s) and remarks |
|---|---|---|
| K. marxianus strains | ||
| DMKU3-1042 | Isolated in Thailand | 27 |
| NCYC587 | ATCC 36907 | 11, 31, 33 |
| NCYC1429 | ATCC 12424 | 25, 48 |
| NCYC2791 | CBS712; type strain | 30 |
| RAK3605 | ura3 | DMKU3-1042 FOA+ |
| RAK3626 | ura3 Ade− [ScURA3] | RAK3605 transformant |
| RAK3627 | ura3 lys1::ScURA3 | RAK3605 transformant |
| RAK4687 | ura3 ScTDH3p-TAA-ScURA3 | RAK3605 transformant |
| S. cerevisiae strains | ||
| BY4704 | MATaade2::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 | 10 |
| BY4700 | MATaura3Δ0 | 10 |
| BY4713 | MATaleu2Δ0 | 10 |
| RAK3599 | MATa/α ura3Δ0/+ +/leu2Δ0 | BY4700 × BY4713 |
| RAK3603 | MATaura3Δ0::ScTDH3p-TAA- PGKter-ScURA3 his3Δ1 leu2Δ0 met15Δ0 | This study |
| NCYC3233 | Isolated from fermented ethanol broth in Brazil |
NCYC, National Center for Yeast Collection, United Kingdom.
Ethanol fermentation and analytical methods.
Forty milliliters of YP plus 5% glucose in 250-ml flasks was inoculated with an overnight YPD culture to an initial optical density at 600 nm of 0.2. Cells were incubated at 30°C or 45°C with shaking at 150 rpm. Supernatant samples were collected by centrifugation and were analyzed directly by high-performance liquid chromatography (Hitachi, Tokyo, Japan) using a Shim-pack SPR-Pb column (Shimadzu, Kyoto, Japan) to determine sugar and ethanol concentrations. Deionized water was used as the mobile phase with a flow rate of 0.6 ml/min; the column temperature was set at 80°C.
Cell growth was analyzed using a TVS062CA biophotorecorder (Toyo Seisakusho, Ltd., Chiba, Japan). To detect α-amylase activity, transformed cells were spotted on starch medium plates and incubated at 28°C for 1 day. The starch plates were then exposed to iodine vapor to detect the halo produced by starch degradation.
Ura− mutant isolation.
K. marxianus DMKU3-1042 was grown overnight in 2 ml of YPD, centrifuged, washed once with sterile water, and resuspended in 1 ml of sterile water. Aliquots (200 μl) of cells were then spread on 5-FOA plates and exposed to UV for 30 s. Colonies were picked after 2 days at 28°C and tested for a uracil requirement by assessing growth on MM with and without uracil.
PCR amplification and primers.
DNA fragments used for transformation were amplified by PCR using a KOD Plus kit (Toyobo, Osaka, Japan) according to the manufacturer's instructions. The PCR was initiated at 94°C for 1 min, followed by 30 cycles of the following program: 94°C for 20 s, 55°C for 30 s, and 68°C for 3 to 4 min. The size of PCR products was analyzed by agarose gel electrophoresis. Primers used are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′-3′) |
|---|---|
| URA3-40 | ATCAAAGAAGGTTAATGTGGCTGTGG |
| URA3-40c | TTCGTCATTATAGAAATCATTACGAC |
| URA3-300 | GAAGAGTATTGAGAAGGGCAAC |
| URA3-300c | TGTTGTGAAGTCATTGACACAG |
| URA3+720 | GGGAAGGGATGCTAAGGTAGAGGGT |
| URA3+25Down | ACCCGGGAATCTCGGTCGTAATGAT |
| URA3+1054 | GACAGGACTGTAAAGATGGACGCAT |
| URA3+61c | GCTTGGCAGCAACAGGACTAGGATG |
| URA3-35c | TGCAGTTGGGTTAAGAATACTGGGC |
| URA3-400c | TCTACATCAGATAACTTCGGTTTG |
| 15C+9A | CCCCCCCCCCCCCCCAAAAAAAAA |
| TDH3-572 | GCTGTAACCCGTACATGCCCAA |
| LYS1-551 | AAGGCAAGTGAAAGCCATTGCC |
| LYS1+1642 | GTATTGTGGAGCAGTCAGCATC |
| KmLYS1-225Up | CTTCCTGAAGAATAATTTATGCCGACTTACTGCTTGAAAGTG |
| KmLYS1-606Down | CGGACACTTTCCCTCTAGTTCATGAGCATATTCAATTTGCCC |
The S. cerevisiae ScURA3 (1.7-kb) gene was amplified using chromosomal DNA from BY4704 as the template and primers URA3-300 and URA3-300c. The ScLYS1 gene was amplified using chromosomal DNA from BY4743 as the template with primers LYS1-551 and LYS1+1642. Integration of the ScURA3 at the K. marxianus KmLYS1 locus was confirmed by PCR using KmLYS1-225Up and KmLYS1-606 primers. The ScTDH3 promoter-driven Aspergillus oryzae α-amylase gene, TAA, was integrated at the ura3Δ0 locus in BY4741 to generate strain RAK3623 (K. Cha-aim, H. Hoshida, and R. Akada, unpublished data). An ScTDH3p-TAA-ScURA3 DNA was amplified using genomic DNA from RAK3623 as the template and primers TDH3-572 and URA3-300c for the transformation of K. marxianus.
Chromosomal DNA isolation.
Cells from a 1.5-ml YPD culture were collected by centrifugation and washed once with sterile water. The cells were suspended in 300 μl of SET buffer (1.2 M sorbitol in 20 mM Tris-HCl, pH 7.5, and 10 mM EDTA) to which 20 μl of zymolyase solution (3 mg/ml zymolyase 100T [Seikagaku Corp., Tokyo, Japan] in 0.9 ml SET, mixed with 0.1 ml β-mercaptoethanol) was added. The mixture was incubated at 37°C for 30 min, after which 50 μl of 10% sodium dodecyl sulfate (SDS) and 200 μl of phenol-chloroform-isoamylalcohol (Sigma-Aldrich, MO) were added. After centrifugation, the upper layer containing chromosomal DNA was transferred to a microtube, and DNA was precipitated with ethanol. The precipitated chromosomal DNA was then washed once with 70% ethanol and dissolved in 100 μl of sterile water.
Transformation.
Yeast transformation was performed by a lithium acetate method (15). Yeast cells were grown overnight in YPD, diluted 1:10 in 10 ml of fresh YPD, and allowed to grow 5 h at 28°C with shaking. The cells were then collected by centrifugation, washed once with sterile water, and suspended in 100 μl of sterile water. Fifty microliters of the cell suspension was then mixed with 115 μl of 60% polyethylene glycol 3350, 5 μl of 4 M lithium acetate, 15 μl of sterile water, 10 μl of 10 mg/ml carrier DNA, and 5 μl of PCR product. The mixture was vortexed for 30 s, incubated at 42°C for 40 min, and spread on uracil dropout plates.
S. cerevisiae centromere/autonomously replicating sequence (ARS) plasmid pRS316 (41) containing ScURA3 as a marker was used for K. marxianus transformations. pRS316 (4.8 kb) was linearized by digestion with SmaI (New England Biolabs, MA).
Southern blot hybridization.
Chromosomal DNA from BY4704, K. marxianus DMKU3-1042, a K. marxianus ura3 mutant (RAK3605), and ScURA3 transformants of the ura3 mutant was isolated and digested with BamHI (Roche Diagnostics GmbH, Mannheim, Germany). The digests were separated on a 0.8% agarose gel and transferred to a positively charged nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany) as described previously (38). ScURA3 amplified from BY4704 chromosomal DNA with primers URA3-40 and URA3-40c was used as a hybridization probe. The probe was labeled using the digoxigenin high prime DNA labeling and detection kit II (Roche Applied Science, IN). Hybridization was performed overnight at 42°C with gentle shaking. The blot was washed twice for 5 min at 25°C in a wash solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] containing 0.1% SDS) as described previously (38). Further washes were carried out twice for 15 min at 65°C in a preheated solution of 0.5× SSC containing 0.1% SDS. Blocking of the blots, antibody reaction, removal of the unbound conjugate, and signal generation were performed according to the manufacturer's instructions. The membrane was exposed to an LAS-1000 imaging system (Fuji Photo Film Co., Ltd., Tokyo, Japan) in order to capture the signal.
Isolation and sequencing of ScURA3 flanking sequences.
For screening of auxotrophic mutants generated by ScURA3 integrative transformation, transformants were replica plated to MM and uracil dropout medium. Colonies that failed to grow on MM were picked and their growth was tested on MM supplemented with adenine, histidine, leucine, methionine, tryptophan, or lysine to identify the nutrient requirement.
To amplify the region flanking ScURA3 in RAK3627, a thermal asymmetric interlaced PCR (TAIL-PCR) method (28) was used with the following modifications. A universal primer, 15C+9A, was used instead of the arbitrary degenerate primer. The primers URA3+61c, URA3-35c, and URA3-400c were used for amplification upstream of ScURA3, and primers URA3+720, URA3+25Down, and URA3+1054 were used for the downstream sequence. The PCR mixture (10 μl) contained 200 pmol of each primer, 1× KOD Plus buffer, 200 μM of each deoxynucleoside triphosphate, 0.2 μM MgSO4, and 0.1 U KOD Plus DNA polymerase. Thermal settings for the TAIL-PCR were annealing and extension temperatures of 60°C and 68°C, respectively. The extension time was changed to 2 min in the primary and secondary PCRs. In the tertiary PCR, the reaction was initiated at 94°C for 1 min followed by 30 cycles of the following program: 94°C for 20 s, 50°C for 1 min, and 68°C for 2 min. In the primary TAIL-PCR, 4 ng of RAK3627 genomic DNA template and 15C+9A and URA3+720 or URA3+61c primers were used. One microliter of a 100-fold dilution of the primary PCR product in a total 10-μl mixture was used as a template for the secondary TAIL-PCR with primers 15C+9A and URA3+25Down or URA3-35c. The secondary PCR product was loaded on a 0.7% (wt/vol) agarose gel, the band was excised, and the PCR product was isolated. The PCR product was then used as a template with primers 15C+9A and URA3+1054 or URA3-400c for the tertiary TAIL-PCR. The PCR product of the tertiary PCR was purified using a purification cartridge with a 30,000-molecular-weight cutoff (Takara Bio Inc., Shiga, Japan). The DNA samples were sequenced using a BigDye Terminator cycle sequencing kit (Applied Biosystems, CA) according to the manufacturer's instructions.
RESULTS
Growth and ethanol production by K. marxianus and S. cerevisiae strains.
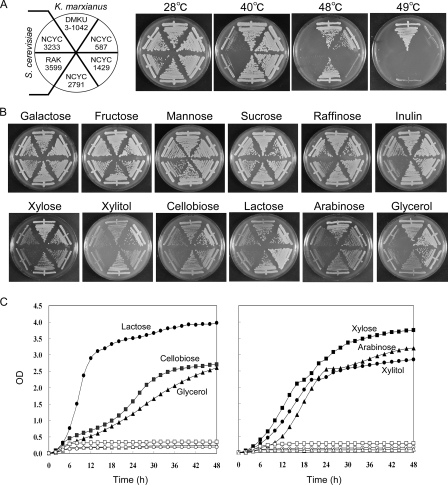
We compared thermotolerant growth levels, utilizable carbon sources, and ethanol production levels of K. marxianus strains DMKU3-1042 (27), NCYC587 (ATCC 36907) (31, 33), NCYC1429 (ATCC 12424) (48), and NCYC2791 (CBS712, K. marxianus type strain) (30) and S. cerevisiae strains NCYC3233 and RAK3599 (diploid laboratory strain with no special nutritional requirements). The growth levels of these six strains were examined at 28°C, 40°C, 48°C, and 49°C (Fig. 1A). All K. marxianus strains except NCYC1429 grew at temperatures of less than 48°C but only the DMKU3-1042 strain grew at 49°C. We also examined the growth of 15 K. marxianus strains obtained from NBRC (NITE Biological Resource Center, Japan) at 48°C. The DMKU3-1042 strain showed the best growth property (data not shown). In contrast, neither S. cerevisiae strain grew at ≥40°C.
FIG. 1.
Growth properties of K. marxianus and S. cerevisiae strains. (A) Strains of K. marxianus and S. cerevisiae were streaked on YPD plates and incubated at the indicated temperatures for 2 days. (B) Strains were streaked as described for panel A and grown at 28°C on MM supplemented with 2% of the indicated sugars. (C) K. marxianus DMKU3-1042 (closed symbols) and S. cerevisiae NCYC3233 (open symbols) were grown in liquid MM supplemented with 2% lactose (circles), cellobiose (squares), or glycerol (triangles) for the graph on the left and with xylitol (circles), xylose (squares), or arabinose (triangles) for the graph on the right. OD, optical density.
Next, the utilization of a wide variety of carbon sources was tested (Fig. 1B and C). All K. marxianus and S. cerevisiae strains utilized galactose, fructose, mannose, sucrose, raffinose, and inulin for growth (Fig. 1B, top). However, in contrast to S. cerevisiae, K. marxianus utilized xylose, xylitol, cellobiose, lactose, arabinose, and glycerol (Fig. 1B, bottom). This growth phenotype in liquid medium was also confirmed (Fig. 1C). The growth test for K. marxianus DMKU3-1042 and S. cerevisiae NCYC3233 in liquid MM supplemented with 2% of each carbon source indicated that K. marxianus DMKU3-1042 was able to utilize xylose, xylitol, cellobiose, lactose, arabinose, and glycerol for growth (Fig. 1C).
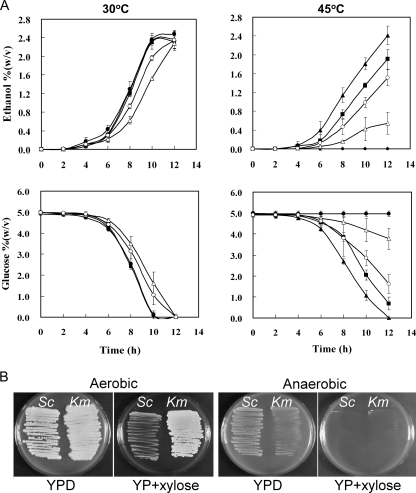
Ethanol production and glucose consumption were also examined at 30°C and 45°C (Fig. 2A). All K. marxianus and S. cerevisiae strains exhibited similar levels of ethanol production and glucose consumption at 30°C (Fig. 2A, left). However, when the ethanol fermentation was carried out at 45°C, S. cerevisiae did not grow and thus did not produce ethanol, but the K. marxianus strains produced ethanol (Fig. 2A, right). Among the four strains of K. marxianus, DMKU3-1042 converted glucose to ethanol faster than the others at 45°C. From these results, we conclude that DMKU3-1042 had the fastest ethanol productivity at high temperatures among the six strains examined.
FIG. 2.
Ethanol fermentation. (A) Ethanol production (top) and glucose consumption (bottom) by S. cerevisiae NCYC3233 (closed circles) and K. marxianus strains DMKU3-1042 (closed triangles), NCYC587 (closed squares), NCYC1429 (open triangles), and NCYC2791 (open circles) in YP medium supplemented with 5% glucose at 30°C (left) and 45°C (right). (B) Growth of K. marxianus DMKU3-1042 (Km) and S. cerevisiae NCYC3233 (Sc) on YPD and YP supplemented with 2% xylose (YP+xylose) under aerobic or anaerobic conditions (3 or 5 days, respectively).
Ethanol production by K. marxianus strains grown on xylose as a sole carbon source was also tested, but very little ethanol was observed. Similar results were obtained for DMKU3-1042 when cellobiose, arabinose, and lactose were used as sole carbon sources (data not shown). We next tested the anaerobic growth of K. marxianus when xylose was the sole carbon source (Fig. 2B). K. marxianus DMKU3-1042 was found to be unable to grow on YP-plus-xylose medium under anaerobic conditions, indicating that the anaerobic fermentation of xylose is not a native trait of K. marxianus.
Development of a K. marxianus transformation system by use of linear DNA.
In order to develop a transformation system for K. marxianus, spontaneous uracil auxotrophs were sought by 5-FOA selection (9). Colonies that arose on 5-FOA plates were streaked onto MM and MM supplemented with uracil (MM-plus-uracil) plates. One mutant that grew on MM plus uracil but not on MM was designated RAK3605 and used as a transformation host.
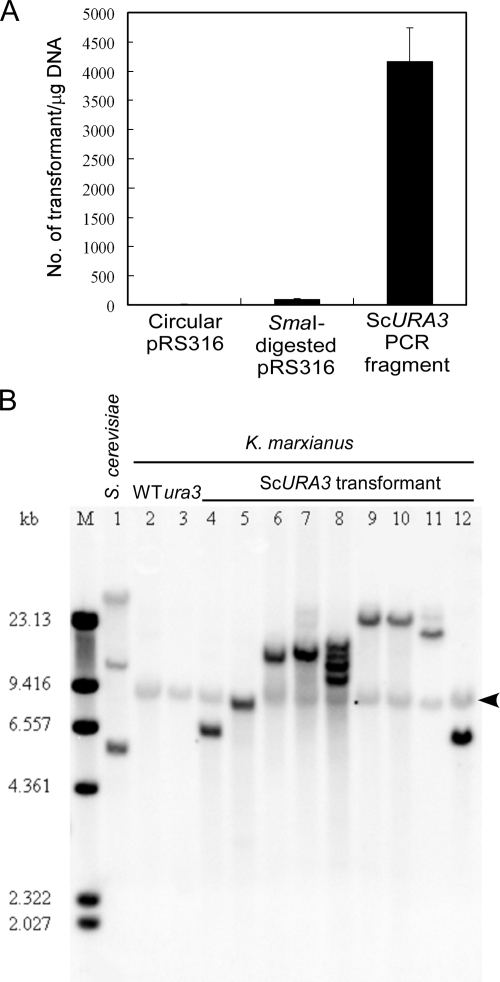
To confirm that the mutation in RAK3605 was actually in the ura3 gene, we attempted to complement the strain with S. cerevisiae URA3, because ScURA3 has been shown to complement a K. marxianus ura3 mutant (21). Initially, S. cerevisiae replicative plasmid pRS316, carrying a ScURA3 marker, was used directly, but no transformants were obtained even at a DNA amount as high as 1 μg, suggesting that the S. cerevisiae ARS cannot function in K. marxianus. Plasmid pRS316 was then linearized with SmaI prior to the transformation, which yielded 90 colonies/μg DNA (Fig. 3A). Transformation with the PCR-amplified ScURA3 from S. cerevisiae genomic DNA was tested next. Surprisingly, transformants were obtained at a frequency of 4.2 × 103 colonies/μg DNA (Fig. 3A).
FIG. 3.
Transformation of a K. marxianus ura3 mutant with a linear DNA of S. cerevisiae URA3 (ScURA3). (A) The K. marxianus ura3 mutant was not transformed with the intact ScURA3 plasmid (pRS316) but with SmaI-digested pRS316 (4.8 kb) and the ScURA3 PCR fragment (1.7 kb). (B) Southern blot hybridization of chromosomal DNA of S. cerevisiae BY4704 (lane 1), K. marxianus wild-type (WT) DMKU3-1042 (lane 2), ura3 mutant RAK3605 (lane 3), and nine ScURA3 transformants from strain RAK3605 (lanes 4 to 12). Chromosomal DNA was digested with BamHI, run on a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with a digoxigenin-labeled ScURA3 probe. The bands indicated by an arrowhead are likely the authentic K. marxianus URA3 that cross-hybridizes with ScURA3. M, DNA size marker.
Because the ScURA3 transformants were stable even after growth in YPD medium (data not shown), we anticipated that the ScURA3 had integrated into a K. marxianus chromosome. Integration was subsequently demonstrated by Southern hybridization using ScURA3 as a probe (Fig. 3B). Hybridization with ScURA3 revealed one faint ∼9-kb band in all K. marxianus strains tested, suggesting that the band represents the authentic cross-hybridizing KmURA3 gene. In addition to this faint band, most ScURA3 transformants had single additional bands of various sizes, indicating single and random integration events. In one transformant (Fig. 3B, lane 8), multiple integrations at different loci were observed.
Insertional mutagenesis by linear ScURA3 transformation.
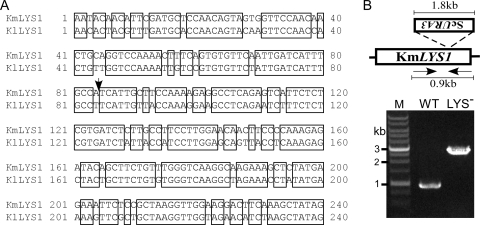
If ScURA3 DNA could insert randomly into K. marxianus chromosomes during transformation, we reasoned that the procedure could be used for insertional mutagenesis, much like transposon mutagenesis (24). Therefore, a screen for auxotrophic mutants was undertaken following transformation with ScURA3. Among several thousand ScURA3 transformants, two were found to be unable to grow on MM. One was subsequently found to be Ade− (RAK3626) and the other Lys− (RAK3627). To identify the integration site of ScURA3 in RAK3627, upstream and downstream regions flanking the insertion were isolated by a modified TAIL-PCR method. The sequence of the flanking regions was compared to a K. lactis database (13). The nucleotide sequence of the flanking 240 bp was 81.7% identical to that of KlLYS1. ScURA3 was found to have inserted into the LYS1 open reading frame without generating any additional nucleotide changes (Fig. 4A).
FIG. 4.
Screening for auxotrophic mutants following transformation with ScURA3. (A) Sequence flanking the ScURA3 insertion was obtained by TAIL-PCR (KmLYS1) and was found to be highly homologous to that of K. lactis (KlLYS1). An arrow indicates the insertion site. (B) PCR amplification using genomic DNA from DMKU3-1042 (wild type [WT]) and the Lys− mutant RAK3627 (LYS−) as templates confirmed the disruption of KmLYS1 by ScURA3. ScURA3 was inserted in an inverse direction relative to KmLYS1. The absence of a wild-type band in the lys1 disruptant suggests a haploid chromosomal constitution.
For further confirmation, primers KmLYS1-225Up and KmLYS1-606Down were designed according to the obtained sequence to amplify a PCR fragment containing 225 bp upstream and 606 bp downstream of the insertion site. As shown in Fig. 4B, a band of 831 bp corresponding to the wild-type LYS1 region and a single band of 2.6 kbp corresponding to the ScURA3 insert are apparent. This result also suggested that K. marxianus DMKU3-1042 is haploid, because there was no 0.8-kb band corresponding to the wild-type allele in the lys1::ScURA3 strain. If it is diploid, two bands, one containing the insert and other the wild type, should have appeared (18). Further, complementation of the lys1 mutant with ScLYS1 was tested. The ScLYS1 gene was amplified from S. cerevisiae by PCR and used directly to transform the K. marxianus lys1 strain. Lys+ transformants were obtained, indicating that functional complementation by the S. cerevisiae gene was feasible in K. marxianus.
Expression of Aspergillus α-amylase gene under the control of the ScTDH3 promoter in K. marxianus.
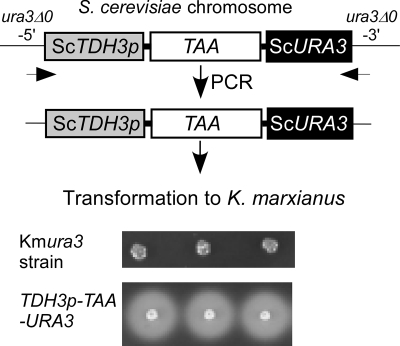
Based on the complementation results, we expected that S. cerevisiae promoters would also be functional in K. marxianus. Therefore, an expression cassette consisting of the Aspergillus oryzae α-amylase (TAA) gene under the control of the S. cerevisiae TDH3 promoter (ScTDH3p) was constructed at the ura3Δ0 locus in S. cerevisiae. Amylase expression in S. cerevisiae was confirmed (data not shown). A PCR fragment containing ScTDH3p-TAA-ScURA3 was amplified from the S. cerevisiae chromosome and directly used to transform the K. marxianus RAK3605 ura3 strain (Fig. 5). The appearance of clear zones around colonies of Ura+ transformants after exposure to iodine vapor indicated functional α-amylase expression in K. marxianus (Fig. 5). When a promoterless TAA-ScURA3 cassette was used for transformation, only 3% (38/1,235) of the colonies had halos (data not shown), suggesting fortuitous insertion downstream of authentic promoters. These results indicate that K. marxianus strains expressing foreign genes can be constructed by simple transformation using S. cerevisiae promoters and open reading frames.
FIG. 5.
Construction of an α-amylase expression strain in K. marxianus by transformation with a linear DNA. An ScTDH3 promoter-driven TAA (Aspergillus oryzae α-amylase gene) was constructed at the URA3 locus of S. cerevisiae and the PCR-amplified construct was then used without additional manipulation to transform a K. marxianus ura3 mutant. The appearance of clear zones around colonies indicates that the α-amylase gene was expressed under the control of the S. cerevisiae TDH3 promoter in K. marxianus.
DISCUSSION
K. marxianus is a well-known thermotolerant yeast species. Its ability to produce ethanol by fermentation at high temperature and its use of a wide range of carbon sources make this species attractive for industrial applications (14). While high-temperature growth and fermentation by K. marxianus strains have been previously reported (3, 6, 20, 27, 42), with maximum growth temperatures varying between 47°C and 52°C (14), to our knowledge, no comparative studies performed under the same experimental conditions have been published. Although a maximum growth temperature of 52°C was reported for K. marxianus IBM (5), this value is likely to be dependent on the experimental conditions and growth medium used. The results of the present study clearly show that NCYC587 and NCYC2791 grew at 48°C but that only DMKU3-1042 grew at 49°C on a YPD plate (Fig. 1A). All K. marxianus strains produced ethanol at 45°C, but the highest productivity was that of DMKU3-1042 (Fig. 2A). All K. marxianus strains tested shared similar abilities to use nonglucose carbon sources. Strain DMKU3-1042 utilized lactose, cellobiose, glycerol, xylose, arabinose, and xylitol similarly to the other K. marxianus strains and unlike S. cerevisiae. These parallel comparisons, undertaken under identical conditions, indicate that DMKU3-1042 is the most suitable strain for high-temperature growth and fermentation among the K. marxianus strains tested.
Recently, bioethanol production from lignocellulosic biomass has become an increasingly attractive process, because cellulose is economical, abundant, and renewable. However, because of the high pentose content of lignocellulosic biomass, (i.e., xylose and arabinose), the use of conventional non-pentose-fermenting S. cerevisiae strains alone will not be sufficient to produce all the ethanol that potentially can be obtained. This recognition has stimulated extensive studies on recombinant S. cerevisiae strains capable of utilizing xylose and arabinose (8, 16, 39, 44, 47). The natural ability of K. marxianus to ferment xylose and arabinose is one important advantage it has over S. cerevisiae. Indeed, direct ethanol production from the fermentation of d-xylose by K. marxianus strains has been reported (29, 46). However, under the conditions used in the present study, we were unable to confirm efficient ethanol production from d-xylose by all tested strains. We also tested whether the strains could grow on YP-plus-xylose medium under anaerobic conditions. While the strains were able to grow anaerobically on YPD, none could grow on YP plus xylose. These results suggest that the production of ethanol from xylose by K. marxianus may not be possible or may require unknown conditions. Nevertheless, the natural ability of K. marxianus to use cellobiose, xylose, and arabinose for growth is certainly an attractive trait. Molecular genetic analysis of the native xylose utilization pathway in K. marxianus, based in part on tools developed in the present study, should help to explain the physiological impediments to the efficient bioconversion of xylose to ethanol.
With respect to a genetic engineering approach, appropriate host strains and vectors are generally required for transformations and gene manipulation. Vectors used for transformation of eukaryotic microorganisms are typically constructed as Escherichia coli shuttle plasmids containing a species- or genus-specific ARS, even in K. marxianus (4, 7, 21, 22). For example, plasmids containing a K. lactis ARS (8, 12, 24) or a 1.6-μm circular plasmid from K. drosophilarum (16) have been used to transform K. marxianus (17). In the present study, K. marxianus could not be transformed with the intact S. cerevisiae plasmid pRS316, containing an S. cerevisiae CEN6-ARS4 sequence, indicating that this S. cerevisiae ARS cannot function in K. marxianus. However, transformation with the linearized pRS316 plasmid and PCR-amplified ScURA3 yielded transformants. Southern blot hybridization with the ScURA3 probe indicated that ScURA3 inserted randomly into the K. marxianus genome. These results demonstrate that K. marxianus DMKU3-1042 is an excellent host for efficient random integration of nonhomologous DNA sequences.
This finding was verified by mutagenesis using a linear, PCR-generated ScURA3 to obtain insertion mutations via transformation. Two auxotrophic mutants (Ade− and Lys− strains) were identified from among several thousand transformants by screening on MM. The Lys− strain was analyzed further. The DNA sequence flanking ScURA3 confirmed that the insertion had occurred in the K. marxianus LYS1 gene. This kind of random integration is typical of transposons, some of which have been modified and used extensively in S. cerevisiae for large-scale insertion mutagenesis (24, 37). This suggests the possibility that PCR-amplified linear DNA instead of a functional transposon may be used for efficient insertional mutagenesis in K. marxianus.
Moreover, an insertion of ScURA3 at the lys1 locus in K. marxianus provided a means for determining the haploid status of K. marxianus DMKU3-1042. The ploidy of other K. marxianus strains has not been determined. Steensma et al. (43) indicated that some K. marxianus strains, including CBS6556, were haploid because they mated with one another. Subsequently, Ribeiro et al. (36) suggested a diploid status based on results of an experiment involving gene disruption and subsequent PCR. Our interpretation of the LYS1 integration results presented in the present study is that K. marxianus DMKU3-1042 is haploid. In K. marxianus NBRC1777, the disruption of URA3, LEU2, and TRP1 led to the detection of the disrupted alleles but no detection of the wild-type alleles, indicating that this strain is also haploid (19). Haploidy may predominate in native K. marxianus strains.
The expression of foreign genes in K. marxianus, e.g., the lactate dehydrogenase gene from Bacillus megaterium (34), the endo-β-1,4-glucanase gene from Aspergillus niger, the cellobiohydrolase and β-glucosidase genes from Thermoascus aurantiacus (19), and the β-glucuronidase gene from E. coli (4), has been reported. All of these genes were initially manipulated in E. coli plasmids prior to their introduction into K. marxianus by transformation. In the present study, we used a PCR-amplified DNA for transformation and expression without additional manipulation (Fig. 5). The use of linear DNA for transformation eliminates time-consuming plasmid construction, including restriction digestions, ligations, cloning in E. coli, plasmid isolation, and purification. The possibility of critical genes being disrupted following integrative transformation can be minimized by analyzing a number of independent transformants.
In the present study, we used S. cerevisiae genes, namely, ScURA3 and ScLYS1, as selectable markers and an S. cerevisiae promoter, ScTDH3p, for expressing a foreign gene in K. marxianus. The results suggest that gene function is conserved between K. marxianus and S. cerevisiae. Native selectable markers and promoters have also been cloned and used for gene manipulation in K. marxianus (4, 19). We speculate here that many S. cerevisiae genes (12) and probably many in K. lactis (13) as well will be functional in K. marxianus. We anticipate that this evolutionary conservation and transformability using linear DNA will facilitate the development of genetic engineering and molecular biological analyses of K. marxianus.
In conclusion, with respect to bioethanol production, the nonconventional yeast K. marxianus offers several advantages lacking for S. cerevisiae. Notably, K. marxianus is able to grow and ferment at high temperature, has the natural ability to use xylose, arabinose, and cellobiose, and can be transformed efficiently via nonhomologous manner with linear DNA. For these reasons, we anticipate that the yeast K. marxianus will be a promising choice for cost-effective ethanol production and an alternative host for molecular genetic studies.
Acknowledgments
We thank Daisuke Suehiro, Shiho Iwai, Sophon Boonlue, and Yukie Misumi for their competent technical assistance and Kazunobu Matsushita for his helpful discussions. We acknowledge the technical expertise of the DNA Core Facility of the Center for Gene Research, Yamaguchi University.
S. Nonklang and K. Cha-aim gratefully acknowledge receipt of Ph.D. fellowships from the Scientific Cooperation Program between the Japan Society for the Promotion of Science (JSPS) and the National Research Council of Thailand (NRCT). This work was supported in part by the New Energy and Industrial Technology Development Organization (NEDO) and by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), Japan.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Akada, R., T. Kitagawa, S. Kaneko, D. Toyonaga, S. Ito, Y. Kakihara, H. Hoshida, S. Morimura, A. Kondo, and K. Kida. 2006. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast 23:399-405. [DOI] [PubMed] [Google Scholar]
- 2.Akada, R., Y. Shimizu, Y. Matsushita, M. Kawahata, H. Hoshida, and Y. Nishizawa. 2002. Use of a YAP1 overexpression cassette conferring specific resistance to cerulenin and cycloheximide as an efficient selectable marker in the yeast Saccharomyces cerevisiae. Yeast 19:17-28. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P. J., K. McNeil, and K. Watson. 1986. High-efficiency carbohydrate fermentation to ethanol at temperatures above 40°C by Kluyveromyces marxianus var. marxianus isolated from sugar mills. Appl. Environ. Microbiol. 51:1314-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, M. M., A. Raynal, M. Guerineau, and F. Iborra. 1999. Construction of efficient centromeric, multicopy and expression vectors for the yeast Kluyveromyces marxianus using homologous elements and the promoter of a purine-cytosine-like permease. J. Mol. Microbiol. Biotechnol. 1:347-353. [PubMed] [Google Scholar]
- 5.Banat, I. M., P. Nigam, D. Singh, R. Marchantand, and A. P. McHale. 1998. Ethanol production at elevated temperatures and alcohol concentrations. I. Yeasts in general. World J. Microbiol. Biotechnol. 14:809-821. [Google Scholar]
- 6.Banat, I. M., P. Nigram, and R. Marchant. 1992. Isolation of thermotolerant, fermentative yeasts growing at 52°C and producing ethanol at 45°C and 50°C. World J. Microbiol. Biotechnol. 8:259-263. [DOI] [PubMed] [Google Scholar]
- 7.Bartkeviciute, D., R. Siekstele, and K. Sasnauskas. 2000. Heterologous expression of the Kluyveromyces marxianus endopolygalacturonase gene (EPG1) using versatile autonomously replicating vector for a wide range of host. Enzyme Microb. Technol. 26:653-656. [DOI] [PubMed] [Google Scholar]
- 8.Becker, J., and E. Boles. 2003. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl. Environ. Microbiol. 69:4144-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 10.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 11.de Sanchez, S. B., and F. J. Castillo. 1980. Effect of pH on the growth of Kluyveromyces fragilis on deproteinized whey. Acta Cient. Venez. 31:24-26. [PubMed] [Google Scholar]
- 12.Dujon, B. 1996. The yeast genome project: what did we learn? Trends Genet. 12:263-270. [DOI] [PubMed] [Google Scholar]
- 13.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44.15229592 [Google Scholar]
- 14.Fonseca, G. G., E. Heinzle, C. Wittmann, and A. K. Gombert. 2008. The yeast Kluyveromyces marxianus and its biotechnological potential. Appl. Microbiol. Biotechnol. 79:339-354. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 16.Hahn-Hagerdal, B., K. Karhumaa, M. Jeppsson, and M. F. Gorwa-Grauslund. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147-177. [DOI] [PubMed] [Google Scholar]
- 17.Hang, Y. D., E. E. Woodams, and L. E. Hang. 2003. Utilization of corn silage juice by Kluyveromyces marxianus. Bioresour. Technol. 86:305-307. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto, S., M. Ogura, K. Aritomi, H. Hoshida, Y. Nishizawa, and R. Akada. 2005. Isolation of auxotrophic mutants of diploid industrial yeast strains after UV mutagenesis. Appl. Environ. Microbiol. 71:312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, J., Y. Wang, H. Kumagai, and H. Tamaki. 2007. Construction of thermotolerant yeast expressing thermostable cellulase genes. J. Biotechnol. 130:114-123. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, D. B., N. J. Tudroszen, and C. J. Moye. 1984. The effect of temperature on the kinetics of ethanol production by a thermotolerant strain of Kluyveromyces marxianus. Biotechnol. Lett. 6:1-6. [Google Scholar]
- 21.Iborra, F. 1993. High efficiency transformation of Kluyveromyces marxianus by a replicative plasmid. Curr. Genet. 24:181-183. [DOI] [PubMed] [Google Scholar]
- 22.Iborra, F., and M. M. Ball. 1994. Kluyveromyces marxianus small DNA fragments contain both autonomous replicative and centromeric elements that also function in Kluyveromyces lactis. Yeast 10:1621-1629. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Kumar, A., M. Seringhaus, M. C. Biery, R. J. Sarnovsky, L. Umansky, S. Piccirillo, M. Heidtman, K. H. Cheung, C. J. Dobry, M. B. Gerstein, N. L. Craig, and M. Snyder. 2004. Large-scale mutagenesis of the yeast genome using a Tn7-derived multipurpose transposon. Genome Res. 14:1975-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laloux, O., J. P. Cassart, J. Delcour, J. Van Beeumen, and J. Vandenhaute. 1991. Cloning and sequencing of the inulinase gene of Kluyveromyces marxianus var. marxianus ATCC 12424. FEBS Lett. 289:64-68. [DOI] [PubMed] [Google Scholar]
- 26.Lertwattanasakul, N., K. Sootsuwan, S. Limtong, P. Thanonkeo, and M. Yamada. 2007. Comparison of the gene expression patterns of alcohol dehydrogenase isozymes in the thermotolerant yeast Kluyveromyces marxianus and their physiological functions. Biosci. Biotechnol. Biochem. 71:1170-1182. [DOI] [PubMed] [Google Scholar]
- 27.Limtong, S., C. Sringiew, and W. Yongmanitchai. 2007. Production of fuel ethanol at high temperature from sugar cane juice by a newly isolated Kluyveromyces marxianus. Bioresour. Technol. 98:3367-3374. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y.-G., N. Mitsukawa, T. Oosumi, and R. Whittier. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8:457-463. [DOI] [PubMed] [Google Scholar]
- 29.Margaritis, A., and P. Bajpai. 1982. Direct fermentation of d-xylose to ethanol by Kluyveromyces marxianus strains. Appl. Environ. Microbiol. 44:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martini, A., H. J. Phaff, and S. A. Douglass. 1972. Deoxyribonucleic acid base composition of species in the yeast genus Kluyveromyces van der Walt emend. van der Walt. J. Bacteriol. 111:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mrak, E. M., H. J. Phaff, R. H. Vaughn, and H. N. Hansen. 1942. Yeasts occurring in souring figs. J. Bacteriol. 44:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozmihci, S., and F. Kargi. 2007. Comparison of yeast strains for batch ethanol fermentation of cheese-whey powder (CWP) solution. Lett. Appl. Microbiol. 44:602-606. [DOI] [PubMed] [Google Scholar]
- 33.Passador-Gurgel, G. C., S. A. Furlan, J. K. Meller, and R. Jonas. 1996. Application of a microtitre reader system to the screening of inulinase nulinase-producing yeasts. Appl. Microbiol. Biotechnol. 45:158-161. [Google Scholar]
- 34.Pecota, D. C., V. Rajgarhia, and N. A. Da Silva. 2007. Sequential gene integration for the engineering of Kluyveromyces marxianus. J. Biotechnol. 127:408-416. [DOI] [PubMed] [Google Scholar]
- 35.Rajoka, M. I., M. Ferhan, and A. M. Khalid. 2005. Kinetics and thermodynamics of ethanol production by a thermotolerant mutant of Saccharomyces cerevisiae in a microprocessor-controlled bioreactor. Lett. Appl. Microbiol. 40:316-321. [DOI] [PubMed] [Google Scholar]
- 36.Ribeiro, O., A. K. Gombert, J. A. Teixeira, and L. Domingues. 2007. Application of the Cre-loxP system for multiple gene disruption in the yeast Kluyveromyces marxianus. J. Biotechnol. 131:20-26. [DOI] [PubMed] [Google Scholar]
- 37.Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heidtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413-418. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sedlak, M., and N. W. Ho. 2001. Expression of E. coli araBAD operon encoding enzymes for metabolizing l-arabinose in Saccharomyces cerevisiae. Enzyme Microb. Technol. 28:16-24. [DOI] [PubMed] [Google Scholar]
- 40.Shimoda, C., A. Itadani, A. Sugino, and M. Furusawa. 2006. Isolation of thermotolerant mutants by using proofreading-deficient DNA polymerase delta as an effective mutator in Saccharomyces cerevisiae. Genes Genet. Syst. 81:391-397. [DOI] [PubMed] [Google Scholar]
- 41.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh, D., P. Nigam, I. M. Banat, I. M. Manta, R. Marchant, and A. P. McHale. 1998. Ethanol production at elevated temperatures and alcohol concentrations. II. Use of Kluyveromyces marxianus IBM3. World J. Microbiol. Biotechnol. 14:823-834. [Google Scholar]
- 43.Steensma, H. Y., F. C. M. de Jongh, and M. Linnekamp. 1988. The use of electrophoretic karyotypes in the classification of yeasts: Kluyveromyces marxianus and K. lactis. Curr. Genet. 14:311-317. [Google Scholar]
- 44.van Maris, A. J., A. A. Winkler, M. Kuyper, W. T. de Laat, J. P. van Dijken, and J. T. Pronk. 2007. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv. Biochem. Eng. Biotechnol. 108:179-204. [DOI] [PubMed] [Google Scholar]
- 45.Vaughan-Martini, A., G. Rosini, and A. Martini. 1988. Killer sensitivity patterns as a tool for the fingerprinting of strains within the yeast species Kluyveromyces lactis and K. marxianus. Biotechnol. Tech. 2:293-296. [Google Scholar]
- 46.Wilkins, M. R., M. Mueller, S. Eichling, and I. M. Banat. 2008. Fermentation of xylose by the thermotolerant yeast strains Kluyveromyces marxianus IBM2, IBM4 and IBM5 under anaerobic conditions. Process. Biochem. 43:346-350. [Google Scholar]
- 47.Wisselink, H. W., M. J. Toirkens, M. del Rosario Franco Berriel, A. A. Winkler, J. P. van Dijken, J. T. Pronk, and A. J. van Maris. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl. Environ. Microbiol. 73:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Workman, W. E., and D. F. Day. 1983. Purification and properties of the β-fructofuranosidase from Kluyveromyces fragilis. FEBS Lett. 160:16-20. [DOI] [PubMed] [Google Scholar]
- 49.Zafar, S., and M. Owais. 2006. Ethanol production from crude whey by Kluyveromyces marxianus. Biochem. Eng. J. 27:295-298. [Google Scholar]