Abstract
Within an isogenic microbial population in a homogenous environment, individual bacteria can still exhibit differences in phenotype. Phenotypic heterogeneity can facilitate the survival of subpopulations under stress. As the gram-positive bacterium Lactobacillus plantarum grows, it acidifies the growth medium to a low pH. We have examined the growth of L. plantarum microcolonies after rapid pH downshift (pH 2 to 4), which prevents growth in liquid culture. This acidification was achieved by transferring cells from liquid broth onto a porous ceramic support, placed on a base of low-pH MRS medium solidified using Gelrite. We found a subpopulation of cells that displayed phenotypic heterogeneity and continued to grow at pH 3, which resulted in microcolonies dominated by viable but elongated (filamentous) cells lacking septation, as determined by scanning electron microscopy and staining cell membranes with the lipophilic dye FM4-64. Recovery of pH-stressed cells from these colonies was studied by inoculation onto MRS-Gelrite-covered slides at pH 6.5, and outgrowth was monitored by microscopy. The heterogeneity of the population, calculated from the microcolony areas, decreased with recovery from pH 3 over a period of a few hours. Filamentous cells did not have an advantage in outgrowth during recovery. Specific regions within single filamentous cells were more able to form rapidly dividing cells, i.e., there was heterogeneity even within single recovering cells.
Lactobacillus plantarum is a widespread gram-positive bacterium commonly found in fermented foods. L. plantarum has applications as a starter culture, as a probiotic, and in the delivery of therapeutics (15, 24, 41). L. plantarum occupies a broad range of niches by virtue of its ability to utilize a broad array of carbohydrates as growth substrates and to grow and survive at low pHs. Growth of L. plantarum results in acidification of the medium, generally via the production of lactic acid. Both lactic acid and pH stresses on L. plantarum strain WCFS1 have been studied, for example, by continuous culture followed by transcriptome profiling or by the measurement of intracellular pH (9, 24, 39). The ability of Lactobacillus species to maintain a moderate intracellular pH under acid conditions contributes to survival at low pHs. A number of responses are implicated in low-pH survival, including proton export by the FoF1-ATPase, repair or protection of cell components, activation of global regulators, and alterations in the cell surface (8, 9, 33, 43). Adaptation to low pH occurs within Lactobacillus species and can protect against other stresses, and vice versa, although the degree of cross-protection is often strain specific (9). Additionally, in some gram-positive bacteria, sensitivity to pH stress depends on cell density (13) and/or growth phase (7).
Studies of the response of Lactobacillus species to pH stress to date have concentrated on measuring the average response of the population (e.g., references 3 and 8). However, it is clear that for many stresses, there is considerable variation within a genetically identical population, even within a homogeneous environment (2, 6). The heterogeneity of stress response is of particular interest in cases in which we wish to kill bacteria or inhibit their growth. Phenotypic heterogeneity makes judging the optimal effective dosage of an antimicrobial treatment difficult. It may also be relevant in cases in which microbial survival under fluctuating stresses is desired. One method of studying phenotypic heterogeneity is to directly or indirectly follow the growth or survival of individual cells (12, 16, 23, 26, 28, 34, 36, 37) during imposition of stress and during recovery. Also, a heterogeneous response to stress can be revealed by biphasic kill curves (5, 11). More recent approaches have used flow cells and digital image analysis to identify subpopulations such as “persister” cells with increased resistance to antibiotics (4). Recently, imaging of microcolonies grown on porous aluminum oxide (PAO) has been used to quantify heterogeneity in the salt stress response of Bacillus cereus (10). This method has been used during exposure to a stressing agent and also in recovery, particularly during the outgrowth of a CFU over the first few hours after exposure to stress.
A common response to stress is a change in cell morphology, particularly filamentation, in which cell growth (in particular, elongation) continues but septation (i.e., division) is inhibited. Filamentation occurs in response to antibiotics, desiccation, high salinity, irradiation and other forms of DNA damage, prophage induction, temperature or pH extremes, and organic solvents, as well as in human infections and in biofilms (14, 32, 35, 36). The production of heterologous proteins in Lactobacillus casei also induces filamentous cells (1). Despite this change being such a widespread phenomenon, the fitness advantage of this change in morphology to the bacterium is often unclear. Filamentation is commonly, but not exclusively, linked to the SOS response. The SOS response is known to be subject to phenotypic heterogeneity (11). Many of the other features of the SOS response, including an elevated mutation rate and stimulation of plasmid transfer, directly contribute to increased fitness for the stressed microorganisms (14). Filamentation during microbial stress recovery in the food industry is of interest. Filamentation may have survival value to microorganisms during processing, and the viable count for CFU may not accurately assess the danger posed by the contamination of foods by filamentous microorganisms (19). The latter point is relevant if a highly elongated cell can rapidly divide into a large number of progeny. In this scenario, an elongated cell with greater biomass could be more effective in immediate outgrowth than a shorter cell, despite both being potentially counted as a single CFU.
In this study, we used cell and microcolony imaging to quantify the heterogeneity of L. plantarum WCFS1 microcolonies in response to rapid acidification, particularly in the recovery of the survivors.
MATERIALS AND METHODS
Culture at low pHs on PAO.
MRS-Gelrite plates were made by combining double-strength MRS medium (Oxoid, United Kingdom) with an equal volume of molten 2% (wt/vol) Gelrite (Merck) and pouring immediately (22, 30). Agar MRS plates were gelled with 1.5% (wt/vol) agar (Oxoid). The pH of MRS medium was adjusted with HCl and checked with two pH indicator papers: Panpeha, with a range from pH 0 to 14 (Sigma, The Netherlands), and Acilit reading from pH 0 to 6 (Merck). The accuracy of pH papers was assessed against those of standards prepared from MRS medium (31) adjusted to the appropriate pH with HCl and measured using a PHM 82 pH meter (Radiometer, Denmark). For microcolony culture and imaging, sterile strips of PAO, sold as Anopore (20, 21), were placed on the MRS-Gelrite or agar plates and inoculated with 104 to 106 CFU cm−2 of L. plantarum WCFS1. Strips were inoculated from a mid-log-phase culture grown in MRS medium under anaerobic conditions. The initial pH of the MRS medium was 6.5; the pH of the mid-log-phase culture varied from 5 to 5.5. Plates were incubated under anaerobic conditions in Anaerocult A bags (Merck) at 37°C from 1 day to 2 weeks.
Survival curves at pH 3.
Mid-log-phase anaerobic cultures of L. plantarum in MRS medium (pH 5 to 5.5) were diluted into liquid medium at pH 3 at densities of ca. 109 and 107 CFU ml−1. At intervals, samples were taken and a viable count was determined on pH 6.5 MRS agar at 37°C with incubation for 36 h. Additionally, the pH of the medium was checked during sampling (as described for medium preparation) and samples were taken for observation by microscopy.
Recovery from low pH monitored by transmission microscopy of growing cells.
A 1.5-mm-thick film of MRS-Gelrite at the appropriate pH was poured on a sterile microscope slide with spacers, and a second slide was set on top to ensure a flat upper surface, which was removed after gelling. After 3 days of culture on PAO at pH 3, cells from discrete microcolonies were sampled using a sterile toothpick and the microcolony was viewed by transmission light microscopy. Cells sampled with a toothpick were inoculated onto this MRS gel film. A coverslip was placed over the inoculum, and the rest of the slide was sealed with plastic film (Euroshopper, The Netherlands) to limit desiccation. The slides were maintained at 37°C on a DC60 heated microscope stage (Linkham Scientific, United Kingdom). Cells were imaged periodically by transmission microscopy (1,000× optical magnification with an oil immersion lens). Imaging was performed with minimal illumination (<10 s per picture), with pictures captured every 20 to 30 min over a period of up to 4 h. Similar experiments were performed by transferring L. plantarum from microcolonies grown on PAO strips on pH 6.5 MRS plates to pH 6.5 MRS-Gelrite or agar slides. For each condition, the outgrowth of at least 20 CFU was measured in at least six separate experiments.
SEM.
Scanning electron microscopy (SEM) of microcolonies was performed as previously described (20). For stress recovery experiments, strips of PAO with cells cultured for 2 to 3 days at pH 3 were transferred to MRS agar plates at pH 6.5 and incubated for 1 to 3 h under anaerobic conditions at 37°C before fixation.
Staining of cells for fluorescence microscopy.
The Live/Dead staining system (Invitrogen, The Netherlands) was used to assess the membrane integrity of L. plantarum cells cultured on PAO (18, 20) by dual staining with Syto9 and propidium iodide (PI). Membranes and nucleic acids of cells isolated from growing microcolonies were stained by adding the lipophilic dye F4-64 (40) (Invitrogen, The Netherlands) at 10 μg ml−1 and Syto9 at (1 μg ml−1) in MRS medium (pH 6.5). The cells were incubated at room temperature for 20 min and then imaged by oil immersion fluorescence microscopy after spotting 5-μl aliquots onto agar gel-pad slides.
Image capture and analysis.
Image acquisition was performed with a charge-coupled-device camera mounted on an Olympus BX41 microscope. Cell and microcolony dimensions were analyzed from TIFF images by using the open source software ImageJ version 1.37v (http://rsb.info.nih.gov/ij/) via the method described for the analysis of B. cereus microcolonies on PAO (10). Cell length was measured by tracing the cell from pole to pole. Cell areas excluded intercellular areas. Both image capture and subsequent processing were performed as previously described (10, 20).
Calculations and statistical analysis.
Calculation of the heterogeneity used log10 transformed microcolony areas. Variances and means were calculated for each time point as previously described (10). Pearson correlation coefficients (r) and coefficients of determination (r2) were calculated using the Vassar statistics server (R. Lowry, VassarStats [http://faculty.vassar.edu/lowry/VassarStats.html]).
RESULTS
Culture of L. plantarum microcolonies at low pH on PAO.
L. plantarum WCFS1 was cultured on strips of PAO supported by MRS medium solidified with Gelrite. Gelrite was used as an agar substitute because the conventional agars and agaroses tested (from several manufacturers) were found to gel poorly at pH values below 3.5. Gelrite is more widely known for its thermal stability (22) but in this work also proved useful to gel MRS medium at pH 2 and above. The culturability of L. plantarum on MRS medium with 1% (wt/vol) Gelrite pH 6.5 was compared to MRS agar (1.5% wt/vol) by a viable count from a late-log-phase culture, with all growth performed under anaerobic conditions. Culturability of L. plantarum on MRS-Gelrite was identical to that of L. plantarum on MRS agar. MRS-Gelrite yielded visible colonies of a similar size to those formed on agar. It was therefore concluded that Gelrite is a suitable matrix for the growth of L. plantarum WCFS1.
Strips of PAO were used to facilitate rapid changes in the environment of the microcolonies simply by moving the strip to a new agar or Gelrite base. Because PAO is highly porous and the strips are 60-μm thick, the change in medium experienced by cells on the surface is rapid; generally it occurred within seconds or minutes (21). This method allowed staining of cells in situ or shifts in the pH (21). PAO is an oxide of aluminum and as such is extremely stable at low pH (20). A combination of these techniques allowed microcolonies to be grown, stressed, and visualized despite the acidic conditions. L. plantarum was grown anaerobically in MRS broth until mid-log phase (pH 5), then rapidly subjected to a change in pH by plating on PAO on MRS-Gelrite (pH range, 2.5 to 6), and then incubated under anaerobic conditions. Staining with Syto9 and PI allowed visualization of microcolonies, which in some cases appeared incapable of growing to visible colonies. The growth of at least a fraction of the plated CFU was observed at a pH value as low as 2.5 (Fig. 1A). The majority of the cells (>90%) in these microcolonies stained preferentially with Syto9, suggesting good membrane integrity and likely viability (Fig. 2). At pH 3, growth of a subpopulation of 1 to 2% of the inoculum formed viable microcolonies of up to 100 cells over a 2- to 4-day period. The majority of cells that did not grow (>95%) stained preferentially with PI, suggesting a compromised membrane and probable loss of viability. This level of PI staining occurred a few hours after plating on PAO. Comparison of the microcolony area of the PI-stained cells grown at pH 3 with the average microcolony area (Syto9-PI staining) of the inoculum suggested there was no significant difference (a P value of <0.05 by Student's t test for >3,000 cells). Therefore, most of the cells inoculated at pH 3 failed to grow, and the preferential PI staining suggested they were dead or damaged. The microcolonies that did grow after 2 days were dominated by filamentous cells (Fig. 2). The arrangement of cells was also unusual for the survivors: not only were the cells often highly elongated but they were growing in entangled or aligned groups. In contrast, L. plantarum WCFS1 cultured on PAO on MRS medium (starting pH of 6.5) formed microcolonies of up to 100 cells (2 to 3 μm long) as a tightly packed monolayer and only then stacked up to form a second layer (data not shown). Growth of cells at a starting pH of 6.5 had a doubling time of 51 min on PAO on MRS. Imaging of cells inoculated at pH 3 suggested that, just considering the growing subpopulation, the doubling time was in excess of 3 h.
FIG. 1.
Effect of low pH on growth and survival of L. plantarum WCFS1. (A) Percentages of CFU showing growth after pH downshift from pH 5 to pH, determined after plating from liquid culture onto PAO on Gelrite plates at the indicated pH, are shown. (B) Kill curve after pH downshift in liquid culture from pH 5 to 3, assayed by a viable count onto pH 6.5 plates. Open squares, pH downshift for cells at high density; filled squares, pH downshift for cells diluted 100-fold; filled triangles, control cells resuspended to high density in pH 6.5 MRS.
FIG. 2.
Fluorescence microscopy images of L. plantarum WCFS1 grown at pH 3 for 2 days. (A) Overview. Scale bar, 100 μm. The growing microcolonies are composed mainly of Syto9 (blue/green)-stained filamented and unfilamented cells with a small number of PI-stained cells (red). More numerous unfilamented and PI-stained bacteria representing CFU composed of nonviable cells with a compromised membrane are also visible. The figure shows the highest plating density to emphasize the heterogeneity of survival; the isolation of cells by use of toothpicks was from lower-density samples. (B) Image similar to that in panel A but at higher magnification. The scale bar from panel A indicates 20 μm when applied to this panel.
There appeared to be a basal level of phenotypic heterogeneity within the wild-type population that permitted growth of some cells despite the rapid imposition of an acidic environment and its continued presence over several days. This conclusion was supported by an experiment in which cells from 10 microcolonies that had grown at pH 3 were picked with a sterile toothpick after 3 days, and hundreds of cells were recovered. These isolates were inoculated into pH 6.5 MRS liquid medium and cultured at 37°C under anaerobic conditions until an optical density at 600 nm of 0.5 was reached. Each liquid culture was then used to reinoculate fresh PAO strips, which were incubated on pH 3 MRS-Gelrite as described above. A fraction (0.9 to 3.3%) of these inoculations formed microcolonies after 3 days. This was essentially the same frequency of microcolony formation at pH 3 as seen for the parental population. Coupled with the high frequency of colony outgrowth (1 to 2%), these results suggest that stable pH-resistant mutants were not likely to be founding these microcolonies.
Cells from a late-log-phase culture of L. plantarum (optical density at 600 nm, >10; pH 4) were also plated and cultured on PAO under anaerobic conditions. As with the mid-log-phase inoculum, only a subpopulation of the CFU viable at pH 6.5 formed microcolonies at pH 3 to 3.5. However, after 1 day of culture on PAO, while the percentage growing as microcolonies was similar to the mid-log-phase inoculum, the average microcolony area was smaller (twofold). Additionally, the filamentation in the growing microcolonies of the late-log-phase culture was less extreme than that in the mid-log-phase culture (on average, two- to threefold shorter). After 2 and 3 days incubation, the mid-log-phase culture showed a threefold increase in the percentage of growing microcolonies compared to the value for the late-log-phase culture. This was apparently due to cells with a long lag phase now starting to divide.
Mid-log-phase cells, cultured under anaerobic conditions (pH 5.5), were resuspended in MRS medium at pH 4 (2 h) for exposure to an intermediate level of acid stress. These cells were then plated to PAO on pH 3 MRS-Gelrite. Pretreatment at pH 4 increased the relative percentage of microcolonies formed by two- to fivefold (up to 10%) suggesting an adaptive response. This effect was not seen when cells grown overnight in MRS medium (already at pH 4) were resuspended in fresh pH 4 MRS medium before being plated on PAO.
Inactivation of L. plantarum at low pH in liquid culture.
The decline in viability of L. plantarum was examined after a downshift from pH 5.0 to pH 3.0 in liquid culture by plating at pH 6.5 (Fig. 1B). Despite rapid transfer to pH 3 MRS, the decline in viability was relatively slow; indeed, for some days it was only marginally faster than a stationary-phase culture resuspended in fresh MRS medium. Irrespective of the concentration of cells, there was no shift in the pH of the growth medium and no detectable growth at pH 3. This suggests that survival in liquid culture was not facilitated by the cells making the bulk environment more alkaline. Cell density did not appear to affect survival; a 100-fold dilution of the culture in pH 3 MRS medium yielded a kill curve similar to that of the more concentrated sample (Fig. 1B).
Recovery of individual cells from acid stress.
The ability to visualize microcolonies growing at pH 3 was exploited to allow the selective isolation of these cells. Cell isolation was performed with a sterile toothpick, and low-power light microscopy was used to guide the tip to its intended target. The cell density used was 50-fold lower than the density used for the experiment whose results are shown in Fig. 2, to avoid cross-contamination. Three approaches were taken to analyze these cells before and during recovery: (i) SEM imaging of the cell surface, (ii) visualization of membranes by fluorescence microscopy using the lipophilic dye FM4-64, and (iii) monitoring of growth using transmission microscopy of cells cultured on MRS-Gelrite-covered microscope slides (described in the three sections below).
(i) SEM imaging of growth at pH 3 and recovery.
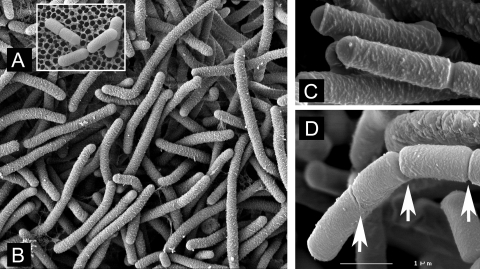
SEM imaging of microcolonies in situ reinforced the observations made by fluorescence microscopy. The elongated cells growing at pH 3 on PAO had no external trace of septation and had an unusually rough lateral surface with smoother domed surfaces at both poles (Fig. 3B and C). The rough surface resembled that caused by lactate stress; cells grown at pH 4.8 with lactate in continuous culture had a similar appearance (39). Cells growing a higher pH also tended to have smoother poles, but the contrast was less extreme, as the main body of the cell was smoother (data not shown).
FIG. 3.
SEM images of cells from microcolonies of acid-stressed L. plantarum WCFS1 grown on PAO and subsequently during recovery. (A) Cells grown at pH 6.5. (B) Filamented cells grown at pH 3 displaying a rougher cell surface without obvious septation. (C) Cells grown at pH 3 that were subsequently allowed to recover for 1 h at pH 6.5. (D) Cells grown at pH 3 that were subsequently allowerd to recover for 2 h at pH 6.5. Arrows show septation; here, the left-hand end of a filamented cell, where cell division is in progress and the cell surface is becoming smoother, is shown. The scale bar in panel D indicates 1 μm for panels C and D and 6 μm for A and B.
Recovery from pH stress was monitored over a 4-h period after transfer of cells from microcolonies and culture for 3 days at pH 3 on pH 6.5 MRS-Gelrite or agar. This was done by moving PAO strips to the higher-pH medium, further incubating them under anaerobic conditions, and then fixing them in situ and imaging them by SEM. After 1 to 2 h, new septation was observed; this was generally asymmetric in that only part of a filamented cell in recovery showed any sign of cell division and it was generally not at the midpoint. Septation was often abnormal, for example the broad band seen in Fig. 3C and the left-hand groove seen in Fig. 3D are typically not observed in unstressed cells grown at higher pH. After 2 h, the recovering cells started to lose the rougher cell surface that was typical of growth at pH 3.
(ii) Imaging of membranes during septation by staining with FM4-64.
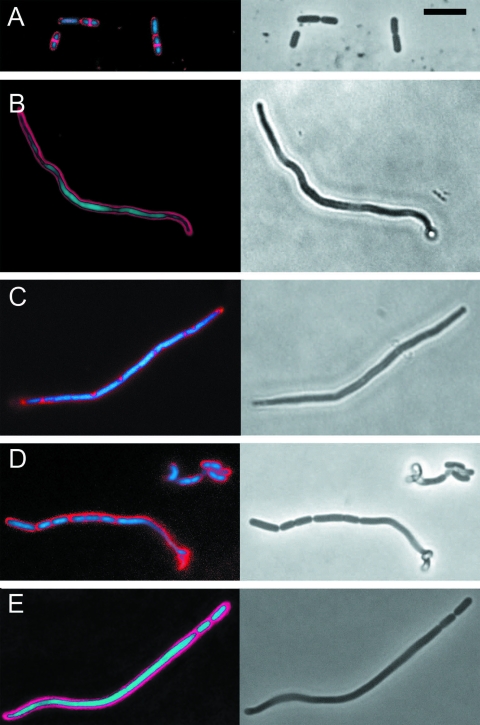
In order to visualize the cell membrane and reveal possible septation points, cells were stained with the lipophilic dye FM4-64 and imaged by fluorescence microscopy. Cells grown at pH 6.5 showed normal division and FM4-64 stained the cytoplasmic membrane, particularly in dividing cells (Fig. 4A). The staining characteristics were similar to those of other gram-positive bacteria, such as Bacillus subtilis, treated with this dye (40). Most filamentous cells (>85%) growing at pH 3 showed an absence of FM4-64 staining membrane across the cell (Fig. 4B). However, a subpopulation (<15%) gave a staining pattern that may indicate membrane inclusion bodies and/or abortive septation (Fig. 4C). In the latter group, bodies staining with FM4-64 were seen but usually failed to completely cross the cell, and hence cell division was not observed.
FIG. 4.
Staining filamented cells with FM4-64 (red) and Syto9 (blue/green) with imaging by fluorescence microscopy (left panels) and transmission microscopy (right panels). Scale bar (in panel A), 5 μm (all panels). (A) L. plantarum WCFS1 inoculated onto PAO and cultured on pH 6.5 Gelrite, then picked by a toothpick, stained, and imaged. Nascent septation points are seen in three of the five cells as bright FM4-64 staining across the dividing cells in regions where Syto9 staining was absent. (B) Typical filamented cell from culture on pH 3 after 2 days. FM4-64 staining was consistent with aseptate cells. (C) Example of a less abundant subpopulation (<20%) from 2 days growth at pH 3, stained with FM4-64. Staining was generally more irregular and punctuate than that for putative septa from cells grown at high pH. (D) Example of pH 3 cultured cells after 2.5-h recovery at pH 6.5. The filamentous cell (lower left) is dividing predominantly from the left-hand end; the irregularity at the right-hand end is a twisted region partially out of focus. (E) Further example of the cells shown in panel D; division primarily at the right-hand end.
We also studied membrane septation in cells that were recovered by isolating filamentous cells cultured on PAO strips on pH 3 MRS medium by means of a toothpick targeted by microscopy, followed by anaerobic incubation in pH 6.5 liquid MRS medium and FM4-64 staining. This analysis supported the conclusions reached by SEM. Only certain sections of filamentous cells during recovery showed a membrane-staining pattern that was consistent with septation (Fig. 4D and E). Again, it was rare (2%; 200 cells) to see an elongated cell bisected by membrane, and simultaneous nascent septation all along a filamentous cell (i.e., dividing into cells of normal length) was not observed.
(iii) Recovery from pH 3 by monitoring cell growth using light microscopy.
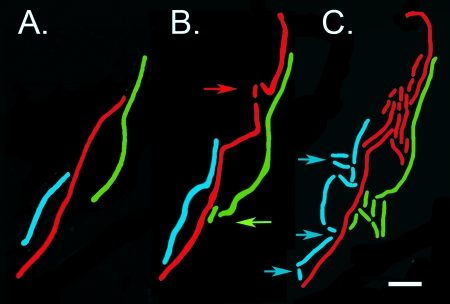
Recovery of L. plantarum from pH 3 was also assessed after cells were picked with a toothpick from microcolonies (after 3 days) and set onto microscope slides coated with a layer of MRS medium solidified with Gelrite or agar. These cells were cultured on a heated microscope stage at 37°C over a period of several hours, with periodic imaging by transmission microscopy. Incubation of 200 recovered cells for periods up to 4 h indicated that 96% of the cells recovered were viable. For a more detailed analysis, stacks of images were created from a time series taken over a few hours, and the fates of individual cells and their progeny were followed through several rounds of division. Qualitatively, these observations supported the results obtained by FM4-64 staining and SEM in terms of the way cells divided upon incubation at pH 6.5, i.e., recovery of filamented cells was generally by outgrowth of a specific portion of the cell and not by symmetrical division (Fig. 5). The difference in the outgrowth potential at different points along a filament was large; for example, for the cell shaded red in Fig. 5, the first progeny cell of normal length divided several times ahead of any other outgrowth. However, the flanking regions of the cell immediately adjacent to those that were growing rapidly were not dead or completely inactive. Measurements of cell elongation from time-lapse pictures suggested that these regions were also elongating but at least fivefold more slowly. These observations suggest that there was intracellular heterogeneity in recovery from acid stress, that regions within a filament have an advantage in forming new cells when the pH of their environment shifts from pH 3 to 6.5. Consistent with this view, a rapid return to normal-length cells (1 to 3 μm) evenly along a filamented cell was never seen (for 200 cells).
FIG. 5.
Example of the elongation and division of filamentous cells imaged by transmission light microscopy during recovery from pH 3. Individual cells and their corresponding progeny are shaded (red, green, and blue). Arrows indicates where division first occurred for each cell. Scale bar, 5 μm. Times at which the images were captured are 0 min (A), 100 min (B), and 200 min (C).
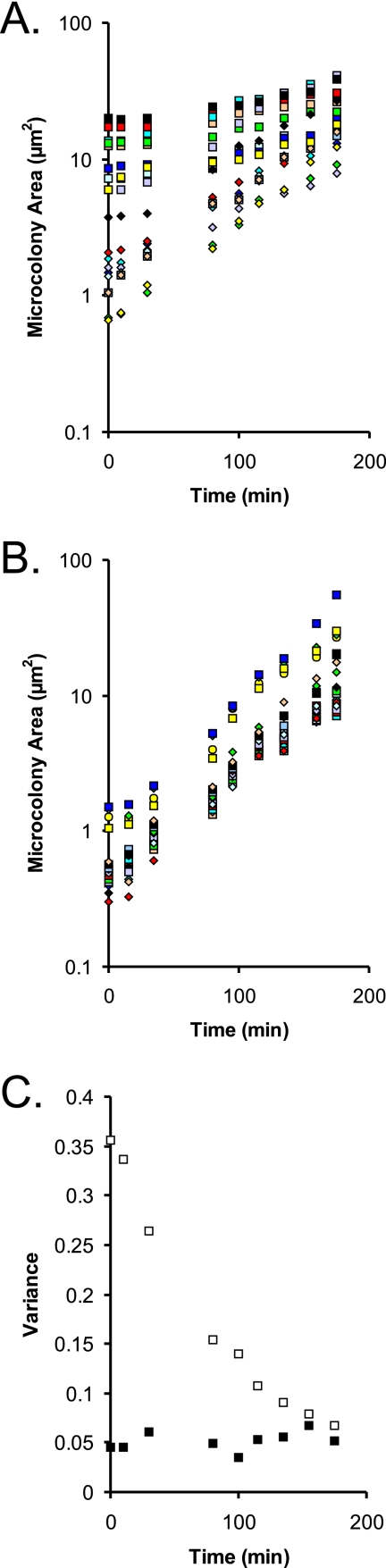
Outgrowth was quantified by measuring and summing the areas of cells making up microcolonies and calculating growth rates as has been previously described for B. cereus grown on PAO (10). There was a marked tendency for the longer cells to grow more slowly during the period immediately following recovery (coefficient of determination r2 = 0.84; n = 20). Smaller cells (1 to 3 μm long, typical of exponential growth of L. plantarum under anaerobic conditions in MRS broth) doubled their area more rapidly than highly filamented cells because, as described above, only a specific region of a filamented cell recovered as quickly. There was also a negative relationship between cell length and the time until the first division (lag time), but the correlation was less strong than the correlation of cell length and doubling time (r2 = 0.34; n = 20). For the growth of single cells into microcolonies, when we plotted log10 of microcolony area against time, the recovery of the population was seen as a tendency for the convergence of the microcolony areas (Fig. 6A). When the variance in microcolony area was plotted from this data set, it was clear that the heterogeneity of the growing microcolonies was decreasing with recovery time (Fig. 6C). These trends compare with the growth of a population that was not in recovery, in which the variance was not changing significantly (Fig. 6B). Effectively, the population became more homogeneous when returning to normal, unstressed exponential growth. Filamented and unfilamented cells appeared to have convergent growth paths during recovery, ultimately tending toward similar-size colonies composed of unfilamented cells.
FIG. 6.
Quantification of growth and recovery of L. plantarum WCFS1 cultured on MRS-Gelrite on warmed microscope slides, tracked during growth by transmission microscopy, and quantified by calculation of microcolony areas. (A) Individual growth curves during recovery from low pH. The outgrowth of 20 CFU is shown. These cells were grown for 2 days on PAO at pH 3 and then moved by use of a toothpick onto MRS-Gelrite at pH 6.5 and imaged during growth. (B) Results of a control experiment with 20 CFU of L. plantarum transferred after 2 days growth on PAO (at a starting pH of 6.5) to MRS-Gelrite at pH 6.5. (C) Variance within microcolony areas of each of the two populations plotted against recovery time, calculated from log10 microcolony areas. Open squares represent data for cells recovering from pH 3 (data calculated as described for panel A), and filled squares represent data for control cells (data from the results described for panel B).
DISCUSSION
In this study, we evaluated population heterogeneity in L. plantarum in response to growth at low pH and recovery after exposure to acidic conditions. Our results show that within a genetically homogeneous population of L. plantarum WCFS1, there is an acid-resistant subpopulation that both survives and grows despite a rapid shift from pH 5 to pH 3 and subsequent maintenance in this hostile environment. This subpopulation comprised 1 to 2% of a mid-log-phase culture and could be cultured at pH 3 on a strip of porous ceramic (PAO) placed on semisolid MRS medium. At low pH, Gelrite proved advantageous in supporting the PAO and supplied it with nutrients, being more stable at low pH values than agar. The PAO was also acid resistant and permitted staining and manipulation of the bacteria cultured on the surface with minimal disturbance so that microcolonies could be imaged, with rapid exchange of substrates, nutrients, or reagents with the bulk phase (21). The occurrence of a resistant and growing subpopulation is not due to mutation, as was shown by isolation of the growing microcolonies; after reculture of these isolates, populations had wild-type acid resistance. Microcolonies growing at pH 3 were found to be dominated by highly elongated cells able to exclude PI and therefore with membranes of good integrity. SEM imaging of the cell surface and FM4-64 staining of membranes suggested that the majority of such cells (>80%) were aseptate. The possibility of more subtle localization of future septation points cannot be excluded (38, 40), but there were clearly no dividing membranes and in most cases no trace of constriction on the cell surface.
The acid-resistant subpopulation was taken as the starting point for a second set of experiments that focused on recovery from acid stress. Many survivors of low-pH stress became elongated during sustained growth at an acidic pH. After return of the microorganism to a less acidic growth environment, detectable septation and the creation of new cells developed preferentially at specific points along a filamented cell. Division never occurred simultaneously all along such an elongated cell. The early septation events appeared localized and were usually asymmetric (i.e., not bisecting the cell). While there was differential growth within a filamented cell, the regions that were not rapidly dividing were not completely inactive, as limited elongation was still observed over a period of a few hours in these less productive regions.
Several authors have argued that filamented cells are a confounding factor in viable counting within the food industry (19, 32). A serious error in viable counting may occur if a filamented cell was already multiply septate and could therefore form a large number of viable cells with a single simultaneous round of division. However, this is clearly not the only model for recovery, and we have shown here that the increased biomass of a filament is not an advantage in the return to relatively unstressed growth. If there is survival value of highly elongated cells in this situation, this suggests it is more likely to be in viability or growth under stress.
For some species, asymmetric division is the norm. For many rod-shaped bacteria (e.g., Escherichia coli or B. subtilis), vegetative cell division normally precisely bisects the cell, and this is also observed with L. plantarum under favorable growth conditions. Under stress, division can shift from a symmetric to an asymmetric mode, e.g., in an extreme case for B. subtilis sporulation (40), and return to symmetric division upon spore outgrowth. Relatively little is known about septation in L. plantarum. However, a knockout mutation of the alanine racemase gene in this organism results in an unusual lytic phenotype compared to B. subtilis (38). Taken with the observation that the regular and symmetric recovery from filamentation is not found, unlike the model proposed for Listeria (19), this suggests there may be interesting features of cell division yet to be discovered in L. plantarum that contrast with other gram-positive rods.
There are a number of possible explanations for differential growth within a single filamentous cell, including the following. (i) The least damaged region of a filamented cell may be dividing. Alternatively, if it contains multiple genomes, growth may occur in the region containing the most viable genome or other cellular components required for survival. (ii) The rest of the filamented cell may be “sacrificed” supporting the growth of the dividing cell. (iii) A single filamented cell may apply multiple survival strategies, leading to a high fitness in a fluctuating environment. Cellular regions growing slowly may be more stress resistant, providing a survival advantage if the stress returns. Other parts growing more rapidly may be able to exploit the new environment that is favorable for growth. Whatever the case, when L. plantarum returns to rapid, unstressed growth, a decrease in the heterogeneity is observed. This may be explained by the fact that in a homogeneous environment that is favorable for growth, there may only one good cell shape for optimal fitness, whereas under stressful conditions, a high level of phenotypic heterogeneity yields the highest benefits in terms of fitness (2, 6).
The mechanism(s) allowing the survival and growth of a small fraction of the population during acid stress is not clear. However, it is known that individual bacteria do vary in their membrane potential and internal pH. It may be that this heterogeneity contributes to some individuals surviving and growing at pH 3 (42). Given recent improvement in tracking and imaging subpopulations in flow cells and other situations (4), it should be possible to address these questions experimentally, potentially integrating measurements of intracellular pH. In E. coli, it is often variation in global regulators that mediates variation in the phenotypic heterogeneity of stress resistance (27). For example, the PhoU global negative regulator in E. coli mediates both a persister response to antibiotics and other stresses, including low pH (29). L. plantarum WCFS1 has two putative PhoU homologues encoded by its genome (17, 24; data not shown). Additionally, it has been shown recently that, in L. plantarum strain BAA-793, the principal sigma factor (RpoD) can mediate the heterogeneity of pH stress (25). This study used (among other conditions) HCl-adjusted MRS medium at pH 3.85 and visible colony size to determine the variation in growth. Our work used a lower pH and smaller microcolony size than this work as the readout. In the study by Klein-Marcushaumer and Stephanopoulous (25), mutations within the rpoD gene were shown to alter the phenotypic variation. Increasing this form of population diversity proved a good point for strain improvement. We suggest that microcolony assays may have a similar potential but are possible under more extreme stresses under which visible colonies rarely form.
In summary, multiple levels of heterogeneity exist within pH-stressed cultures of L. plantarum. In terms of growth at low pH, there is clearly phenotypic heterogeneity resulting in the appearance of subpopulations of cells able to grow in acidic conditions. In recovery, there is intracellular heterogeneity, at least within the more elongated cells. Filamentous cells recover through asymmetric cell division and are not at an advantage in outgrowth compared to shorter cells.
Acknowledgments
This work was funded by Top Institute Food and Nutrition project C-015.
We thank Michiel Kleerebezem, Maria Marco, and Marcel Zwietering for constructive comments on the manuscript.
Footnotes
Published ahead of print on 24 October 2008.
REFERENCES
- 1.Aires, K. A., A. M. Cianciarullo, S. M. Carneiro, L. L. Villa, E. Boccardo, G. Perez-Martinez, I. Perez-Arellano, M. L. S. Oliveira, and P. L. Ho. 2006. Production of human papillomavirus type 16 L1 virus-like particles by recombinant Lactobacillus casei cells. Appl. Environ. Microbiol. 72:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, S. V. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4:577-587. [DOI] [PubMed] [Google Scholar]
- 3.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balaban, N. Q., J. Merrin, J. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1628. [DOI] [PubMed] [Google Scholar]
- 5.Bigger, W. B. 1944. Treatment of staphylococcal infections with penicillin. Lancet 244:497-500. [Google Scholar]
- 6.Booth, I. R. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19-30. [DOI] [PubMed] [Google Scholar]
- 7.Browne, N., and B. C. A. Dowds. 2002. Acid stress in the food pathogen Bacillus cereus. J. Appl. Microbiol. 92:404-414. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: response of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Angelis, M., and M. Gobbetti. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106-122. [DOI] [PubMed] [Google Scholar]
- 10.den Besten, H. M. W., C. J. Ingham, J. E. T. van Hylckama Vlieg, M. Beerthuyzen, M. H. Zwietering, and T. Abee. 2007. Quantitative analysis of population heterogeneity of the adaptive salt stress response and outgrowth of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 73:4797-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhar, N., and J. D. McKinney. 2007. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 10:30-38. [DOI] [PubMed] [Google Scholar]
- 12.Elfwing, A., Y. LeMarc, J. Baranyi, and A. Ballagi. 2004. Observing growth and division of large numbers of individual bacteria by image analysis. Appl. Environ. Microbiol. 70:675-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francois, K., F. Devlieghere, K. Smet, A. R. Standaert, A. H. Geeraerd, J. F. Van Impe, and J. Debevere. 2005. Modelling the individual cell lag phase: effect of temperature and pH on the individual cell lag distribution of Listeria monocytogenes. Int. J. Food Microbiol. 100:41-53. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. SOS responses and DNA damage tolerance in prokaryotes, p. 407-464. In E. C. Friedberg, G. C. Walker, and W. Siede (ed.), DNA repair and mutagenesis. ASM Press, Washington, DC.
- 15.Fuentes, S., M. Egert, M. Jimenez-Valera, M. Monteoliva-Sanchez, A. Ruiz-Bravo, and H. Smidt. 2008. A strain of Lactobacillus plantarum affects segmented filamentous bacteria in the intestine of immunosuppressed mice. FEMS Microbiol. Ecol. 63:65-72. [DOI] [PubMed] [Google Scholar]
- 16.Guillier, L., P. Pardon, and J. C. Augustin. 2005. Influence of stress on individual lag time distributions of Listeria monocytogenes. Appl. Environ. Microbiol. 71:2940-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, S., K. Lewis, and M. Vulic. 2008. The role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718-27226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haugland, P. 1999. Handbook of fluorescent probes and research chemicals, 7th ed. Molecular Probes, Eugene, OR.
- 19.Hazeleger, W. C., M. Dalvoorde, and R. R. Beumer. 2006. Fluorescence microscopy of NaCl-stressed, elongated Salmonella and Listeria cells reveals the presence of septa in filaments. Int. J. Food Microbiol. 112:288-290. [DOI] [PubMed] [Google Scholar]
- 20.Ingham, C. J., M. van den Ende, P. C. Wever, and P. M. Schneeberger. 2006. Rapid antibiotic sensitivity testing and trimethoprim-mediated filamentation of clinical isolates of the Enterobacteriaceae assayed on a novel porous culture support. J. Med. Microbiol. 55:1511-1519. [DOI] [PubMed] [Google Scholar]
- 21.Ingham, C. J., A. Sprenkels, J. Bomer, D. Molenaar, A. van den Berg, J. E. T. van Hylckama Vlieg, and W. M. de Vos. 2007. The micro-Petri dish: a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. USA 46:18217-18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansson, P. E., B. Lindberg, and P. A. Sandford. 1983. Structural studies of gellan gum an extracellular polysaccharide elaborated by Pseudomonas elodea. Carbohydr. Res. 124:135-139. [Google Scholar]
- 23.Kelly, C. D., and O. Rahn. 1932. The growth rate of individual bacterial cells. J. Bacteriol. 23:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleerebezem, M. J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein-Marcuschamer, D., and G. Stephanopoulos. 2008. Assessing the potential of mutational strategies to elicit new phentypes in industrial strains. Proc. Natl. Acad. Sci. USA 105:2319-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutalik, Z., M. Razaz, A. Elfwing, A. Ballagi, and J. Baranyi. 2005. Stochastic modelling of individual cell growth using flow chamber microscopy images. Int. J. Food Microbiol. 105:177-190. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K., H. G., K. Pi, and Y. J. Choi. 2008. The effect of low pH on protein expression by the probiotic bacterium Lactobacillus reuiteri. Proteomics 8:1624-1630. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., J. A. Odumeru, M. Griffiths, and R. C. McKellar. 2006. Effect of environmental stresses on the mean and distribution of individual cell lag times of Escherichia coli O157:H7. Int. J. Food Microbiol. 110:278-285. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., and Y. Zhang. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, C. C., and L. R. Casida. 1984. Gelrite as a gelling agent in media for the growth of thermophilic microorganisms. Appl. Environ. Microbiol. 47:427-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marco, M. L., R. S. Bongers, W. M. de Vos, and M. Kleerebezem. 2007. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattick, K. M., F. Jørgensen, J. D. Legan, M. B. Cole, J. Porter, H. M. Lappin-Scott, and T. J. Humphrey. 2000. Survival and filamentation of Salmonella enterica serovar Enteritidis PT4 and Salmonella enterica serovar Typhimurium DT104 at low water activity. Appl. Environ. Microbiol. 66:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald, L. C., H. P. Fleming, and H. M. Hassan. 1990. Acid tolerance of Leuconostoc mesenteroides and Lactobacillus plantarum. Appl. Environ. Microbiol. 56:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKellar, R. C., and A. Hawke. 2006. Assessment of distributions for fitting lag times of individual cells in bacterial populations. Int. J. Food Microbiol. 106:169-175. [DOI] [PubMed] [Google Scholar]
- 35.Mellefont, L. A., T. A. McMeekin, and T. Ross. 2005. Viable count estimates of lag time responses for Salmonella typhimurium M48 subjected to abrupt osmotic shifts. Int. J. Food Microbiol. 105:399-410. [DOI] [PubMed] [Google Scholar]
- 36.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. The cell cycle, p. 389-417. In F. C. Neidhardt, J. L. Ingraham, and M. Schaechter (ed.), Physiology of the bacterial cell. M. Sinauer Associates, Sunderland, MA.
- 37.Niven, G. W., T. Fuks, J. S. Morton, S. A. C. G. Rua, and B. M. Mackey. 2006. A novel method for measuring lag times in division of individual bacterial cells using image analysis. J. Microbiol. Methods 65:311-317. [DOI] [PubMed] [Google Scholar]
- 38.Palumbo, E., C. F. Favier, M. Deghorain, P. S. Cocconcelli, C. Grangette, A. Mercenier, E. E. Vaughan, and P. Hols. 2004. Knockout of the alanine racemase gene in Lactobacillus plantarum results in septation defects and cell wall perforation. FEMS Microbiol. Lett. 233:131-138. [DOI] [PubMed] [Google Scholar]
- 39.Pieterse, B., R. J. Leer, F. H. J. Schuren, and M. T. J. van der Werf. 2005. Unravelling the multiple effects of lactic acid stress on Lactobacillus plantarum by transcription profiling. Microbiology 151:3881-3894. [DOI] [PubMed] [Google Scholar]
- 40.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y.-L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramiah, K., C. A. van Reenen, and L. M. T. Dicks. 2008. Surface-bound proteins of Lactobacillus plantarum 423 that contribute to adhesion of Caco-2 cells and their role in competitive exclusion and displacement of Clostridium sporogenes and Enterococcus faecalis. Res. Microbiol. 159:470-475. [DOI] [PubMed] [Google Scholar]
- 42.Shabala, L., T. McMeekin, B. B. Bjørn Budde, and H. Siegumfeldt. 2006. Listeria innocua and Lactobacillus delbrueckii subsp. bularicus employ different strategies to cope with acid stress. Int. J. Food Microbiol. 110:1-7. [DOI] [PubMed] [Google Scholar]
- 43.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie van Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]