Abstract
Many marine sponges, hereafter termed high-microbial-abundance (HMA) sponges, harbor large and complex microbial consortia, including bacteria and archaea, within their mesohyl matrices. To investigate vertical microbial transmission as a strategy to maintain these complex associations, an extensive phylogenetic analysis was carried out with the 16S rRNA gene sequences of reproductive (n = 136) and adult (n = 88) material from five different Caribbean species, as well as all published 16S rRNA gene sequences from sponge offspring (n = 116). The overall microbial diversity, including members of at least 13 bacterial phyla and one archaeal phylum, in sponge reproductive stages is high. In total, 28 vertical-transmission clusters, defined as clusters of phylotypes that are found both in adult sponges and their offspring, were identified. They are distributed among at least 10 bacterial phyla and one archaeal phylum, demonstrating that the complex adult microbial community is collectively transmitted through reproductive stages. Indications of host-species specificity and cospeciation were not observed. Mechanistic insights were provided using a combined electron microscopy and fluorescence in situ hybridization analysis, and an indirect mechanism of vertical transmission via nurse cells is proposed for the oviparous sponge Ectyoplasia ferox. Based on these phylogenetic and mechanistic results, we suggest the following symbiont transmission model: entire microbial consortia are vertically transmitted in sponges. While vertical transmission is clearly present, additional environmental transfer between adult individuals of the same and even different species might obscure possible signals of cospeciation. We propose that associations of HMA sponges with highly sponge-specific microbial communities are maintained by this combination of vertical and horizontal symbiont transmission.
Sponges (phylum Porifera) are evolutionarily ancient Metazoa whose origin dates back about 600 million years to the Precambrian (26). With 7,000 formally described species and an estimated 15,000 extant species, sponges are among the most diverse marine invertebrate groups and are important components of all aquatic habitats, including freshwater environments, tropical reefs, and even the deep sea (19). Despite an enormous range of shapes, colors, and sizes, all sponges possess a relatively simple body plan which is adapted to a filter-feeding lifestyle (4). Large volumes of seawater are pumped through canals embedded in an extracellular matrix (55), termed the mesohyl, and microorganisms and small unicellular eukaryotes are taken up from the seawater with high efficiency, leaving the expelled water essentially sterile (37, 54).
Numerous sponges live in permanent and close associations with microorganisms, and many of them host phylogenetically diverse populations of microbes (15, 17, 49). These microorganisms are located mainly extracellularly in the sponge mesohyl in high concentrations, contributing up to 40% of the sponge's biomass and exceeding microbial seawater concentrations by 2 to 4 orders of magnitude (13, 53). Such host species are therefore termed bacteriosponges or high-microbial-abundance (HMA) sponges, in contrast to low-microbial-abundance (LMA) sponges that lack large and complex microbial consortia within their mesohyl (15, 38). The total phylogenetic microbial diversity of HMA sponges is extremely high and includes representatives of up to 16 bacterial phyla and both major archaeal lineages (49). Despite this microbial complexity, there exists a high degree of uniformity among the microbial communities from taxonomically and geographically disparate HMA sponges. Many microbial phylotypes were repeatedly detected in different sponges but not in seawater. The respective microbes were therefore defined as sponge specific (16). Collectively, these microbes possess diverse metabolic pathways (e.g., for photosynthesis, sulfate reduction, and nitrification) and their involvement in sponge metabolism suggests a beneficial interaction (3, 18, 56). Additionally, those associations were shown to be permanent and stable, with low temporal and spatial variability in the microbial communities (17).
To maintain stable and specific symbioses, the presence of symbionts in each generation must be assured. In the symbioses of many insects, including carpenter ants, tsetse flies, and aphids, as well as of certain marine clams, the microbial symbionts are strictly vertically transmitted through reproductive stages, resulting in cospeciation (20, 31, 36). Similarly, horizontal symbiont transmission between single adult individuals or environmental acquisition can also be highly specific and maintain obligate symbioses, sometimes also resulting in parallel evolution. Examples are hydrothermal vent tubeworms, which rely on their chemoautotrophic symbionts (34), and the sepiolid squid-vibrio symbiosis (33).
The first evidence for vertical transmission in sponges came from electron microscopy studies (references 6 and 9 and references cited therein; 25, 44, 52). To date, microbes have been detected in oocytes, embryos, and larvae of more than 20 oviparous sponges (which release gametes) and viviparous sponges (which release fully developed larvae) (9, 42). Recently, a culture-based study successfully isolated the same alphaproteobacterial strain from adult and embryo samples of the Caribbean sponge Mycale laxissima, and vertical transmission of this bacterium was suggested by the authors (8). Additional evidence for vertical transmission comes from molecular studies that allow the phylogenetic identification of microbes within sponge reproductive stages and comparison with the microbial community of the adult. Using Cyanobacteria-specific primers, Oren et al. (35) found a Synechococcus-related sequence type in Diacarnus erythraenus adult sponges and their embryos. However, so far only three molecular studies investigated the total microbial diversity in sponge offspring. Complex bacterial consortia were described for each of the species M. laxissima, Ircinia felix, and Corticium sp. (8, 45, 46, respectively). Sharp et al. (46) also described archaea within Corticium sp. embryos and verified the presence of microbes by fluorescence in situ hybridization (FISH). Additionally, a complex microbial community was described for the first time in I. felix juvenile sponges which were obtained after larval settlement experiments (45).
To provide insights into the phylogenetic diversity of bacteria and archaea in sponge reproductive stages, a comprehensive 16S rRNA gene survey was performed with >250 sequences obtained from eight sponges representing six orders, two geographic regions, and both oviparous and viviparous modes of reproduction. Furthermore, the mechanistic aspects of vertical transmission were investigated in the oviparous Caribbean sponge Ectyoplasia ferox using a combination of electron and fluorescence microscopy. E. ferox was chosen because its early reproductive stages are obtainable, allowing mechanistic investigations from an early developmental stage onward. Based on the phylogenetic and mechanistic results, we propose a symbiont transmission model for HMA sponge symbioses, including a combination of vertical and horizontal symbiont transmission to maintain the association with sponge-specific, though not species-specific, microbial consortia.
MATERIALS AND METHODS
Sponge collection.
The sponges Agelas conifera, Agelas wiedenmayeri, Ectyoplasia ferox, Smenospongia aurea, and Xestospongia muta were collected by scuba diving off the coast of Key Largo, Florida, in June 2002, June and August 2004, and May 2005 (Table 1). Additional material from E. ferox was collected by scuba diving offshore of French Wells, Bahamas, in July 2003. The samples were taken at depths of 5 to 30 m, transferred in ziplock bags containing seawater to the surface, and kept on ice in a cooler until further processing (within 4 to 5 h). Offspring samples (oocytes, embryos, and larvae) were collected in larval traps using the methodology of Lindquist et al. (27). Additionally, tissue samples of the corresponding adult individuals were taken. After the adult samples were cut into small pieces, the adult and offspring materials were washed in sterile filtered seawater, fixed either in 96% ethanol or in 2.5% glutaraldehyde-double-distilled water (ddH2O), and stored at either −20 or 4°C. The field settlement experiments were performed with a portion of the captured S. aurea larvae as described by Schmitt et al. (45). Briefly, larvae were enclosed in plastic containers (∼75 ml in volume), which were placed at a 9-m-deep reef site. These containers consisted of a plastic frame supporting a covering of 200-μm nylon mesh to allow water exchange. Juveniles were recovered 1 to 3 days postsettlement and preserved as described above.
TABLE 1.
Compilation of all available 16S rRNA gene sequences from sponge reproductive stages until December 2007
| Host sponge (order)a | Collection site(s) (latitude/longitude) | Mode of reproduction | Sequence source(s)c | No. of offspring sequences | Reference |
|---|---|---|---|---|---|
| HMA sponges | |||||
| Agelas conifera (Agelasida) | Key Largo, FL (25°02′N/80°24′W) | Oviparous | 16S rRNA libraryd | 5 | This study |
| Agelas wiedenmayeri (Agelasida) | Key Largo, FL (25°02′N/80°24′W) | Oviparous | DGGE | 7 | This study |
| Corticium sp. (Homosclerophorida) | Palau islands (07°23′N/134°38′E) | Viviparous | 16S rRNA library | 18 | 46 |
| Ectyoplasia ferox (Poecilosclerida) | Key Largo, FL (25°02′N/80°24′W) | Oviparous | DGGE | 28 | This study |
| Bahamas islands (24°32′N/75°55′W) | |||||
| Ircinia felix (Dictyoceratida) | Key Largo, FL (25°02′N/80°24′W) | Viviparous | DGGE | 71 | 45 |
| 16S rRNA libraryd | 1 | This study | |||
| Smenospongia aurea (Verongidab) | Key Largo, FL (25°02′N/80°24′W) | Viviparous | DGGE | 19 | This study |
| Xestospongia muta (Haplosclerida) | Key Largo, FL (25°02′N/80°24′W) | Oviparous | 16S rRNA library | 76 | This study |
| LMA sponges | |||||
| Mycale laxissima (Poecilosclerida) | Key Largo, FL (25°02′N/80°24′W) | Viviparous | Isolates, 16S rRNA library | 27 | 8 |
All sponges belong to the class Demospongiae (classification after Systema Porifera by Hooper and van Soest [19]).
Reclassified after the paper by Schmitt et al. (43).
Sequences were obtained using universal 16S rRNA primers either by the construction of 16S rRNA libraries or by the excision of DGGE bands.
The 16S rRNA library was constructed using group-specific primers.
Extraction of DNA and 16S rRNA gene library construction.
Adult samples that had been fixed in 96% ethanol were air dried, ground with a mortar and pestle in liquid nitrogen, and DNA extracted using the Fast DNA Spin kit for soil (Q-Biogene, Heidelberg, Germany) in accordance with the manufacturer's instructions. Reproductive material containing oocytes and embryos was also air dried and transferred into 150 μl ddH2O, and DNA was extracted by heating the samples in a water bath for 10 min at 100°C. This same methodology was used to obtain DNA from collections of larvae and juveniles (three, five, or seven individuals pooled immediately after collection). The DNA solutions were then used directly as templates (0.01 to 1.0 μg) in the following PCR.
The universal primers 27f and 1492r (23) were used for the PCR amplification of bacterial 16S rRNA genes. The primer pairs POR389f/POR1130r (12), 11f/Ntspa865r (47), and Arch21f/Arch958r (7) were used for the specific PCR amplification of the candidate phylum “Poribacteria,” Nitrospira, and Archaea 16S rRNA genes, respectively. Negative controls (PCRs without DNA templates) were included for each 16S rRNA gene amplification. Cycling conditions on a Mastercycler gradient (Eppendorf, Hamburg, Germany) were as follows: an initial denaturing step at 96°C for 5 min; 30 cycles of denaturing at 96°C for 1 min; primer annealing at 54, 64, 60, and 56°C for 1 min; and elongation at 72°C for 90, 50, 90, and 50 s for universal, Poribacteria, Nitrospira, and Archaea PCRs, respectively. The PCR program ended with a final extension step at 72°C for 5 min.
DGGE.
DNA was extracted as described above. The universal 16S rRNA gene primers 341f with GC-clamp and 907r (32) were used for the PCR amplification of bacterial 16S rRNA genes. The cycling conditions were as follows: initial denaturing step at 95°C for 5 min, 30 cycles of denaturing at 95°C for 1 min, primer annealing at 54°C for 1 min and elongation at 72°C for 45 s, followed by a final extension step at 72°C for 10 min. Denaturing gradient gel electrophoresis (DGGE) was performed with a Bio-Rad DCode universal mutation detection system (Bio-Rad, München, Germany) on a 10% (wt/vol) polyacrylamide gel in 1× Tris-acetate-EDTA using a 0 to 90% denaturing gradient; 100% denaturant corresponded to 7 M urea and 40% (vol/vol) formamide. Electrophoresis was performed for 6 h at 150 V and 60°C. The gels were stained for 30 min in Sybr gold (Molecular Probes) and scanned on a Typhoon 8600 scanner (Amersham Biosciences). Selected bands were excised with an ethanol-sterilized scalpel and incubated in 25 μl ddH2O overnight at 4°C. A total of 4 μl of eluted DNA was subsequently used for reamplification with primers 341f and 907r using the PCR conditions described above.
Cloning, sequencing, and phylogenetic analysis.
The PCR products from the excised DGGE bands and for 16S rRNA gene libraries were ligated into the pGEM-T Easy vector (Promega) and transformed by electroporation into competent Escherichia coli XL 1-Blue cells. Plasmid DNA was isolated by standard miniprep procedures (40), and the correct insert size was verified using agarose gel electrophoresis following restriction digestion. The clones from the 16S rRNA gene libraries were characterized by single digestions with the restriction endonucleases HaeIII and Sau3AI (restriction fragment length polymorphism analysis). The clones with identical restriction patterns were grouped together, and one random clone from each group was chosen for sequence analysis. Sequencing was performed on an ABI 377XL automated sequencer (Applied Biosystems), and the sequences were edited with the ContigExpress tool in the Vector NTI suite 6.0 (InforMax, Inc.). The sequences were checked for chimeras with the program Pintail (2) and for other amplification and sequencing artifacts. Following the removal of chimeras from the data set, the percentage similarities (p-distances) between the sequences from the same sponge developmental stage (adult, oocyte, embryo, larva, and juvenile) within each species were determined with the editor Align, and those with identities above 99% were grouped together into operational taxonomic units. Only one randomly chosen sequence per operational taxonomic unit was used for further analysis. The sequences obtained in this study, all the available sequences from sponge reproductive material in GenBank (http://www.ncbi.nlm.nih.gov), and the appropriate reference sequences (all nearest BLAST matches including sequences from adult sponges and, moreover, representatives of the respective phylum) were aligned automatically with ClustalX (51), and the alignment was subsequently corrected manually in Align. Phylogenetic trees were constructed with the ARB software package (28). Initially, neighbor-joining (Jukes-Cantor correction) and maximum parsimony trees were calculated with nearly full-length sequences (>1,250 bp) and 100 pseudoreplicates. Subsequently, partial sequences were added to the trees without changing the topology by the use of the parsimony-interactive method in ARB.
TEM.
Adult samples that had been fixed in 2.5% glutaraldehyde-ddH2O were cut into small cubes of about 1 mm3. All samples (adults, material containing oocytes and embryos, larvae, and juveniles) were then washed five times in cacodylate buffer (50 mM, pH 7.2), fixed in 2% osmium tetroxide for 90 min, washed again five times in ddH2O, and incubated overnight in 0.5% uranyl acetate. After being dehydrated in an ethanol series (30, 50, 70, 90, 96, and 3 times at 100% for 30 min each), the samples were incubated three times for 30 min in 1× propylene oxide, maintained overnight in 1:1 (vol/vol) propylene oxide-Epon 812 (Serva), incubated twice for 2 h in Epon 812, and finally embedded in Epon 812 for 48 h at 60°C. The samples were then sectioned with an ultramicrotome (OM U3; C. Reichert, Austria) and examined by transmission electron microscopy (TEM) (Zeiss EM 10; Zeiss, Germany).
FISH.
Small pieces (ca. 5 mm3) of the ethanol-preserved E. ferox offspring material (oocytes/embryos embedded in a mucous sheath) were incubated in 50 and 30% ethanol for 3 min each, transferred into 1× phosphate-buffered saline for 3 min, and frozen in Tissue-Tek cryo-embedding medium (Miles Laboratories, Inc., Naperville, IL) at −80°C for at least 2 days. Frozen E. ferox offspring material was directly placed in cryo-embedding medium and kept at −80°C. For hybridization, pieces were sectioned with a cryomicrotome (Mikrom, Walldorf, Germany), placed on microscope slides, and air dried. After fixation in 4% paraformaldehyde (final concentration) overnight at 4°C in a humid chamber, the samples were washed twice in 1× phosphate-buffered saline for 5 min each, heat fixed at 42°C, dehydrated in an ethanol series (50, 75, and 100% for 5 min each), and air dried. FISH was performed in an isotonically equilibrated humid chamber at 46°C in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 8.0], 0.01% sodium dodecyl sulfate) and probe-dependent amounts of formamide (25 to 50%) for 4 h. The Cy3-labeled probe mix EUB I-III (5) was used at concentrations of 2 to 7.5 ng/μl, whereas the fluorescein isothiocyanate-labeled EUK516 probes (1) were used at concentrations of 15 to 50 ng/μl. Additionally, 5 μl of a DAPI stock solution (100 μg/ml) was added to each reaction 30 min prior to stopping the hybridization. The hybridization buffer was gently rinsed off, and the slides were incubated at 48°C in washing buffer (0.7 M NaCl, 20 mM Tris-HCl [pH 8.0], 50 mM EDTA, 0.01% sodium dodecyl sulfate) for 30 min. The washing buffer was rinsed off with ice-cold dH2O, and the slides were air dried in the dark and mounted in Citifluor (Citifluor Ltd., London, United Kingdom). Visualization was achieved with a confocal laser scanning microscope (LSM 510; Zeiss MicroImaging).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences were deposited in EMBL/GenBank/DDBJ under accession numbers EF159732 to EF159941 and EU273899 to EU273914.
RESULTS
Phylogenetic sequence analysis.
In total, 218 bacterial 16S rRNA gene sequences were obtained in this study: 6 from A. conifera (3 adult, 3 offspring), 8 from A. wiedenmayeri (1 adult, 7 offspring), 33 from E. ferox (7 adult, 26 offspring), 2 from I. felix (1 adult, 1 offspring), 61 from S. aurea (42 adult, 19 offspring), and 108 from X. muta (32 adult, 76 offspring). Additionally, six archaeal 16S rRNA gene sequences were recovered from the sponges A. conifera (2 adult, 2 embryo) and E. ferox (2 embryo). These sequences, together with 18 sequences from Corticium sp. offspring (46), 71 from I. felix offspring (45), 27 from M. laxissima offspring (8), and 2 from Luffariella variabilis offspring (48), were used for phylogenetic tree construction.
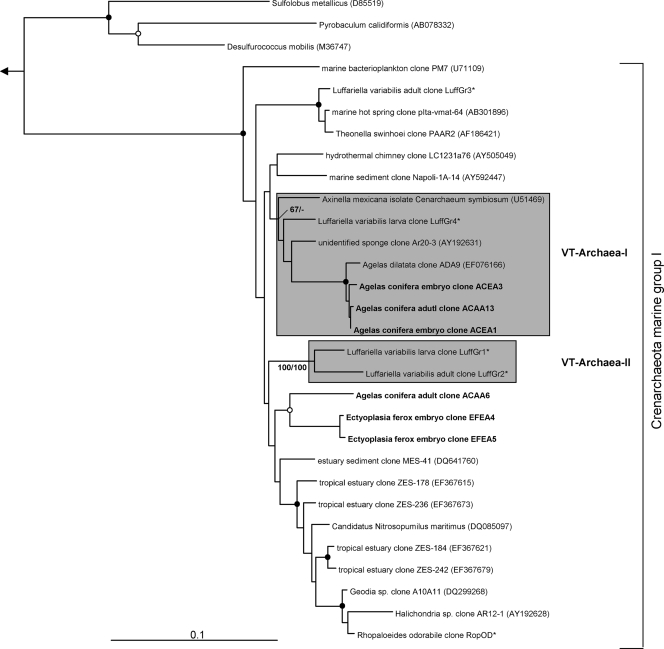
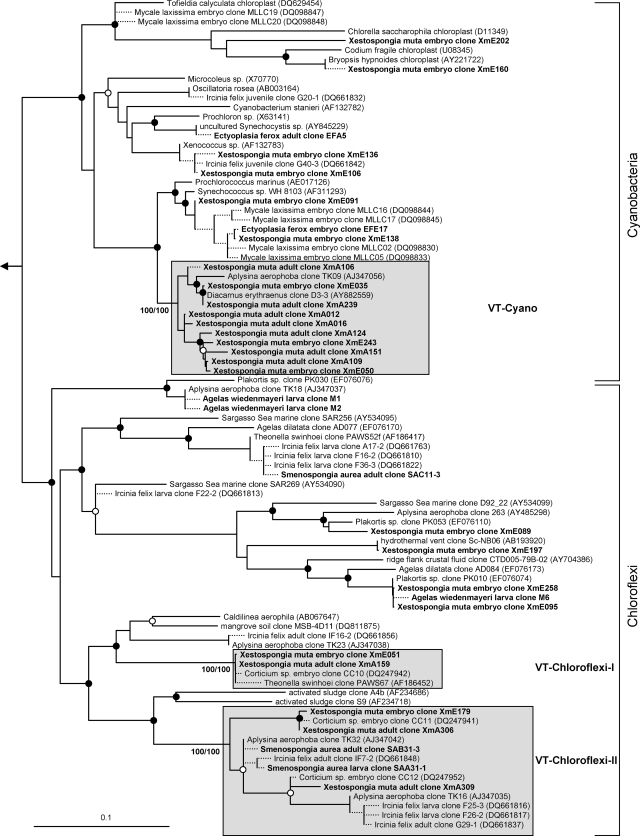
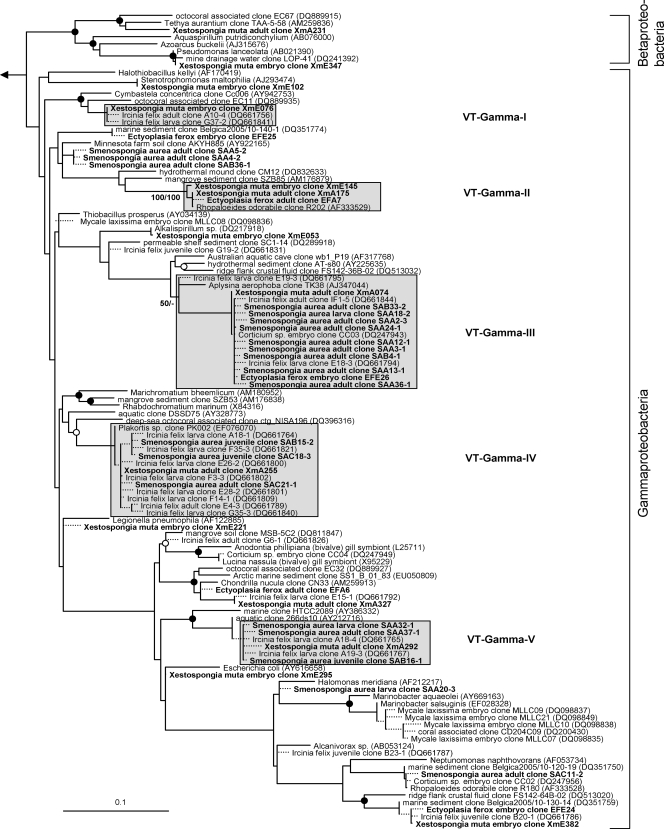
The overall microbial diversity in sponge offspring is high (Fig. 1 to 7). 16S rRNA gene phylotypes are affiliated with the bacterial phyla Acidobacteria (n = 22), Actinobacteria (n = 26), Bacteroidetes (n = 17), Chloroflexi (n = 19), Cyanobacteria (n = 14), Firmicutes (n = 8), Gemmatimonadetes (n = 6), Nitrospira (n = 4), Planctomycetes (n = 6), Proteobacteria (Alpha- [n = 55], Beta- [n = 1], Gamma- [n = 40], and Deltaproteobacteria [n = 8]), and Verrucomicrobia (n = 2), as well as with the bacterial candidate phyla Poribacteria (n = 4) and TM7 (n = 1). An additional cluster of bacterial sequences (n = 8) could not be unambiguously affiliated within the Bacteria (Fig. 6). Archaeal sequences obtained from sponge offspring all fell within marine group I of the Crenarchaeota (n = 6) (Fig. 7).
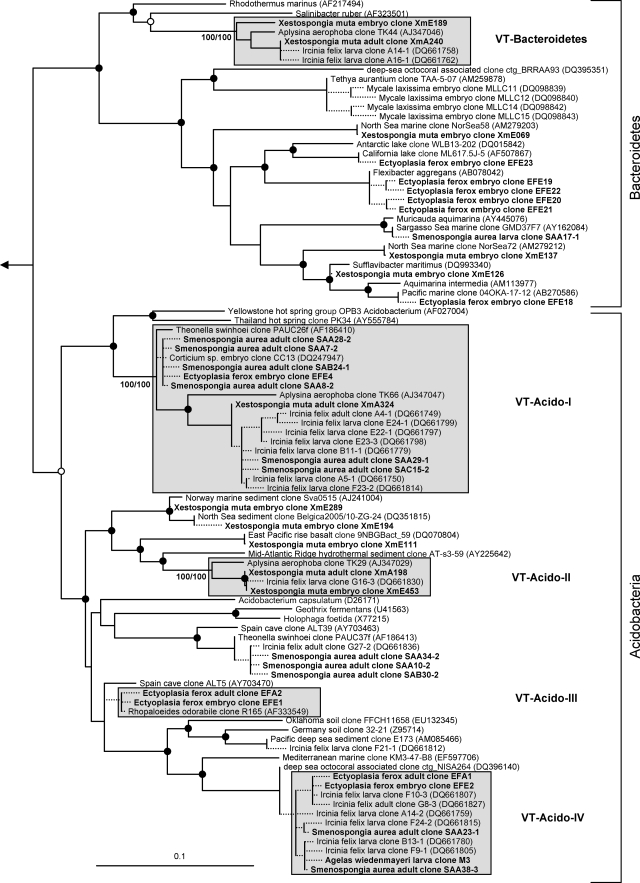
FIG. 1.
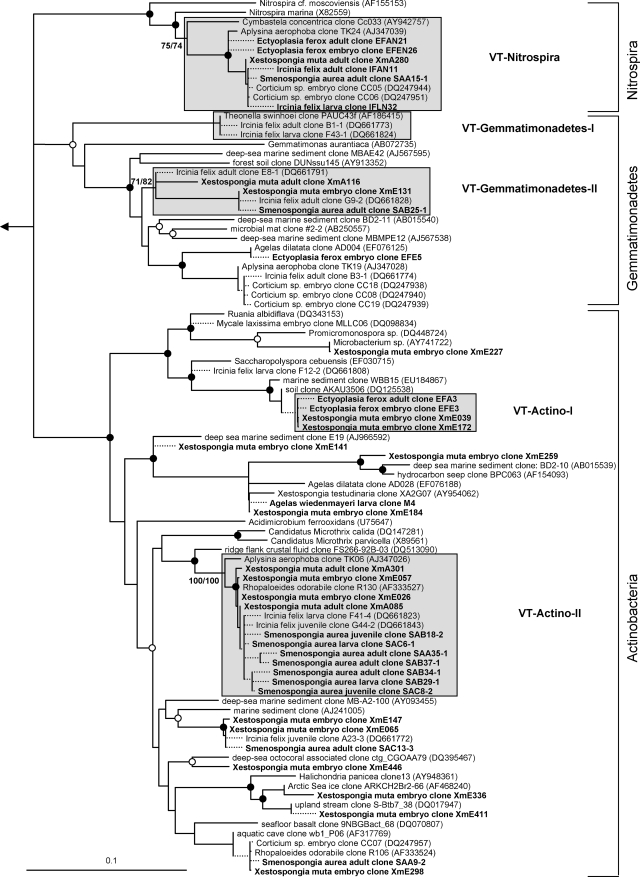
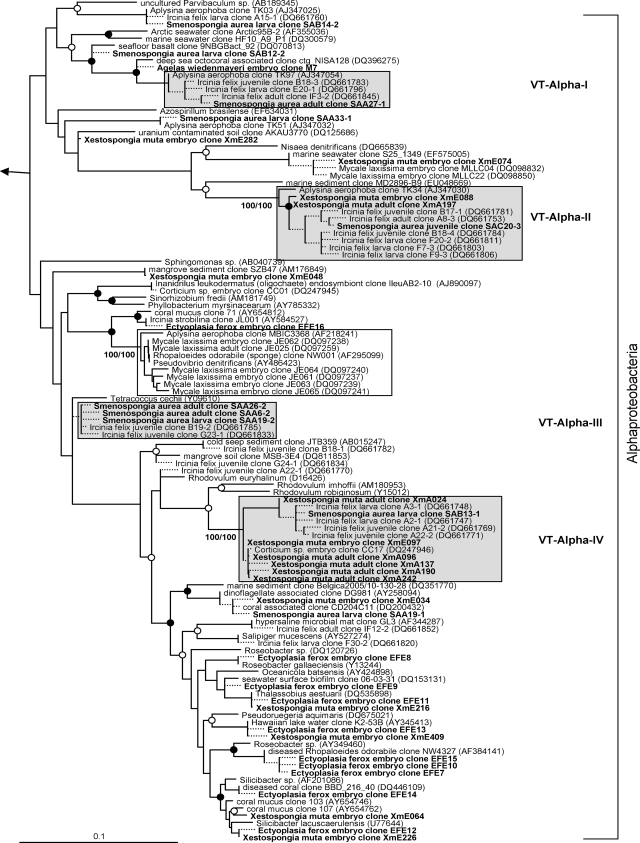
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the bacterial phyla Bacteroidetes and Acidobacteria. Filled and open circles represent neighbor-joining bootstrap values above 95% and 80%, respectively. Neighbor-joining and maximum parsimony bootstrap values are given for each VT cluster. Sequences obtained in this study are given in bold. Short sequences that were added to the tree using the parsimony interactive tool in ARB are indicated by dashed lines. Arrow, to outgroup (Pyrobaculum calidiformis AB078332 and Sulfolobus metallicus D85519). Gray boxes depict VT clusters that contain sequences obtained from adults and offspring of the same species. Scale bar indicates 10% sequence divergence.
FIG. 7.
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the Archaea. Details are the same as those provided for Fig. 1, except the outgroup here consists of Haloferax mediterranei (D11107) and Methanogenium marinum (DQ177344). *, sequences kindly provided by M. Taylor (University of Auckland, New Zealand) prior to publication (48).
FIG. 6.
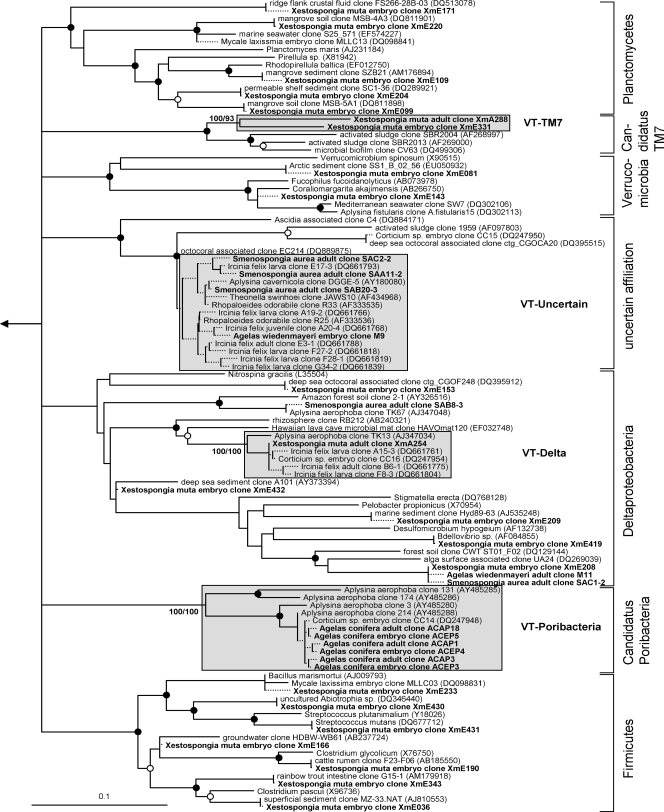
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the bacterial phyla Planctomycetes, Verrucomicrobia, and Firmicutes; with the class Deltaproteobacteria; and with the candidate phyla TM7 and Poribacteria. One group of sequences could not be unambiguously affiliated within the Bacteria and is marked as “uncertain affiliation.” Details are the same as those provided for Fig. 1.
VT clusters.
Sequences from an adult sponge and its offspring that cluster together in the phylogenetic analysis are referred to as vertically transmitted sequence pairs. Together with other sponge-specific sequences, they build clusters which are referred to as vertical-transmission (VT) clusters. Twenty-five VT clusters were found, distributed among 10 different bacterial phyla (including two candidate phyla) (Fig. 1 to 6). One additional VT cluster could not be assigned to any known bacterial phylum and was therefore termed VT-Uncertain (Fig. 6). Within the Archaea, two VT clusters were obtained (Fig. 7). Six clusters are double VT clusters (VT-Acido-IV, VT-Actino-II, VT-Alpha-II, VT-Gamma-III, VT-Gamma-IV, and VT-Nitrospira), and one is a triple VT cluster (VT-Chloroflexi-II), as they contain vertically transmitted sequence pairs of two or three sponge species, respectively. Twenty-nine percent of A. wiedenmayeri, 53% of Corticium sp., 23% of E. ferox, 75% of I. felix, 0% of M. laxissima, 68% of S. aurea, and 23% of X. muta offspring sequences belong to VT clusters.
TEM.
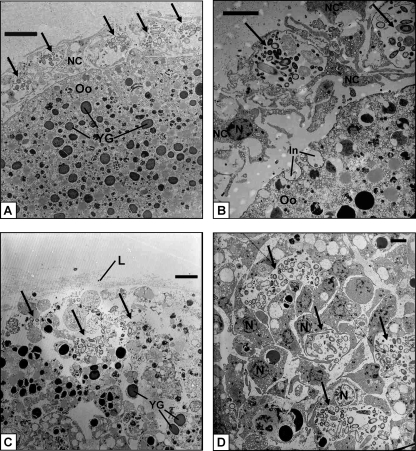
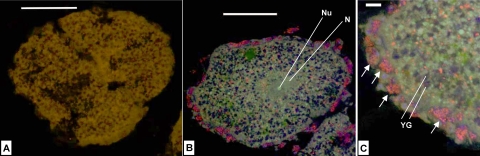
E. ferox released orange-colored oocytes which were embedded in a mucous sheath. Electron micrographs of the oocytes revealed the presence of many inclusions equally scattered within the cytoplasm (Fig. 8A) These inclusions vary in electron density and in size and represent, e.g., lipid droplets (1 to 1.5 μm in diameter) and yolk granules (2 to 3 μm in diameter). Microorganisms could not be detected in the oocyte cytoplasm. The oocyte is surrounded by a layer of amoeboid-shaped nurse cells which are 5 to 8 μm in size (Fig. 8A and B). Each nurse cell contains one 1- to 2-μm large nucleus, several smaller vacuoles, and usually one large vesicle with 15 to 20 (sometimes more) microorganisms of different sizes of up to 2.5 μm and various shapes including rod-like and coccus-like cells (Fig. 8B). At this stage of development, there is a clear space between nurse cells and the oocyte. However, the oocyte already shows signs of invagination (Fig. 8B). After 2 days, the oocytes developed into early-stage embryos, which were still embedded in the gelatinous sheath. The embryo consists of many small, densely packed amoeboid-shaped cells that contain one 2- to 3-μm large nucleus each and again diverse inclusions, including yolk granules (Fig. 8C and D). A thin lamella of collagen fibrils surrounds the embryo (Fig. 8C), and nurse cells are no longer present at this stage of development. Clusters of morphologically diverse microorganisms can be found extracellularly within the embryo, mostly located in the periphery.
FIG. 8.
TEM of the E. ferox oocyte (A), a layer of nurse cells surrounding the oocyte (B), the surface area (C), and the peripheral part (D) of an embryo. Arrows indicate clusters of microorganisms. Abbreviations: In, invaginations of oocyte; L, lamella of collagen fibrils; N, nucleus; NC, nurse cells; Oo, oocyte; YG, yolk granules. Scale bars, 5 μm (A and C) and 3 μm (B and D).
FISH.
A cohybridization with the bacterial probe mix EUB I-III and the eukaryotic probe EUK516, in addition to the DNA dye DAPI, was performed on cross-sections of E. ferox oocytes (Fig. 9). The 150- by 100-μm-large oocyte showed no background autofluorescence (Fig. 9A). The interior appeared heterogeneous (Fig. 9B). Eukaryotic ribosomes are present in large numbers as indicated by many green signals. The 4- to 5-μm-large nucleolus is especially prominent in the center of the oocyte within the nucleus, which has a size of 20 μm. Additionally, dark blue, circular signals (about 3 μm in diameter), as well as red signals of various sizes, are present. The oocyte is surrounded by nurse cells which can be identified by their nuclei due to blue DAPI signals and contain clusters of microorganisms represented by red signals (Fig. 9C).
FIG. 9.
FISH of E. ferox oocytes. (A) Negative control; (B) whole oocyte; (C) nurse cell layer. Cohybridization with the Cy3-labeled universal bacterial probe mix EUB I-III (red signals) and the fluorescein isothiocyanate-labeled eukaryotic probe EUK516 (green signals). Additionally, the fluorescent dye DAPI (blue signals) was applied. Arrows indicate clusters of microorganisms. Abbreviations: N, nucleus; Nu, nucleolus; YG, yolk granules. Scale bars, 50 μm (A and B) and 10 μm (C).
DISCUSSION
Molecular microbial diversity survey of sponge reproductive stages.
The first evidence for the vertical transmission of microorganisms in sponges came from electron microscopy studies that documented the presence of microbes in reproductive stages (references 6 and 9 and references cited therein; 25, 44, 52). The application of molecular tools allowed the phylogenetic identification of microorganisms in sponges and their reproductive stages. Prior to this study, the total microbial diversity in sponge offspring was analyzed for only three species in separate studies (8, 45, 46). Here we provide new 16S rRNA gene sequences from five additional sponges and report the first comprehensive phylogenetic analysis including more than 250 16S rRNA gene sequences from sponge reproductive stages.
The observed total phylogenetic diversity of microbes in sponge reproductive stages is high and comprises phylotypes within one archaeal phylum and 13 bacterial phyla (two of them are candidate phyla) including the four classes Alpha-, Beta-, Gamma-, and Deltaproteobacteria (Fig. 1 to 7). These high numbers almost match the known microbial diversity of adult sponges, which comprises two archaeal and 16 bacterial phyla and is predicted to be close to the total microbial diversity in sponges at the phylum level as assessed by the current approaches used (49). Most sponge offspring 16S rRNA gene sequences were derived from X. muta and I. felix, representing 11 and 8 phyla, respectively. However, just 7 sequences from A. wiedenmayeri embryos are affiliated with 4 phyla and 19 sequences from Corticium sp. already fall into 8 phyla. These numbers suggest that the presented data largely reflect the overall microbial diversity in sponge offspring, at least at the phylum level, and they show that equally complex microbial consortia are present in adult sponges and reproductive stages.
Microbes which are vertically transmitted were determined by VT clusters which are defined as phylotypes that are present in adult and offspring samples of the same species. More than two-thirds of the offspring sequences from I. felix and S. aurea and more than half from Corticium sp. fall into VT clusters implying that a large part of the adult microbial community is vertically transmitted in these three species. In A. wiedenmayeri, E. ferox, and X. muta, about one-third of the offspring-derived sequences are within VT clusters. Phylotypes that do not fall within VT clusters may represent environmental contamination, e.g., seawater microbes. However, it is noteworthy that many of the offspring sequences outside of the VT clusters still cluster with sequences obtained from other sponge species. For example, within the Gemmatimonadetes phylum (Fig. 2), three embryo sequences from Corticium sp. (clones CC08, -18, and -19) and one from E. ferox (clone EFE5) cluster with clones from the adult sponges I. felix (clone B3-1), Aplysina aerophoba (clone TK19), and Agelas dilatata (clone AD004). This cluster was not defined as a VT cluster because sequences from adult Corticium sp. and E. ferox are missing. Additionally, all VT clusters except clusters VT-TM7 (Fig. 6) and VT-Archaea-II (Fig. 7) contain additional sequences derived from other sponge species. The general overlap between offspring and adult microbial communities is striking. It is well recognized that HMA sponges share a uniform, sponge-specific microbial community (references 16 and 49 and references cited therein), and the presented data now demonstrate that this uniform microbial community is collectively transmitted through the reproductive stages.
FIG. 2.
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the bacterial phyla Nitrospira, Gemmatimonadetes, and Actinobacteria. Details are the same as those provided for Fig. 1.
In contrast to the results for the HMA sponges discussed above, sequences from M. laxissima embryos tend to cluster much less with other sponge-derived sequences, and none fall within a VT cluster. A recent analysis of the adult microbial community of M. laxissima also showed little overlap with the microbial diversity of HMA sponges but revealed a specific community in M. laxissima that is different from seawater bacterioplankton (30). Interestingly, M. laxissima was recently classified as an LMA sponge based on microscopic observations (44). The phylogenetic microbial signature of both the adult sponge and embryos that differ from the signature of HMA sponges supports this classification. M. laxissima might contain its own specific microbial community that is vertically transmitted. Notable is a bacterial isolate that was cultivated from the adults and embryos of M. laxissima. The respective sequence was also recovered from a 16S rRNA gene library of M. laxissima embryos and clusters with the marine alphaproteobacterium Pseudovibrio denitrificans (Fig. 4). The same phylotype was previously detected by cultivation-dependent and -independent approaches in many other sponges but also in corals and marine environments (41, 49). It cannot be discounted that the Pseudovibrio-like bacterium was actually taken up by sponges from the seawater. However, because it is widespread among sponges and appears to be vertically transmitted in M. laxissima, it probably represents a true sponge symbiont.
FIG. 4.
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the bacterial class Alphaproteobacteria. Details are the same as those provided for Fig. 1. The cluster containing the isolate from M. laxissima adults and embryos is indicated by an unshaded box.
Mechanism of vertical transmission in E. ferox.
The phylogenetic results demonstrated vertical transmission in eight species representing six different orders and two modes of reproduction (ovipary and vivipary) (Table 1). The passage of symbionts through reproductive stages appears to be common and widespread among sponges. However, differences in the mechanism of symbiont transmission, e.g., between oviparous and viviparous species, are almost unknown (but see references 9 and 29). In this study, we conducted a combined electron and fluorescence microscopic examination of microbes within oocytes and embryos of the Caribbean sponge E. ferox. This oviparous species was chosen because oocytes can be collected, allowing the examination of vertical transmission from the early reproductive stages onward.
The oocyte is large and represents a late stage of gamete development. As fertilization was not observed, this unicellular stage could possibly also be a zygote. Typically, the oocyte contains many inclusions, including yolk and lipids, which mostly serve as nutrient reserves during the gamete and embryo growth periods (Fig. 8). The oocyte is metabolically highly active, as demonstrated by the large number of eukaryotic ribosomes throughout the cytoplasm (Fig. 9B). The nucleus is located in the center of the oocyte and contains several nucleoli (data not shown) of which one is visible in Fig. 9B as a bright green signal due to the synthesis of new rRNA. The oocyte is encircled by a layer of nurse cells (Fig. 8A and B). Nurse cells play a major role in the nutrition, and consequently the growth, of oocytes. The transfer of substances from nurse cells to oocytes via cytoplasmic bridges and/or direct phagocytosis of the whole nurse cell by oocytes has been described for several viviparous species (e.g., see references 11 and 22) as well as oviparous species (e.g., see references 14 and 24). In this developmental stage of E. ferox, the nurse cells have not yet made contact with the oocyte, but the phagocytotic activity of the gamete is already visible (Fig. 8B). The presence of microorganisms within nurse cells was shown in electron micrographs (Fig. 8A and B) and further confirmed by clusters of bacterial-specific red signals encircling the oocyte in fluorescence micrographs (Fig. 9B and C). Single red signals were also observed within the oocyte in FISH analyses but were absent in the negative control (Fig. 9A). These signals might correspond to microbes. However, as microbes within the oocyte were never detected in the electron micrographs, the identity of these red signals remains unclear.
The embryonic stage of development that is represented by Fig. 8C and D is characterized by many densely packed sponge cells that include lipids, yolk reserves, and other inclusions. The embryo is surrounded by a lamella of collagen fibrils. Such filamentous structures were previously described, e.g., the enveloping embryos of Spongia sp. and Hippospongia lachne (22) as well as Latrunculia magnifica (21). Ilan (21) suggested that the fibrils are secreted by a layer of sponge cells. However, the origin as well as the function of this lamella in E. ferox remains unknown. Clusters of microorganisms are present extracellularly within the embryo (Fig. 8D).
E. ferox, like all other sponges, lacks reproductive organs, and gamete production occurs within the mesohyl of adult sponges. Based on the microscopic observations, the following mechanism of microbial transmission in E. ferox is proposed: during the establishment of oocytes, nurse cells take up adult microorganisms from the mesohyl via phagocytosis. Presumably, this uptake occurs mainly at random. However, nurse cell phagocytosis of cyanobacteria in the cortex region and subsequent transport to the developing eggs deeper within the sponge were observed in Chondrilla australiensis (52), suggesting a possible control of symbiont uptake by these cells. Afterwards, the reproductive propagules, along with associated nurse cells that now form a cell layer around the oocytes, become embedded in a mucous sheath that is produced and then released by the sponge. The same mode of reproduction was observed to occur in other sponges, including X. muta, A. conifera, and A. wiedenmayeri (N. Lindquist, personal observation). Microorganisms are then either transferred from nurse cells to oocytes or, at a later stage, to the developing embryo. In the majority of oviparous species as well as in at least one viviparous sponge, the transfer of microbes appears to occur directly as the microbes are taken up by the oocyte itself from the adult mesohyl (reference 9 and references cited therein). In E. ferox and some other sponges, such as Chondrosia reniformis (24), the transfer takes place indirectly because the microbes are translocated through nurse cells to the progeny.
Transmission model for symbioses of HMA sponges and complex microbial consortia.
Cospeciation between sponges and microbes is often difficult to prove due to the complexity of the microbial consortia and/or the generally conservative character of some phylogenetic markers such as the 16S rDNA gene. Based on largely congruent phylogenies, cospeciation was demonstrated for the filamentous cyanobacterium Oscillatoria spongeliae and dictyoceratid sponges (39, 50) and for an alphaproteobacterial symbiont and halichondrid sponges (10). However, in both cases, at least one host-switching event occurred. Our data demonstrate the transmission of a highly sponge-specific microbial community via reproductive stages. However, the phylogenetic trees show no indication of host-species specificity or cospeciation.
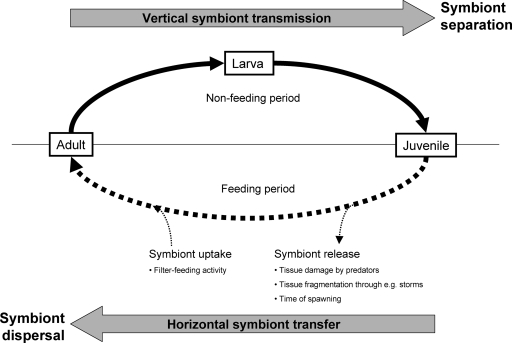
Based on our phylogenetic results and the mechanistic details described for E. ferox, we therefore propose the following symbiont transmission model for associations of HMA sponges and their microbial consortia (Fig. 10): during sponge reproduction, gametes or later reproductive stages take up microbes from the adult mesohyl microbial community. Thereby, at least in some species, including E. ferox, nurse cells are involved in microbial incorporation and might actually represent the important factor fostering selection of this association. Sponge larvae are unable to feed and might therefore be regarded as a closed system with no exchange with the environment. Vertical transmission therefore causes the separation of symbionts in different sponges which might eventually result in genetic differentiation and would ultimately lead to cospeciation. After the settlement of sponge larvae and their subsequent metamorphosis into juveniles, the filter-feeding life phase begins, allowing the uptake of seawater microbes. In this phase, the horizontal transfer of symbionts between host individuals or environmental acquisition from seawater appears to be possible, at least to some degree, and evidence from sponges was recently discussed in detail by Taylor and colleagues (49). Symbionts might be released by sponges due to tissue damage by predators and environmental factors (currents and storms) or during spawning periods. The uptake of symbionts could possibly occur during filter-feeding periods of the host. It might also be conceivable that sponge larvae acquire microbial symbionts that colonize the outer surface of the larvae, after the release of larvae from the adult sponge. Such symbiont uptake and release mechanisms would result in symbiont dispersal among host individuals and even species, thus obscuring the pattern of cospeciation in phylogenetic trees (Fig. 10).
FIG. 10.
Proposed symbiont transmission model in HMA sponges. Adult mesohyl microbial consortia are vertically transmitted through nonfeeding reproductive stages, resulting in symbiont separation (solid line). During the feeding life stage of sponges, occasional horizontal symbiont transfer occurs, leading to symbiont dispersal among sponge individuals and even species (dashed line). Uniformly shared, sponge-specific microbial consortia are maintained by this combination of vertical and horizontal transmission.
The importance of vertical transmission lies in the efficient passage of symbionts to the next generation. In HMA sponges, vertical transmission represents a bottleneck in the sense that only members of the specific adult microbial community, but apparently no seawater microorganisms, are transferred through reproductive stages. Horizontal transmission acts against symbiont separation and specialization. Instead of leading to cospeciation, this mechanism might maintain a uniformly shared microbial community in HMA sponges. We propose that a combination of vertical and horizontal symbiont transmission maintains the associations of HMA sponges with highly sponge-specific, complex microbial communities.
FIG. 3.
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the bacterial phyla Cyanobacteria and Chloroflexi. Details are the same as those provided for Fig. 1.
FIG. 5.
Evolutionary distance tree calculated with 16S rRNA gene sequences affiliated with the bacterial classes Beta- and Gammaproteobacteria. Details are the same as those provided for Fig. 1.
Acknowledgments
We gratefully acknowledge the staff of the University of North Carolina at Wilmington's NOAA National Undersea Research Center in Key Largo, FL, for their exceptional assistance during the field work. We especially thank M. Taylor (University of Auckland, New Zealand) for kindly providing 16S rRNA gene archaeal sequences prior to publication and for valuable comments on the manuscript. We also thank J. Cowart (University of North Carolina at Wilmington) for sponge sampling and M. Meinhold and C. Gernert (University of Wuerzburg, Germany) for technical assistance in the lab.
This work was supported by Deutsche Forschungsgemeinschaft grants HE3299/1-1 and 1-2 to U.H. and UNCW/NURC and NSF Chemical Oceanography Program grants (NA03OAR4300088 and OCE 0351893/OCE 0531422, respectively) to N.L. and C. S. Martens.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, K., S. Schmitt, and U. Hentschel. 2008. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ. Microbiol. 10:2942-2955. [DOI] [PubMed] [Google Scholar]
- 4.Brusca, R. C., and G. J. Brusca. 1990. Phylum Porifera: the sponges, p. 181-210. In A. D. Sinauer (ed.), Invertebrates. Sinauer Press, Sunderland, MA.
- 5.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 6.deCaralt, S., M. J. Uriz, and R. H. Wijffels. 2007. Vertical transmission and successive location of symbiotic bacteria during embryo development and larva formation in Corticium candelabrum (Porifera: Demospongiae). J. Mar. Biol. Assoc. UK 87:1693-1699. [Google Scholar]
- 7.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enticknap, J. J., M. Kelly, O. Peraud, and R. T. Hill. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ereskovsky, A. V., E. Gonobobleva, and A. Vishnyakov. 2005. Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarca). Mar. Biol. 146:869-875. [Google Scholar]
- 10.Erpenbeck, D., J. A. J. Breeuwer, H. C. van der Velde, and R. W. M. van Soest. 2002. Unravelling host and symbiont phylogenies of halichondrid sponges (Demospongiae, Porifera) using a mitochondrial marker. Mar. Biol. 141:377-386. [Google Scholar]
- 11.Fell, P. E. 1969. The involvement of nurse cells in oogenesis and embryonic development in the marine sponge, Haliclona ecbasis. J. Morphol. 127:133-150. [Google Scholar]
- 12.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 14.Gallissian, M. F., and J. Vacelet. 1976. Ultrastructure de quelques stades de l'ovogénèse des spongiaires du genre Verongia (Dictyoceratida). Ann. Sci. Nat. Zool. 18:381-404. [Google Scholar]
- 15.Hentschel, U., L. Fieseler, M. Wehrl, C. Gernert, M. Steinert, M. Horn, and J. Hacker. 2003. Microbial diversity of marine sponges, p. 59-88. In W. E. G. Müller (ed.), Molecular marine biology of sponges. Springer, Heidelberg, Germany. [DOI] [PubMed]
- 16.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, F., O. Larsen, V. Thiel, H. T. Rapp, T. Rape, W. Michaelis, and J. Reitner. 2005. An anaerobic world in sponges. Geomicrobiol. J. 22:1-10. [Google Scholar]
- 19.Hooper, J. N. A., and R. W. M. van Soest. 2002. Systema Porifera. A guide to the classification of sponges. Plenum Publishers, New York, NY.
- 20.Hurtado, L. A., M. Mateos, R. A. Lutz, and R. C. Vrijenhoek. 2003. Coupling of bacterial endosymbionts and host mitochondrial genomes in the hydrothermal vent clam Calyptogena magnifica. Appl. Environ. Microbiol. 69:2058-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilan, M. 1995. Reproductive biology, taxonomy, and aspects of chemical ecology of Latrunculiidae (Porifera). Biol. Bull. 188:306-312. [DOI] [PubMed] [Google Scholar]
- 22.Kaye, H. 1991. Sexual reproduction in four Caribbean commercial sponges. II. Oogenesis and transfer of bacterial symbionts. Invertebr. Reprod. Dev. 19:13-24. [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, London, United Kingdom.
- 24.Levi, C., and P. Levi. 1976. Embryogenese de Chondrosia reniformis (Nardo), demosponge ovipare, et transmission des bacteries symbiotiques. Ann. Sci. Nat. Zool. 18:367-380. [Google Scholar]
- 25.Levi, C., and A. Porte. 1962. Ètude au microscope électronique de l'éponge Oscarella lobularis Schmidt et de sa larve amphiblastula. Cah. Biol. Mar. 3:307-315. [Google Scholar]
- 26.Li, C. W., J. Y. Chen, and T. E. Hua. 1998. Precambrian sponges with cellular structures. Science 279:879-882. [DOI] [PubMed] [Google Scholar]
- 27.Lindquist, N., R. Bolser, and K. Laing. 1997. Timing of larval release by two Caribbean demosponges. Mar. Ecol. Prog. Ser. 155:309-313. [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maldonado, M. 2007. Intergenerational transmission of symbiotic bacteria in oviparous and viviparous demosponges, with emphasis on intracytoplasmically-compartmented bacterial types. J. Mar. Biol. Assoc. UK 87:1701-1713. [Google Scholar]
- 30.Mohamed, N. M., J. J. Enticknap, J. E. Lohr, S. M. McIntosh, and R. T. Hill. 2008. Changes in bacterial communities of the marine sponges Mycale laxissima on transfer into aquaculture. Appl. Environ. Microbiol. 74:1209-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran, N. A., and P. Baumann. 2000. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3:270-275. [DOI] [PubMed] [Google Scholar]
- 32.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology, manual 3.4.4. Kluwer, Dordrecht, The Netherlands.
- 33.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nussbaumer, A. D., C. R. Fisher, and M. Bright. 2006. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441:345-348. [DOI] [PubMed] [Google Scholar]
- 35.Oren, M., L. Steindler, and M. Ilan. 2005. Transmission, plasticity and the molecular identification of cyanobacterial symbionts in the Red Sea sponge Diacarnus erythraenus. Mar. Biol. 148:35-41. [Google Scholar]
- 36.Peek, A. S., R. A. Feldman, R. A. Lutz, and R. C. Vrijenhoek. 1998. Cospeciation of chemoautotrophic bacteria and deep sea clams. Proc. Natl. Acad. Sci. USA 95:9962-9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiswig, H. 1974. Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14:231-249. [Google Scholar]
- 38.Reiswig, H. 1981. Partial carbon and energy budgets of the bacteriosponge Verongia fistularis (Porifera: Demospongiae) in Barbados. Mar. Ecol. 2:273-293. [Google Scholar]
- 39.Ridley, C. P., P. R. Bergquist, M. K. Harper, D. J. Faulkner, J. N. A. Hooper, and M. G. Haygood. 2005. Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem. Biol. 12:397-406. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Scheuermayer, M., S. Pimentel-Elardo, L. Fieseler, L. Grozdanov, and U. Hentschel. 2006. Microorganisms of sponges: phylogenetic diversity and biotechnological potential, p. 289-312. In P. Proksch and W. E. G. Müller (ed.), Frontiers in marine biotechnology. Horizon Bioscience, Norfolk, United Kingdom.
- 42.Schmitt, S. 2007. Vertical microbial transmission in Caribbean bacteriosponges. Ph.D. thesis. University of Wuerzburg, Wuerzburg, Germany.
- 43.Schmitt, S., U. Hentschel, S. Zea, T. Dandekar, and M. Wolf. 2005. ITS-2 and 18S rRNA gene phylogeny of Aplysinidae (Verongida, Demospongiae). J. Mol. Evol. 60:327-336. (Erratum, 61:148-150.) [DOI] [PubMed] [Google Scholar]
- 44.Schmitt, S., M. Wehrl, N. Lindquist, J. B. Weisz, and U. Hentschel. 2007. Morphological and molecular analyses of microorganisms in Caribbean reef adult sponges and in corresponding reproductive material, p. 561-568. In M. Custodio, G. Lobo-Hajdu, E. Hajdu, and G. Muricy (ed.), Porifera research: biodiversity, innovation & sustainability. Museo National, Rio de Janeiro, Brazil.
- 45.Schmitt, S., J. B. Weisz, N. Lindquist, and U. Hentschel. 2007. Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp, K. H., B. Eam, D. J. Faulkner, and M. G. Haygood. 2007. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73:622-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siripong, S., J. J. Kelly, D. A. Stahl, and B. E. Rittmann. 2006. Impact of prehybridization PCR amplification on microarray detection of nitrifying bacteria in wastewater treatment plant samples. Environ. Microbiol. 8:1564-1574. [DOI] [PubMed] [Google Scholar]
- 48.Steger, D., P. Ettinger-Epstein, S. Whalan, U. Hentschel, R. DeNys, M. Wagner, and M. W. Taylor. 2008. Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ. Microbiol. 10:1087-1094. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thacker, R. W., and S. Starnes. 2003. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar. Biol. 142:643-648. [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usher, K. M., J. Kuo, J. Fromont, and D. C. Sutton. 2001. Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461:15-23. [Google Scholar]
- 53.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 54.Wehrl, M., M. Steinert, and U. Hentschel. 2007. Bacterial uptake by the marine sponge Aplysina aerophoba. Microb. Ecol. 53:355-365. [DOI] [PubMed] [Google Scholar]
- 55.Weisz, J. B., N. Lindquist, and C. S. Martens. 2008. Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155:367-376. [DOI] [PubMed] [Google Scholar]
- 56.Wilkinson, C. R. 1983. Net primary productivity in coral reef sponges. Science 219:410-412. [DOI] [PubMed] [Google Scholar]