Abstract
A genomic comparison of Yarrowia lipolytica and Saccharomyces cerevisiae indicates that the metabolism of Y. lipolytica is oriented toward the glycerol pathway. To redirect carbon flux toward lipid synthesis, the GUT2 gene, which codes for the glycerol-3-phosphate dehydrogenase isomer, was deleted in Y. lipolytica in this study. This Δgut2 mutant strain demonstrated a threefold increase in lipid accumulation compared to the wild-type strain. However, mobilization of lipid reserves occurred after the exit from the exponential phase due to β-oxidation. Y. lipolytica contains six acyl-coenzyme A oxidases (Aox), encoded by the POX1 to POX6 genes, that catalyze the limiting step of peroxisomal β-oxidation. Additional deletion of the POX1 to POX6 genes in the Δgut2 strain led to a fourfold increase in lipid content. The lipid composition of all of the strains tested demonstrated high proportions of FFA. The size and number of the lipid bodies in these strains were shown to be dependent on the lipid composition and accumulation ratio.
The oleaginous yeast Yarrowia lipolytica is one of the most extensively studied “nonconventional” yeasts. This yeast is able to accumulate large amounts of lipids (in some cases, more than 50% of its dry weight [DW]) (26). Several technologies including different fermentation configurations have been applied for single-cell oil production by strains of Y. lipolytica grown on various agro-industrial by-products or wastes (i.e., industrial fats, crude glycerol, etc.) (10, 23). Potential applications of these processes are targeting the production of reserve lipids with a specific structure and/or composition. These include (i) oils enriched in essential polyunsaturated FA as potential nutritional complements, (ii) lipids showing composition similarities to cocoa butter, and (iii) nonspecific oils destined for use as renewable starting materials for the synthesis of biofuels. Accordingly, increased research interest has focused on genetic and metabolic engineering approaches to “construct” yeast strains to be used as cell factories for storing and producing huge amounts of oil with an attractive FA composition (10).
Y. lipolytica is able to utilize hydrophobic substrates (e.g., alkanes, FA, and oils) efficiently as a sole carbon source (5, 10). Its superior capacity to accumulate lipids when grown on these substrates is probably related to protrusions formed on cell surfaces, facilitating the uptake of hydrophobic substrates from the medium (21). Internalized aliphatic chains can be either degraded for growth requirements or accumulated in an unchanged or a modified form (10). Storage molecules such as triglycerides (TAG) and steryl esters (SE) are not suitable for integration into phospholipid bilayers. Therefore, they cluster to form the hydrophobic core of so-called lipid bodies (LB). Originally, LB were considered only as a depot of neutral lipids which can be mobilized under starving conditions. However, the view of the LB as a simple storage compartment, excluded from any metabolic process, had to be revised since many of its proteins were identified as enzymes involved in lipid metabolism, especially in TAG synthesis and degradation. Recently, a lipid binding protein identified in Y. lipolytica LB (2, 9) was shown to be involved in lipid trafficking between cytoplasm and LB. This indicates that LB are probably capable of accommodating free FA (FFA) as well (2, 6, 21, 23).
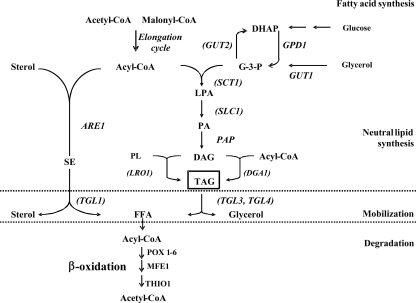
In yeasts, TAG synthesis follows the Kennedy pathway (15). FFA are activated to coenzyme A (CoA) and used for the acylation of the glycerol backbone to synthesize TAG. In the first step of TAG assembly, glycerol-3-phosphate (G3P) is acylated by G3P acyltransferase (SCT1) to lysophosphatidic acid (LPA), which is then further acylated by LPA acyltransferase (SLC1) to phosphatidic acid (PA). This is followed by dephosphorylation of PA by PA phosphohydrolase (PAP) to release diacylglycerol (DAG). In the final step, DAG is acylated either by DAG acyltransferase (DGA1 with acyl-CoA as an acyl donor) or by phospholipid DAG acyltransferase (LRO1 with glycerophospholipids as an acyl donor) to produce TAG (Fig. 1 and Table 1) (8, 28).
FIG. 1.
Overview of different pathways involved in FA synthesis and storage and degradation of neutral lipids. Synthesis of FA (Acyl-CoA) is catalyzed by the FA synthesis complex from the basic blocks acetyl-CoA and malonyl-CoA. Acyl-CoA can be stored either as SE or as TAG. The synthesis of SE is catalyzed by SE synthases homologous to the human acyl-CoA:cholesterol acyltransferase and their mobilization by the SE hydrolases, releasing sterol and FFA. The synthesis of TAG require acyl-CoA and G3P. G3P could be produced from glycerol or from DHAP. GUT1 encodes a glycerol kinase that converts glycerol to G3P in the cytosol. The product of this reaction can be oxidized to DHAP by the G3PDH encoded by the GUT2 gene. DHAP can enter either glycolysis or gluconeogenesis. G3P could also be used as a skeleton for TAG synthesis. Three acyls are added to the G3P backbone to give TAG, and this process requires four enzymatic steps. First, an acyl is added at the sn-1 position of G3P by a G3P acyltransferase to produce LPA, and then a second acyl is added at the sn-2 position by a 1-acyl G3P acyltransferase to produce PA, which is then dephosphorylated by PAP, yielding DAG. Finally, the third acyl can be added at the sn-3 position either by the acetyl-CoA-dependent pathway (directly from acyl-CoA) or by the acetyl-CoA-independent pathway (from a glycerophospholipid). TAG can be mobilized by the conversion to FFA and DAG upon hydrolysis by TAG lipase. The FFA can then be degraded in the β-oxidation pathway. This pathway requires four enzymatic steps (for a review, see reference 11). In Y. lipolytica, six genes (POX1 to POX6) that code for acyl-CoA oxidases are involved in the second step of β-oxidation (11, 29). Proteins encoded by the genes in parentheses were found to be associated with lipid particles (LB) in Y. lipolytica.
TABLE 1.
Genes involved in FA metabolism in Y. lipolytica and S. cerevisiaea
| Gene | S. cerevisiae name | EC no. | Y. lipolytica ortholog | % Amino acid identityb (% gap) | Function |
|---|---|---|---|---|---|
| GUT1 | YHL032c | EC 2.7.1.30 | YALI0F00484g | 35 (29) | Glycerol kinase |
| GPD1 | YDL022w | EC 1.1.1.18 | YALI0B02948g | 52 (11) | G3P dehydrogenase (NAD+) |
| GPD2 | YDL059w | EC 1.1.1.18 | 61 (11) | G3P dehydrogenase (NAD+) | |
| GUT2 | YIL155c | EC 1.1.99.5 | YALI0B13970g | 44 (11) | G3P dehydrogenase |
| SCT1 | YBL011w | EC 2.3.1.15 | YALI0C00209g | 40 (7) | G3P acyltransferase |
| GPT2 | YKR067w | EC 2.3.1.15 | 28 (7) | G3P acyltransferase | |
| SLC1 | YDL052c | EC 2.3.1.51 | YALI0E18964g | 33 (23) | 1-Acyl-sn-G3P acyltransferase |
| DGA1 | YOR245c | EC 2.3.1.20 | YALI0E32769g | 33 (27) | DAG acyltransferased |
| LRO1 | YNR008w | EC 2.3.1.158 | YALI0E16797g | 45 (4) | Phospholipid:DAG acyltransferased |
| TGL3 | YMR313c | EC 3.1.1.3 | YALI0D17534g | 21 (13) | Triacylglycerol lipase |
| TGL4 | YKR089c | EC 3.1.1.3 | YALI0F10010g | 29 (13) | Triacylglycerol lipase |
| TGL5 | YOR081c | EC 3.1.1.3 | 29 (13) | Triacylglycerol lipase | |
| ARE1 | YCR048w | EC 2.3.1.26 | YALI0F06578g | 30 (7) | Acyl-CoA:sterol acyltransferasec |
| ARE2 | YNR019w | EC 2.3.1.26 | 33 (7) | Acyl-CoA:sterol acyltransferase | |
| TGL1 | YKL140w | EC 3.1.1.13 | YALI0E32035g | 32 (11) | Cholesterol esterase |
| POX1 | YGL205w | EC 6.2.1.3 | YALI0E32835g | 35 (10) | Acyl-CoA oxidasec |
| POX2 | YALI0F10857g | 35 (8) | Acyl-CoA oxidased | ||
| POX3 | YALI0D24750g | 35 (9) | Acyl-CoA oxidased | ||
| POX4 | YALI0E27654g | 33 (0) | Acyl-CoA oxidasec | ||
| POX5 | YALI0C23859g | 35 (0) | Acyl-CoA oxidasec | ||
| POX6 | YALI0E06567g | 35 (2) | Acyl-CoA oxidasec | ||
| MFE1 | YKR009c | EC 4.2.1.74 | YALI0E15378g | 49 (5) | Multifunctional β-oxidation proteinc |
| POT1 | YIL160c | EC 2.3.1.16 | YALI0E18568g | 55 (4) | Peroxisomal oxoacyl thiolasec |
Shown are genes, corresponding S. cerevisiae gene names and EC (Enzyme Commission) numbers, Y. lipolytica orthologs (gene name), and corresponding functions. Bioinformatic data were obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/) and the Genolevures database (http://cbi.labri.fr/Genolevures/).
The comparative analysis was based on pairwise alignments of the full protein sequence predicted by the ClustalX program (http://bips.u-strasbg.fr/fr/Documentation/ClustalX/), and sequence identity or similarity was determined on the basis of this full alignment with Genedoc (http://www.nrbsc.org/gfx/genedoc/). ID, identity. % gap, percentage of residues unaligned within gaps.
Confirmed by functional analysis.
Confirmed by protein characterization.
Degradation of FA in yeasts occurs via the β-oxidation pathway, a multistep process requiring four different enzymatic activities. Enzyme localization is restricted to peroxisomes in yeasts, whereas in mammals these reactions are located in both mitochondria and peroxisomes. Mobilization of accumulated lipids occurs as a consequence of three different metabolic states, (i) during the exponential phase of growth, when storage lipids are used for membrane lipid synthesis to support cellular growth and division, (ii) during stationary phase, when, upon nutrient depletion, FFA are liberated rather slowly from the TAG and subjected to peroxisomal β-oxidation, and (iii) when cells exit starvation conditions, e.g., from stationary phase, and enter a vegetative growth cycle, and upon carbon supplementation, lipid depots are very rapidly degraded to FFA (17).
In this study, we focused on the importance of G3P in TAG formation. Localized in LB (2), it is suspected of playing a crucial role in TAG metabolism (16, 22, 28). There are two different routes to G3P synthesis. In one, G3P is derived from glycerol via glycerol kinase (GUT1). In the second, G3P is directly synthesized from dihydroxyacetone phosphate (DHAP) catalyzed by G3P dehydrogenase (GPD1). This last reaction is reversible, and the DHAP formation from G3P is catalyzed by a second G3P dehydrogenase (G3PDH) isoform encoded by GUT2. In order to stimulate the TAG biosynthetic pathway and increase lipid accumulation in Y. lipolytica, we deleted the GUT2 gene that encodes the G3PDH isoform and showed that its deficiency leads to a strong boost in the final lipid content of this yeast. To further increase lipid content, we prevented the yeast from degrading its lipid stores by abolishing the β-oxidation pathway. This modification was attained by deleting the POX genes that encode the acyl-coenzyme oxidases (Aox), a family of six peroxisomal acyl-coenzyme oxidases encoded by the POX1 to POX6 genes in Y. lipolytica (10, 29). We demonstrate here that combination of GUT2 and POX deficiencies, together with modification of the composition of the growth medium, leads to an altered composition of lipid species and a change in the accumulation ratio of lipids stored in the yeast Y. lipolytica.
MATERIALS AND METHODS
Yeast strains, growth, and culture conditions.
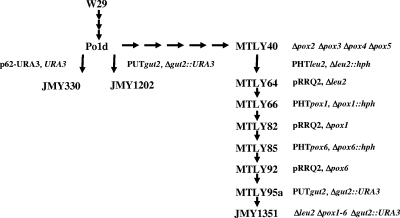
The Y. lipolytica strains used in this study were derived from wild-type (WT) strain Y. lipolytica W29 (ATCC 20460) (Table 2 and Fig. 2). Auxotrophic strain Po1d (Leu− Ura−) was described previously by Barth and Gaillardin (5). Prototrophic strain MTLY37, in which four POX genes that code for acyl-CoA oxidases (Aox) were inactivated, was described by Wang et al. (29). The auxotrophic derivative MTLY40 (Ura−) was obtained by transformation of MTLY37 by using a 1.5-kb PCR fragment carrying the ura3-41 allele, followed by selection of transformants on YNB-5FOA medium (21). The strains with deletions of the GUT2 gene, which codes for G3PDH, and the remaining POX genes are indicated in Table 2, and a schematic presentation of their construction is depicted in Fig. 2. Details of their construction are presented below.
TABLE 2.
E. coli and Y. lipolytica strains and plasmids used in this study
| Strain (host strain) | Genotype or plasmid | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80 dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| JME459 (DH5α) | pBluescript II KS+ (ColE1 ori LacZ bla) | Stratagene |
| JME461 (DH5α) | pRRQ2 (cre ARS68 LEU2 in pBluescript II KS+) | 12 |
| JME508 (DH5α) | 1.6-kb I-SceI fragment containing hph marker, MH cassette | 12 |
| JME507 (DH5α) | 1.3-kb I-SceI fragment containing URA3 marker, MU cassette | 12 |
| JME743 (DH5α) | 1.6-kb PCR fragment containing ylGUT2PT cassette in pCR4Blunt-TOPO | This work |
| JME641 (DH5α) | 1.3-kb PCR fragment containing ylLEU2PT cassette in pDrive | This work |
| MTLE34 (DH5α) | 1.6-kb PCR fragment containing ylPOX1PT cassette in pBluescript KS+ | This work |
| MTLE35 (DH5α) | 1.6-kb PCR fragment containing ylPOX6PT cassette in pBluescript KS+ | This work |
| JME744 (DH5α) | MU cassette in JME743, ylGUT2PUT cassette | This work |
| JME651 (DH5α) | MH cassette in JME641, ylLEU2PHT cassette | This work |
| MTLE36 (DH5α) | MU cassette in MTLE34, ylPOX1PHT cassette | This work |
| MTLE37 (DH5α) | MU cassette in MTLE35, ylPOX6PHT cassette | This work |
| Y. lipolytica strains | ||
| W29 | MATa WT | 1 |
| Po1d | MATaura3-302 leu2-270 xpr2-322 | |
| JMY330 | MATaleu2-270 xpr2-322 | This work |
| MTLY40 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 | 21 |
| MTLY64 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 leu2::hph | This work |
| MTLY66 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 Δleu2 | This work |
| MTLY82 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 Δleu2 pox1::hph | This work |
| MTLY85 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 Δleu2 Δpox1 | This work |
| MTLY94 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 Δleu2 Δpox1 pox6::hph | This work |
| MTLY97 | MATaura3-302 xpr2-322 Δpox2 Δpox3 Δpox4 Δpox5 Δleu2 Δpox1 Δpox6 | This work |
| JMY1202 | MATaura3-302 leu2-270 xpr2-322 Δgut2::URA3 | This work |
| JMY1351 | MATaura3-302 xpr2-322 Δleu2 Δpox1-6 Δgut2::URA3 | This work |
FIG. 2.
Schematic representation of strain construction. Auxotrophic strain Po1d (Leu− Ura−) was derived from WT strain W29. Strain JMY1202, carrying a disrupted G3PDH gene (gut2::URA3), was obtained by transformation of the PUTgut2 cassette into Po1d. Strain MTLY40, which contains four deletions of POX genes that code for acyl-CoA oxidases, was constructed by successive gene disruptions (21, 29). Additional deletions of the two remaining POX genes (POX1 and POX6) were introduced by successive gene disruptions and marker rescue according to Fickers and coworkers (12); i.e., (i) a deletion of the LEU2 gene (leu2::hyg) was introduced into MTLY40; (ii) the marker was rescued after transformation with replicative plasmid pRRQ2, followed by plasmid loss; (iii) the POX1 gene was deleted with the PHTpox1 disruption cassette; (iv) the marker was rescued as in step ii; (v) the POX6 gene was deleted with the PHTpox6 disruption cassette; (vi) the marker was rescued as for step ii; and finally (vii) the G3PDH gene was deleted. Strain JMY1351 contained complete deletions of the six POX genes (Δpox1-6), together with the GUT2 deletion. Control strain JMY330 was obtained by transformation with the p62-URA3 cassette containing the URA3 marker from plasmid JMP62.
Media and growth conditions for Escherichia coli were described by Sambrook et al. (27), and those for Y. lipolytica were described by Barth and Gaillardin (5). Rich medium (YPD), minimal glucose medium (YNB), and minimal medium with Casamino Acids (YNBcas) were prepared as described previously (21). Minimal medium (YNB) contained 0.17% (wt/vol) yeast nitrogen base (without amino acids and ammonium sulfate, YNBww; Difco, Paris, France), 0.5% (wt/vol) NH4Cl, 0.1% (wt/vol) yeast extract (Bacto-DB), and 50 mM phosphate buffer (pH 6.8). Glucose medium YNBD (2%, wt/vol; Merck, Fontenay-sous-Bois Cedex, France) was supplemented with oleic acid from Merck (60% purity), and glycerol medium YNBG (2%, wt/vol; Merck) and oleic acid media YNBDO and YNBOup and were supplemented with oleic acid from Fluka (98% purity). Uracil (0.1 g/liter) and leucine (0.2 g/liter) were added when required. For solid media, 1.5% agar was added. Oleic acid was emulsified by sonication in the presence of 0.02% Tween 40 (21).
The YNBO (YNBD0.5O3) medium used for following optimum accumulation and remobilization is YNB with 0.5% glucose and 3% oleic acid. The YP2D4O3 medium used for optimum lipid accumulation contained yeast extract (1%), proteose peptone (2%), glucose (4%), and oleic acid (3%).
Typically, cultivation was performed as follows. From a YPD plate, a first preculture was inoculated into YPD (15 ml in 50-ml Erlenmeyer flasks, 170 rpm, 28°C, 6 h). Cells from the preculture were used to inoculate the second preculture in YNBD medium (50 ml in 500-ml Erlenmeyer flasks, 170 rpm, 28°C, overnight). For the experimental culture, exponentially growing cells from the overnight culture were harvested by centrifugation and resuspended in fresh YNB medium to an optical density (A600) of 0.5.
To determine cell growth, cultures were centrifuged at 10,000 × g for 10 min and the cell pellet was washed twice with equal volumes of SB solution (9 g/liter NaCl-0.5% bovine serum albumin). Biomass production was determined by measuring A600 and by estimation of the cell DW (drying at 80°C/24 h).
General genetic techniques.
Standard molecular genetic techniques were used throughout this study (27). Restriction enzymes were obtained from Eurogentec S.A. (Liège, Belgium). Yeast cells were transformed by the lithium acetate method (19). Genomic DNA from yeast transformants was prepared as described by Querol et al. (25). PCR amplifications were performed on an Eppendorf 2720 thermal cycler with both Taq (Promega, Madison, WI) and Pfu (Stratagene, La Jolla, CA) DNA polymerases. PCR fragments were purified with a QIAgen Purification Kit (Qiagen, Hilden, Germany), and DNA fragments were recovered from agarose gels with a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). The Staden package of programs (8a) was used for sequence analysis.
Deletion of the GUT2, LEU2, POX1, and POX6 genes, expression of Cre recombinase, and marker excision.
The deletion cassettes were typically generated by PCR amplification according to Fickers and coworker (12). First, the promoter (P) and terminator (T) regions were amplified with Y. lipolytica W29 genomic DNA as the template and the gene-specific P1/P2 and T1/T2 oligonucleotides as primer pairs (Table 3). Primers P2 and T1 contained an extension in order to introduce the IsceI restriction site.
TABLE 3.
Primers used in this studya
| Primer | Sequence (5′→3′) | Restriction site introduced |
|---|---|---|
| G3P-ver1 | GAATGACGGGGGCAACGCAG | |
| G3P-P1 | GCAGATCCACTGTCAAGCCG | |
| G3P-P2 | GCTAGGGATAACAGGGTAATGCGGTAGGAAAGAGAAGTTCCGCG | I-SceI |
| G3P-T1 | GCATTACCCTGTTATCCCTAGCCGGACTATTTCCCCGCAGC | I-SceI |
| G3P-T2 | GCAGCCAGCAGCACGTAGTAG | |
| G3P-ver2 | CAGCAGCCACAAATAGCAGACTGCC | |
| LEU2-P1 | AATCTAGATGGTCACAGTGGAATCATGTTCGTGG | XbaI |
| LEU2-P2 | CATTACCCTGTTATCCCTAGGTTCCATTGTGGATGTGTGTGGTTG | I-SceI |
| LEU2-T3 | CTAGGGATAACAGGGTAATGCTCTGGGTCTGCTGCCCTC | I-SceI |
| LEU2-T4 | AGTAAGCTTAGATCTGTTCGGAAATCAACGGATGCTCAACC | HindIII |
| POX1-ver1 | ATCCAGACCTCCAGGCGGG | |
| POX1-P1 | CATGGAGTGGATCGCTCGAGGACG | |
| POX1-P2 | GCATTACCCTGTTATCCCTAGCCAGGAGGATCGGTGAATGTG | I-SceI |
| POX1-T1 | GCTAGGGATAACAGGGTAATGCCTTGTTCCGAGAAGAGGAGGACG | I-SceI |
| POX1-T2 | CGGCAGTGGCTCACCAAGC | |
| POX1-ver2 | GCTGCGTCTCAATCTGGCGAATG | |
| POX6-ver1 | GCTCAAGAAGGTAGCTGAGTC | |
| POX6-P1 | CCAAGCTCTAAGATCATGGGGATCCAAG | |
| POX6-P2b | GCATTACCCTGTTATCCCTAGCGTTGAGGGACTGTTGAGAGAG | I-SceI |
| POX6-T1b | GCTAGGGATAACAGGGTAATGCGATGAGGAAATTTGCTCTCTTGAGG | I-SceI |
| POX6-T2 | ATCTCGAGATTGGTCCCCTCAAACACAC | |
| POX6-ver2 | CATTAAGTGTCAGATCAGCTCGC |
Underlined sequences correspond to introduced restriction sites. Primers P1, P2, T1, and T2 were used for the construction of disruption cassettes. Primers ver1 and ver2 were used for the verification of gene disruption by PCR amplification of the genomic loci. Primers P2 and T1 contained the sequence of the IsceI endonuclease.
For the GUT2 (G3PDH) gene, primer pairs G3P-P1/G3P-P2 and G3P-T1/G3P-T2 were used. The P and T regions were purified and used for the second PCR. The resulting PT fragment was cloned into pCR4Blunt-TOPO, yielding the JME743 construct. The URA3 marker was then introduced at the IsceI site, yielding the JME744 construct containing the GUT2-PUT cassette.
For the LEU2 gene, primer pairs LEU2-P1/LEU2-P2 and LEU2-T1/LEU2-T2 were used. P1 was designed to introduce an XbaI restriction site at the 5′ end of the P fragment, whereas T2 was designed to introduce a HindIII site. After amplification, the PT PCR fragment was digested with XbaI and HindIII and cloned into pDRIVE at corresponding sites, yielding the JME641 construct. The hph marker was then introduced, giving rise to JME651.
For the POX1 and POX6 genes, the PT sequences were amplified with their specific primers and cloned into Bluescript KS+ as a blunt-ended fragment. After transformation into DH5α on LB agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), white colonies were selected. Strains MTLE34 and MTLE35 contained plasmids carrying the POX1-PT and POX6-PT cassettes, respectively. Strains MTLE36 and MTLE37 contained corresponding plasmids POX1-PHT and POX2-PHT.
The disruption cassettes (PUT and PHT) obtained by PCR were used for transformation by the lithium acetate method (4). Transformants were selected onto YNBcasa and YPDH, respectively. Typically, about 5 ×103 transformants were obtained per μg of PCR fragments. Four to eight transformants were analyzed by PCR with ver1/ver2 primer pairs (Table 3), giving more than 50% of the clones with correct gene disruption, which was confirmed by Southern blotting, and then marker rescue was performed according to Fickers and coworkers (12).
Light microscopy.
For light microscopy investigations, cells from 10 ml of a growing yeast culture were prefixed by addition of 1.34 ml of formaldehyde stock solution (50 mM sodium phosphate buffer, pH 6.8; 0.5 mM MgCl2; 4.8% formaldehyde) and further incubated for 1 h at 28°C with shaking at 250 rpm. Prefixed cells were harvested, resuspended to an A600 of 2.5 in the formaldehyde stock solution, and incubated for 5 h at room temperature. Cells were washed twice with 50 mM sodium phosphate buffer (pH 6.8) and stored in 0.1 M K-Pi buffer (pH 7.5) at an A600 of 2.5 at 4°C until light microscopy observation.
Fluorescence microscopy.
For visualization of LB, Nile red (1 mg/ml solution in acetone; Molecular Bioprobe, Montluçon, France) was added to the cell suspension (1/10, vol/vol) and incubated for 1 h at room temperature. Cells were harvested, washed twice with distilled water, and resuspended in 50 mM sodium phosphate buffer (pH 6.8) to an A600 of 2.5. Microscopy was performed with an Olympus BX 51 fluorescence microscope with a 100× oil immersion objective. Photometrics CoolSNAP software was used for recording the images.
Lipid determination.
Lipids from the equivalent of a culture optical density of 10 were either extracted by the procedure of Folch et al. (13) for thin-layer chromatography (TLC) analysis or directly converted into their methyl esters with freeze-dried cells according to Browse et al. (7) and used for gas chromatography (GC) analysis. Complete transmethylation was verified by the BF3-in-methanol method (3) and on TLC plates. GC analysis of FA methyl esters was performed with a Varian 3900 instrument equipped with a flame ionization detector and a Varian FactorFour vf-23ms column, where the bleed specification at 260°C is 3 pA (30 m, 0.25 mm, 0.25 μm). FA were identified by comparison to commercial FA methyl ester standards (FAME32; Supelco) and quantified by the internal standard method by adding 50 μg of commercial C17:0 (Sigma). A C16:1(n-9) methyl ester standard was purchased from Cayman, France.
LB quantification and analysis.
LB analysis was performed by microscopic observation of a growing cell culture as described above. Nomarski (differential interference contrast) optics were used to record transmission images. LB numbers and volumes at various stages of growth were calculated from images at different focuses. Ten representative cells were selected from light microscopy images to determine LB number and size. This analysis was performed with ImageJ picture processing software (http://rsb.info.nih.gov/ij/).
Lipid fractionation.
Total lipids were fractionated in TAG and FFA for lipid class quantification with an Isolute SPE Aminopropyl column (IST, France) according to reference 18. Column conditioning was performed three times with 3 ml of hexane at a normal flow rate. One milliliter of total lipids extracted by the Folch method in CHCl3 was loaded onto the column, and the fraction of neutral lipids was collected. Total elution of neutral lipids was performed by washing the column three times with 3 ml of CHCl3-isopropanol (2/1). The FFA fraction was collected by washing the column three times with 3 ml of diethyl ether-2% acetic acid at a normal flow rate. Fraction solvent was evaporated under N2 flux, and direct transmethylation was followed for GC analysis (18). TLC plates were used for extraction verification. The efficiency of the procedure was further verified by comparison of the GC profiles of fractionated and unfractionated samples.
TLC separation of lipid classes.
Precoated TLC plates (silica G60, 20 by 20 cm, 0.25-mm thickness) from Merck (Germany) were used. Lipid classes were separated with a two-development solvent system. System A (half-plate migration) consisted of petroleum ether-ethyl ether-acetic acid at 20/20/0.8 (vol/vol/vol). System B (whole-plate migration) consisted of petroleum ether-diethyl ether at 49/1 (vol/vol). A 5% phosphomolybdic acid solution was sprayed onto the plates, and lipid bands were revealed after 10 min at 105°C.
RESULTS
Genomic comparison of lipid accumulation and degradation pathways between Saccharomyces cerevisiae and Y. lipolytica.
Genes involved in lipid accumulation and mobilization pathways in yeast have been recently reviewed (8). In order to compare those genes between S. cerevisiae (a nonoleaginous yeast) and Y. lipolytica (an oleaginous yeast), we used the S. cerevisiae protein sequences at the SGD site (http://www.yeastgenome.org/) and blasted them at the Génolevures site (http://cbi.labri.fr/Genolevures/). Results are summarized in Table 1, including gene names, S. cerevisiae and Y. lipolytica sequence designations, EC numbers, percentages of amino acid identity, and putative enzyme functions. For the first step catalyzed by the glycerol kinase, a single gene (GUT1) was found in both yeasts. The Y. lipolytica homologue was encoded by YALI0F00484g, presenting 35% amino acid identity. However, functional analysis was performed with mutants lacking GUT1 activity in order to confirm gene identity (R. Haddouche et al., unpublished data). The enzyme catalyzing the conversion of G3P to DHAP was encoded by YALI0B13970g in Y. lipolytica (GUT2, 44% amino acid identity). Two genes were present in S. cerevisiae (GPD1 and GPD2) for the conversion of DHAP to G3P as the opposite reaction, while only one gene was found in Y. lipolytica, presenting 52 and 61% amino acid identity, respectively. In S. cerevisiae, two genes, SCT1 and GPT2, were shown to code for proteins showing G3P acyltransferase activity. A single corresponding homologue, highly similar to S. cerevisiae SCT1 (44% amino acid identity; YALI0C00209g), was found in Y. lipolytica.
In both yeasts, a single gene encodes 1-acyl-sn-G3P acyltransferase; the Y. lipolytica homologue was YALI0E18964g (ylSLC1). Similarly, genes that code for DAG acyltransferase (DGA1) and phospholipid:DAG acyltransferase (LRO1) were each encoded by a single gene in both yeasts (YALIOE32769g and YALI0E16797g, respectively). Only one SE synthase (ARE1), homologous to are2p of S. cerevisiae, has been identified as being encoded by YALIOF06578g. Apart from amino acid identity, acyltransferase gene identity was confirmed by functional and enzymatic analyses (unpublished data). Additionally, the overexpression or deletion of the different acyltransferases present in Y. lipolytica gave interesting results regarding enzymatic specificity and TAG formation (A. Beopoulos et al., unpublished data). Three genes that code for triacylglycerol lipases (TGL3, TGL4, and TGL5) are present in S. cerevisiae for the mobilization of TAG, while only two genes were found in Y. lipolytica (ylTGL2 and ylTGL3, encoded by YALY0D17534g and YALI0F10010g, respectively). For the β-oxidation process, Y. lipolytica contains six genes (POX1 to POX6) that code for acyl-CoA oxidases (Aox) with different substrate specificities and different activity levels: Aox3p is specific for short-chain acyl-CoAs, Aox2p preferentially oxidizes long-chain acyl-CoAs, and Aox4p and Aox5p activities do not appear to be sensitive to the length of the aliphatic chain of CoA. Aox1p and Aox6p are specific for the degradation of dicarboxylic acids (F. Thevenieau et al., unpublished data). In contrast, S. cerevisiae contains only one POX gene, presenting 33 to 35% amino acid identity with all Y. lipolytica acyl-CoA oxidases. The multifunctional β-oxidation enzyme and thiolase are each encoded by a single gene in both yeasts. In Y. lipolytica, the multifunctional enzyme is encoded by YALI0E15378g and thiolase is encoded by YALI0E18568g. Several enzymes of the TAG synthesis pathway were observed in the proteome of the LB (2), as shown in Fig. 1.
Construction of the strain with a deletion in the GUT2 gene.
A GUT2-PUT disruption cassette was constructed as described in Materials and Methods (JME744, Table 2). This cassette contains the promoter and terminator regions of the GUT2 gene and allowed the deletion of the complete 1.2-kb region carrying the open reading frame that codes for G3PDH. The GUT2 deletion was first introduced by transformation into Po1d, a derivative of WT strain W29 (Table 2 and Fig. 2). The gene deletion was verified by PCR with primers ver1 and ver2 and Southern blotting (data not shown). One of the strains containing a disrupted copy of GUT2 (Δgut2) was designated JMY1202 (Δgut2::URA3 Leu− Ura+). As a WT control, a strain showing the same auxotrophy was constructed by transformation of Po1d with the URA3 cassette, yielding the construct JMY330 (Leu− Ura+) (Fig. 2 and Table 2).
Growth phenotype dependence on carbon source of a mutant with a deletion of G3PDH (Δgut2).
We first analyzed growth on solid media containing different carbon sources. On glucose medium (YNBD agar), the Δgut2 and WT strains showed similar growth and morphology. As expected, the Δgut2 strain did not grow on glycerol medium, as this substrate can no longer be used in the glycolytic cycle (Fig. 1). On YNBT plates, the Δgut2 strain demonstrated reduced growth (data not shown). Growing the strains in liquid media (YNBD, YNBG, and YNBO) confirmed the results of plate tests (data not shown).
Lipid accumulation in a mutant with a deletion of G3PDH (Δgut2).
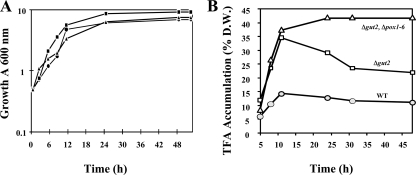
To determine the effect of a GUT2 gene deletion on lipid accumulation and mobilization, control strain JMY330 and mutant strain JMY1202 were grown in glucose-based (YNBD) and oleic acid-glucose-based (YNBD0.5O3) media. The two strains showed similar growth rates on YNBD (data not shown). In contrast, the Δgut2 strain showed a slightly higher growth rate (μmax of 0.470 ± 0.063 h−1) than the control strain (μmax of 0.350 ± 0.027 h−1, Fig. 3A) in YNBD0.5O3. Lipid accumulation increased during the first 11 h in both strains grown on both media. Reaching the stationary growth phase, microbial lipids were mobilized, resulting in a decrease in total FA (TFA) content in both strains. In YNBD, TFA constituted 8.7% of the cell DW of the Δgut2 strain at 11 h, compared to 7.0% for the control, which corresponds to a 1.2-fold increase (data not shown). However, when stationary phase was reached in glucose-based medium (i.e., 24 h after inoculation), both strains showed a tendency to reduce slightly the quantity of their total synthesized lipids. Hence, the TFA level in the Δgut2 strain was at 6.9% (wt/wt), which remains 1.2-fold higher than in the control strain (5.7%). In both instances, the TFA contents were lower compared to the levels reached in the initial growth phases (Fig. 4). In contrast, accumulation of storage lipid was greatly enhanced in the Δgut2 mutant strain when oleic acid was used as a glucose cosubstrate (medium YNBD0.5O3) (Fig. 3B and 4). As in YNBD, the maximum lipid contents were observed at 11 h. At this time point, the Δgut2 strain grown in YNBD0.5O3 contained 37.0% (wt/wt) lipid, compared to only 14.1% TFA content in the WT strain, corresponding to a 2.6-fold increase (data not shown). Microbial lipid turnover was observed in later phases since the TFA content decreased to 12.8 and 31.6% (Fig. 3A and B) in the WT and Δgut2 strains, respectively.
FIG. 3.
Growth (A) and total lipid accumulation (B) in the WT strain and in mutant strains with altered GUT2 and POX genotypes in YNBD0.5O3. The growth of the WT and mutant strains listed in Table 2 was monitored over time (A). TFA accumulation, expressed as a percentage of cell DW, is shown in panel B. Symbols: circles, WT strain; squares, Δgut2 mutant strain; triangles, Δgut2 Δpox1-6 mutant strain. Closed symbols correspond to growth expressed as optical density at 600 nm. Open symbols correspond to TFA content. The results are mean values from three independent experiments. The standard deviations were <10% of the respective values.
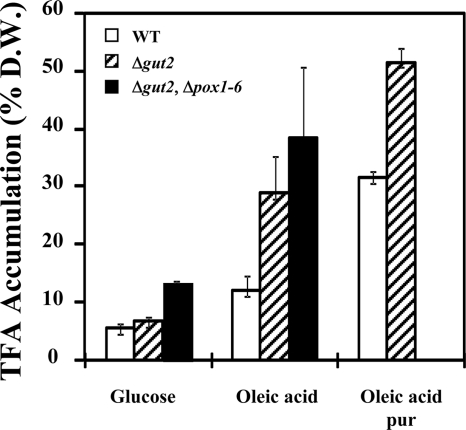
FIG. 4.
TFA content at 24 h. The TFA contents of the WT and mutant strains are expressed as percentages of cell DW at the end of cell cultivation (24 h). Strains were grown on YNBD, YNBD0.5O3, and YNBD0.5O3up. The results represent mean values from three independent experiments. Open bars, WT strain; hatched bars, Δgut2 strain; black bars, Δgut2 Δpox1-6 strain; pur, ultrapure (>98% purity).
Lipid accumulation depends on the FA composition of the medium.
Previous investigations have indicated that during growth of Y. lipolytica strains on hydrophobic substrates, the process of lipid accumulation is dramatically influenced by the FA composition of the culture medium (2, 24). The present study was aimed at evaluation of the potential of lipid accumulation in the WT and Δgut2 strains grown on YNBD0.5O3up medium containing ultrapure oleic acid and comparison of the results with those obtained with medium containing low-purity oleic acid (Fig. 4). Maximum TFA levels observed at 11 h reached 50.0% of the DW of the WT strain and 70.1% of the DW of the Δgut2 mutant (data not shown). After this time point, a decrease of approximately 30% in lipid content was observed in both strains due to lipid remobilization. At 24 h, the TFA content was 31.5 and 51.5% of the DW for the WT and Δgut2 strains, respectively (Fig. 4 and Table 4). For the WT, at this time point the lipid content represents a 2.5-fold increase compared to the lipid accumulation at 24 h in YNBO (12.8% of DW). A similar pattern was observed at 24 h for the Δgut2 strain, with an increase from 31.6 to 51.5%, i.e., a 1.7-fold increase. It may be thus assumed that the FA composition of the culture medium has a crucial effect upon the lipid accumulation process in both the WT and genetically engineered Y. lipolytica strains used in the present study.
TABLE 4.
FA profile and accumulationa
| Strain and medium | Avg FA accumulation (% of TFA)
|
TFA (% of DW) | |||||
|---|---|---|---|---|---|---|---|
| C16:0 | C16:1(n-9) | C16:1(n-7) | C18:0 | C18:1(n-9) | C18:2 | ||
| WT | |||||||
| YNBD | 11.70 | 1.68 | 15.01 | 0.24 | 13.77 | 47.43 | 5.71 |
| YNBD0.5O3 | 6.28 | 1.70 | 0.15 | 0.63 | 63.73 | 23.75 | 12.76 |
| YNBD0.5O3up | 2.71 | 3.65 | 0.13 | 0.23 | 76.68 | 15.25 | 31.50 |
| Δgut2 | |||||||
| Glu | 13.29 | 4.09 | 15.72 | 0.69 | 12.06 | 42.96 | 6.87 |
| YNBD0.5O3 | 7.15 | 14.90 | 0.20 | 0.80 | 56.20 | 20.01 | 31.60 |
| YNBD0.5O3up | 2.00 | 18.25 | 0.17 | 0.36 | 68.77 | 8.42 | 51.53 |
| Δgut2 Δpox1-6 | |||||||
| Glu | 12.90 | 0.16 | 27.30 | 0.25 | 11.56 | 36.16 | 12.89 |
| YNBD0.5O3 | 6.10 | 0.05 | 0.00 | 0.70 | 61.25 | 22.39 | 41.92 |
FA profiles of WT and mutant strains as percentages of the total FA accumulated are shown. Strains were grown on YNB medium with addition of 2% glucose or 3% oleic acid. The values are means from five different experiments at 24 h of culture. The standard deviations were <5% of the values shown. The last column represents the total lipid accumulation as a percentage of the cell DW. Supplements in growth media: glucose, YNBD; oleic acid (65% purity), YNBD0.5O3; ultrapure oleic acid (>98% purity), YNBD0.5O3up.
FA profile of the Δgut2 strain.
The FA profiles of the WT and Δgut2 strains were analyzed during growth in YNBD, YNBD0.5O3, and YNBD0.5O3up media. Quantification of major lipid components at 24 h of culture is shown in Table 4. For WT Y. lipolytica cells cultivated in glucose medium, the major FA in the lipid fraction was linoleic acid [C18:2(n-6), 47%]. Several other FA were present at similar levels of about 10 to 15%. They included palmitic acid (16:0, 11.7%), palmitoleic acid [16:1(n-7), 15%], and oleic acid [18:1(n-9), 13.8%]. Stearic acid (18:0, 0.2%) and (Z)-7-hexadecenoic acid [16:1(n-9), 1.7%] were minor constituents of TFA. It should be mentioned that the 16:1(n-9) FA corresponds to the derivative of oleic acid (18:1) formed after one cycle of β-oxidation. The ratio of C16 to C18 acyl chain FA was 0.46, and the ratio of unsaturated to saturated FA reached 6.5 after 24 h of culture.
For the Δgut2 strain cultivated on glucose-based medium, the profile of total cellular FA was very similar to that of the WT strain, except for the level of 16:1(n-9), which was about 2.5-fold higher (4.1%) compared to that of the WT strain (1.7%; Table 4).
In oleic acid-based medium, the WT preferentially accumulates oleic acid [18:1(n-9), 64%] and linoleic acid [18:2(n-6), 24%]. C18:2 levels increased slightly during growth in both media, while C18:1 levels showed the opposite trend.
Incorporation of oleic acid into complex lipids led to a strong increase in the unsaturated-to-saturated FA ratio. In glucose-based media, the ratio was 6.5 and 5.3 for the WT and Δgut2 strains, respectively. In oleic acid-supplemented medium, this ratio was 12.9 and 11.4, respectively, for the WT and Δgut2 strains. This ratio was even higher for both strains when cultivation was performed with pure oleic acid utilized as a glucose cosubstrate (32 and 40, respectively).
Deletion of GUT2 significantly modified the C16/C18 acyl chain ratio. For the WT, the C16/C18 ratio decreased from 0.46 in glucose to 0.09 in oleic acid medium. In contrast, the Δgut2 strain showed a decrease in this ratio from 0.59 to 0.29.
When the strains were shifted to ultrapure oleic acid (YNBD0.5O3up), the ratio of unsaturated to saturated FA reached a value of 35. An increase in (Z)-7-hexadecenoic acid [16:1(n-9)] content was also observed under these conditions. This FA represented 3.7% of the total lipid content of the WT, twice the level observed in YNBD0.5O3 (Table 4). The concentration of this FA also increased in the mutant, representing more than 18% of the total lipid content. This confirmed that this FA originates from the degradation of oleic acid.
Construction of a Δgut2 Δpox1-6 mutant strain and its growth phenotype.
To confirm that the 16:1(n-9) FA results from oleic acid degradation and to further increase lipid accumulation, the GUT2 deletion was introduced into a strain with deletions of the POX genes that code for the acyl-CoA oxidases. Six POX genes have been identified in the Y. lipolytica genome (10). We previously reported the construction of strain MTLY37, a quadruple mutant carrying Δpox2, -3, -4, and -5 deletions (29). A uracil auxotroph derivative, MTLY40, was constructed as described in Materials and Methods (21) (Fig. 2). In order to delete the two remaining POX genes (POX1 and POX6), POX1-PHT and POX6-PHT disruption cassettes were constructed as described in Materials and Methods. In six successive steps, the final strain, MTLY95a, was obtained, which contained a complete deletion of all six POX genes. The Δgut2 deletion was then introduced, resulting in strain JMY1351 (Δgut2 Δpox1-6) (Fig. 3 and Table 2). This mutant is able to grow neither on glycerol medium, due to the Δgut2 deletion, nor on oleic acid medium, due to the complete block of β-oxidation resulting from the deletion of the six POX genes (Δpox1-6) (data not shown). However, growth on glucose medium was not affected.
Growth, TFA levels, and FA profiles were analyzed during growth in YNBD and YNBD0.5O3 (Fig. 3 and 4 and Table 4). The Δgut2 Δpox1-6 strain showed growth similar to that of the WT strain. Lipid content increased similarly as in the Δgut2 strain during the first 11 h of growth. However, in the subsequent time period, lipid could not be mobilized in the Δgut2 Δpox1-6 strain, in contrast to the other strains, and the TFA level even increased from 35.5 to 41.9% of the cell DW at 24 h of growth on oleic acid medium (Fig. 3B and Table 4). The TFA level remained constant until the end of the cultivation period at 52 h (Fig. 3B). When grown on glucose minimal medium, TFA accumulation at 24 h increased 2.2-fold compared to that of the WT strain and 1.9-fold compared to that of the Δgut2 strain (Fig. 4 and Table 4). The lipid profile was very similar to that of strains previously tested, except for the absence of C16:1(n-9) accumulation, confirming the origin of this isomer in β-oxidation (Table 4). The unsaturated-to-saturated FA ratio increased from 5.7 to 12.3 when the strains were shifted from glucose to oleic acid medium, and the C18 to C16 aliphatic chain ratio increased from 1.19 to 14.3.
LB morphology of Y. lipolytica WT and mutant strains.
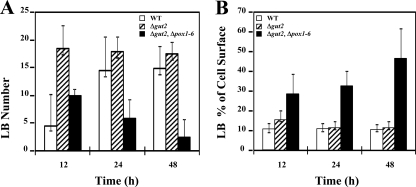
LB can be easily visualized by light microscopy of formaldehyde-fixed cells or by fluorescence microscopy of cells stained with Nile red (see Materials and Methods). No significant variations were observed in either the number or the size of LB formed in strains grown on glucose-based media, where only a few small LB were observed (data not shown). After 24 h of growth on oleic acid medium, the Δgut2 Δpox1-6 strain contained one to three large LB and a few smaller ones (Fig. 5E and F), while high numbers of small LB were found in the Δgut2 strain (Fig. 5C and D). Lower numbers of small-sized LB were formed in the WT strain (Fig. 5A and B). Inspection of Δgut2 Δpox1-6 cells over time revealed numerous small LB in early culture stages, which then probably started to coalesce, resulting in one or a few large LB. The total LB content reached 40 to 60% of the cell area. When the Δgut2 Δpox1-6 strain was shifted to rich oleic acid medium (YP2D4O3), one or two large LB occupied up to 70% of the cell area and cells appeared bigger (data not shown). Overall, a good correlation was observed between the percentages of the LB surface/cell surface ratio and the TFA content measured by GC analysis. The time course of the changes in the number of LB and the area they occupied in the cell is shown in Fig. 6.
FIG. 5.
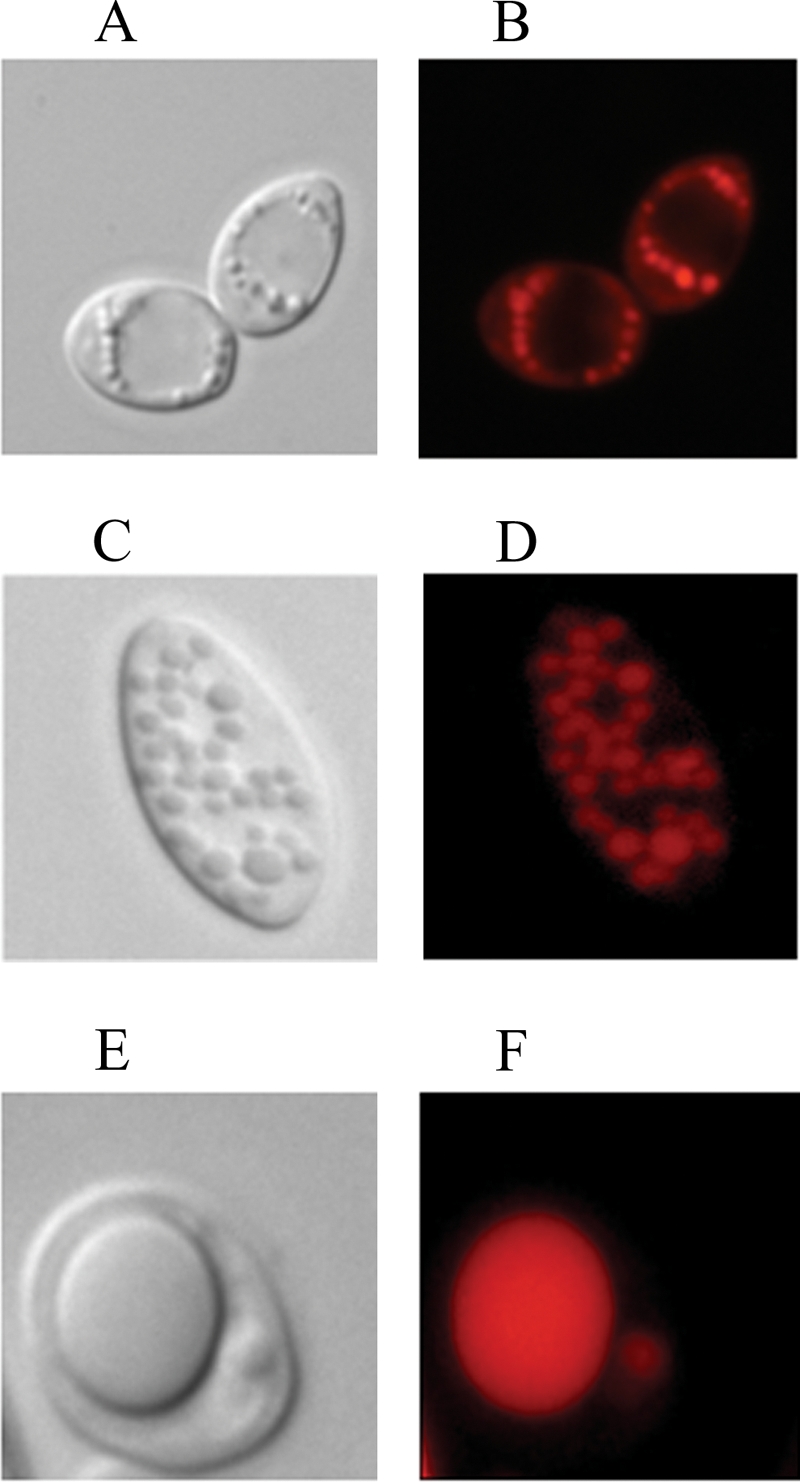
LB phenotypes of mutant strains. LB morphologies of the WT (A, B), Δgut2 (C, D), and Δgut2 Δpox1-6 (E, F) strains are shown. Light microscopy images are shown in panels A, C, and E, and fluorescence microscopy images of Nile red-stained cells are shown in panels B, D, and F. Strains were grown for 24 h on YNBD0.5O3 medium.
FIG. 6.
Changes in LB number and cell area occupied by LB over time. The mean LB counts over time in the WT and mutant strains are shown in panel A. Panel B represents the percentage of the cell area occupied by LB. Strains were grown on YNBD0.5O3 medium. Standard deviations of measurements are also shown. Open bars, WT strain; hatched bars, Δgut2 strain; black bars, Δgut2 Δpox1-6 strain.
Analysis of lipid classes and lipid profiles in Y. lipolytica strains.
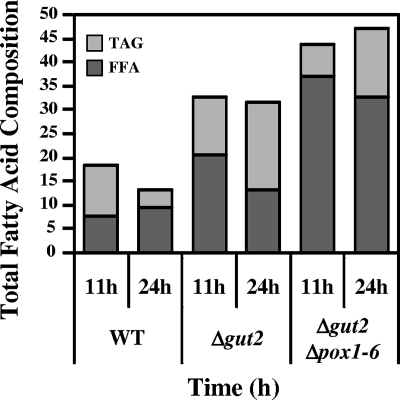
The WT and mutant strains were grown in accumulation medium YP2D4O3, and the total lipid (TFA) content was determined at 11 and at 24 h (Fig. 7). Lipid analysis by TLC revealed that lipids accumulated mainly in the form of FFA and TAG. SE, monoglycerides, and diglycerides were present in very small amounts (data not shown). TFA were further analyzed after SPE fractionation and determination of the proportions of TAG and FFA. WT cells contained 60% TAG and 40% FFA at 11 h after inoculation. After 24 h, the proportions were reversed due to the mobilization of TAG, yielding more FFA (74% of the TFA content). The Δgut2 strain accumulated a similar total amount of lipid at 11 h; however, this strain accumulated more FFA, resulting in 40% of the TFA as TAG and 60% as FFA. In this medium, TFA slightly decreased at 24 h due to lipid mobilization. However, the FA were able to accumulate as TAG, which then represented 58% of the TFA at 24 h. The Δgut2 Δpox1-6 mutant was found to accumulate lipids mainly as FFA, representing 83% of the TFA content at 11 h. Only a small amount of these FFA were stored as TAG in the later phase of growth, thus representing only 31% of the TFA content at 24 h. An analysis of the FA composition of the TAG fraction and of the FFA fraction is shown in Table 5. We observed that TAG in the WT strain and in the Δgut2 Δpox1-6 mutant contained mainly C18:1 and C18:2, while in the Δgut2 strain we also found significant levels of C16:1(n-9). A Low levels of saturated FA were also observed in TAG.
FIG. 7.
TAG and FFA contents as fractions of total lipids. The graph represents the total lipid accumulation over time (11 and 24 h) as a percentage of the DW of cells grown on YP2D4O3 medium. Lipids were fractionated into TAG and FFA by solid-phase extraction, and quantification was done by GC.
TABLE 5.
Lipid class profiles of WT and mutant strainsa
| FA | WT
|
Δgut2
|
Δgut2 Δ pox1-6
|
|||
|---|---|---|---|---|---|---|
| % TTAG | % TFFA | % TTAG | % TFFA | % TTAG | % TFFA | |
| C16:0 | 2.25 | 9.74 | 5.15 | 14.00 | 4.45 | 6.56 |
| C16:1(n-9) | 2.08 | 0.95 | 14.76 | 3.95 | 0.25 | 0.06 |
| C16:1(n-7) | 0.00 | 0.07 | 0.19 | 0.00 | 0.12 | 0.09 |
| C18:0 | 0.62 | 5.62 | 1.10 | 9.77 | 0.72 | 1.94 |
| C18:1(n-9) | 44.92 | 67.71 | 50.16 | 58.64 | 63.58 | 70.31 |
| C18:2(n-6) | 31.32 | 9.73 | 22.54 | 9.60 | 24.93 | 17.26 |
| Total | 81.19 | 93.82 | 94.11 | 96.96 | 94.04 | 96.22 |
Shown are the FFA, TAG, and total lipid profiles of WT and mutant cells grown for 24 h on YNBO3 medium. The columns represent FFA and TAG as percentages of the total FFA (TFFA) and TAG (TTAG), respectively.
DISCUSSION
Genomic comparison and strategy definition.
In yeasts, acyl-CoA can follow two separate pathways during lipid metabolism, either toward SE formation or toward phosphate substitution in the activated glycerol backbone, resulting in TAG formation (glycerol pathway). Lipid profile and genomic comparison of Y. lipolytica and S. cerevisiae strongly suggests that lipid metabolism of Y. lipolytica is oriented toward the glycerol pathway; Y. lipolytica encodes only one homologue of ARE1 sterol acyltransferase, and SE accumulation is negligible, especially compared to that in S. cerevisiae. We believe that in the glycerol pathway, the availability of G3P as the initial substrate for TAG synthesis is crucial for lipid accumulation. G3P can be formed directly from glycerol or from DHAP. The formation of G3P from DHAP is catalyzed by Gut2p. The conversion of DHAP to G3P is catalyzed by a unique G3P dehydrogenase isomer (Gpd1p), while two different genes exist in S. cerevisiae. In the present study, we attempted to increase lipid accumulation by increasing the G3P levels in the cell. Therefore, we deleted the GUT2 gene that codes for the isomer catalyzing the conversion of G3P to DHAP. It is unlikely that another enzyme is capable of catalyzing this step since the Δgut2 mutant strain was unable to grow on glycerol as the sole carbon source. DHAP can be formed only from G3P in glycerol medium, and DHAP is the first precursor of the energy cycle in yeasts. Y. lipolytica contains six genes that code for acyl-CoA oxidases with different substrate specificities and activity levels. This contrasts with the situation in S. cerevisiae, which contains only one corresponding gene. This fact strongly suggests a high specialization of Y. lipolytica in lipid degradation and could contribute to its ability to use the surplus of FFA or alkanes as a carbon source. Y. lipolytica shows a significant mobilization of lipid reserves after its exit from the exponential growth phase. Previously constructed mutant strains of Y. lipolytica lacking the POX2 to POX5 genes (21) showed increased lipid accumulation in the final growth phase. In this study, we identified and deleted the remaining two genes, POX1 and POX6, constructing a Y. lipolytica strain completely incapable of β-oxidation. It is unlikely that other pathways of FA degradation exist in Y. lipolytica as the Δpox1-6 mutant strain shows no remobilization of lipids and lipid analysis revealed no traces of β-oxidation derivatives.
Genetic manipulation of lipid accumulation.
Compared to the WT strain, the Δgut2 mutant demonstrated a vast increase in cellular lipid content in all of the studies carried out so far but especially when oleic acid was used as a (co)substrate in the growth medium. Additionally, a significant increase in the lipid accumulation rate was observed. The maximum quantity of total accumulated lipids was reached at 11 h after inoculation. This also happens to be the transition point from exponential to stationary phase. Typically, accumulation of reserve lipids is the primary anabolic activity in yeast grown on hydrophobic compounds as (co)substrates. Lipid accumulation occurs simultaneously with the formation of nonlipid material, and it is completely independent of the nitrogen availability in the culture medium (1). In the present study, growth on glucose/oleic acid-based media revealed that the Δgut2 mutant strain has a slightly higher growth rate compared to the WT strain, as indicated by the maximum specific growth rate (μmax) values. However, the final growth yield, monitored as A600, was almost the same for both strains and both reached the plateau simultaneously (Fig. 3A). Lipid production was critically affected by the genetic manipulation resulting in significantly higher quantities of total cellular lipids produced by the Δgut2 and Δgut2 Δpox1-6 mutants compared to the WT strain (Fig. 3B and 4). These results provide evidence that G3P availability is of significant importance in the glycerol pathway. Alternatively, G3P may act as a signal on its own, leading to upregulation of FA metabolism (28). Lipid accumulation was also limited by lipid mobilization and a subsequent decrease in storage lipid in the Δgut2 and WT strains. This lipid mobilization occurred in the stationary growth phase in both strains (Fig. 3A and B). In contrast, the Δgut2 Δpox1-6 mutant continued to accumulate lipids in stationary phase, reaching even higher final values due to the absence of mobilization of the accumulated lipid (Fig. 1 and 3A). It should be mentioned that in the case of oleaginous yeasts and molds, biodegradation of the previously produced storage lipid is frequently observed (14, 21). Repression of storage lipid degradation can be achieved by genetic engineering (in the case of the Δgut2 Δpox1-6 mutant), as well as by cultivation conditions, such as using double (or multiply)-limited media for the cultivation of oleaginous microorganisms. In all of these cases, repression of storage lipid breakdown is mediated by inactivation of the enzymes involved in the relevant pathways (21) (i.e., intracellular lipases, enzymes involved in the glyoxylic acid bypass, and acyl-CoA oxidases).
Effect of medium composition on lipid accumulation.
All of the strains analyzed achieved higher lipid accumulation when grown on oleic acid. Compared to the WT strain, the Δgut2 and Δgut2 Δpox1-6 mutants showed three- and fourfold increases in lipid accumulation, respectively. When shifted to ultrapure oleic acid, the Δgut2 mutant accumulated lipids up to more than 70% of its DW. In addition, our results indicate that lipid composition also critically depends on the carbon source used to support lipid accumulation. Y. lipolytica grown on glucose as a carbon source for de novo lipid synthesis preferentially forms C18:2(n-6) FA due to strong expression of endogenous Δ12 desaturase (20). The present study indicates that Y. lipolytica strains grown in oleic acid medium show a positive energy balance toward lipid accumulation. Less energy is used for FA elongation and desaturation reactions. Cells grown in ultrapure oleic acid medium perform only reactions needed for lipid storage and one step of desaturation yielding linoleic acid, resulting in more efficient lipid accumulation. These observations are consistent with previous studies (2) which have shown that the purity and lipid composition of the media directly affected the process of lipid accumulation.
Lipid composition analysis.
Lipid analysis revealed that the Δgut2 mutant stores large amounts of C16:1(n-9), which is a β-oxidation derivative of C18:1. This particular compound reached 20% of the total lipids in cells grown on oleic acid media. In contrast, the WT strain accumulates C16:1(n-9) to less than 3% of its total lipids, regardless of the composition of the medium. Moreover, the Δgut2 Δpox1-6 mutant shows no production of this derivative, which is consistent with complete inhibition of the β-oxidation process. This fact could indicate that G3P levels regulate the expression or the specificity of one or more of the acyl-CoA oxidases involved in lipid catabolism, causing incomplete degradation of the corresponding substrate. Fractionation of accumulated lipids revealed that C16:1(n-9) is found in both the TAG and FFA fractions, suggesting the existence of a transport mechanism carrying the β-oxidation derivatives from the peroxisome to the cytoplasm for TAG acylation. Our results revealed that FFA were the dominant fraction of total cellular lipids in all of the strains studied. In general, when glucose is used as the substrate for lipid synthesis in various oleaginous yeast or molds, neutral lipids accumulate principally as TAG (80 to 90%, wt/wt, of the total lipids). FFA represent only a marginal storage compound under this condition (20). On the other hand, our study showed that growth on medium containing hydrophobic compounds (e.g., FFA) as (co)substrates results in the accumulation of storage lipid containing considerable quantities of FFA (30 to 40%, wt/wt, of the total lipids). Even though FFA in large quantities are considered to be toxic for microbial cells, no lethality or growth deficit was observed. Both mutants showed an increased ratio of TAG to FFA over time, contrary to the WT strain, which had a tendency to reduce TAG (Fig. 7). This observation is consistent with the hypothesis that glycerol is the limiting factor in TAG synthesis by the WT strain. On the other hand, mutant strains accumulate lipids more quickly than they can store them as TAG during exponential growth. This reaction is accelerated upon entry into stationary phase due to increased glycerol availability. The proportion of FFA in the total lipids is even higher in the Δgut2 Δpox1-6 mutant. This could suggest a regulatory mechanism preventing lipid storage in the form of TAG if FA cannot be used as an energy source in the absence of Aox proteins (21). In all of the strains tested, saturated FA are preferably stored as FFA, contrary to C18:2, which, as the final product, is found mostly in the TAG fraction (Table 5).
LB.
Genetic modification of storage lipid metabolism significantly affects the number and form of LB in the strains studied. The WT strain and the Δgut2 mutant tend to form numerous small LB, depending on the accumulation ratio, while Δgut2 Δpox mutant cells show one to three large LB covering 50 to 70% of the cell's cross-sectional area up until the end of cultivation. Since the lipid fraction of the Δgut2 Δpox mutant in this growth phase contains 60% FFA, we may ask if only neutral lipids can be stored in LB. Due to their polar nature and mild detergent capacity, FFA could facilitate LB coalescence, especially since structural proteins such as oleosins and apoproteins were not found in yeast LB (in contrast to plant and mammalian LB). Large LB containing large amounts of FFA should have a specific structure, FFA being at the periphery close to the external phospholipid layer and neutral lipids constituting the central core of the bodies. Alternatively, neutral lipids could be positioned between FFA layers, resulting in a multilayer onion-like structure with a higher storage capacity. Further studies should be performed, including purification and microscopic analysis of LB, to clarify the storage ability of this cellular compartment.
In conclusion, we have proven that lipid accumulation in Y. lipolytica can be strongly influenced by modification of the flux and availability of G3P. The increased proportion of FFA suggests that the acyl-CoA-transferases are the limiting factor in TAG formation. On the other hand, the mobilization of lipid supplies is regulated by the presence of the Aox proteins. Modifications of the POX genotype is useful in preventing lipid degradation and therefore leads indirectly to an increase in lipid accumulation. These modifications make the multiple-deletion mutant Δgut2 Δpox1-6 strain a good candidate for single-cell oil production.
Acknowledgments
A. Beopoulos was partly supported by an ERASMUS fellowship and by an INRA fellowship. Z. Mrozova was the recipient of a Marie Curie training fellowship during her stay at the INRA—Grignon Yeasts of Technological Interest (YETI) training site. We thank Aerospace Valley and Airbus for financial support.
Footnotes
Published ahead of print on 24 October 2008.
REFERENCES
- 1.Aggelis, G., and J. Sourdis. 1997. Prediction of lipid accumulation-degradation in oleaginous micro-organisms growing on vegetable oils. Antonie van Leeuwenhoek 72:159-165. [DOI] [PubMed] [Google Scholar]
- 2.Athenstaedt, K., P. Jolivet, C. Boulard, M. Zivy, L. Negroni, J. M. Nicaud, and T. Chardot. 2006. Lipid particle composition of the yeast Yarrowia lipolytica depends on the carbon source. Proteomics 6:1450-1459. [DOI] [PubMed] [Google Scholar]
- 3.Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, G., J. M. Beckerich, A. Dominguez, S. Kerscher, D. Ogrydziak, V. Titorenko, and C. Gaillardin. 2003. Functional genetics of Yarrowia lipolytica, p. 227-271. In J. H. de Winde (ed.), Functional genetics of industrial yeasts, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 5.Barth, G., and C. Gaillardin. 1996. Yarrowia lipolytica, p. 313-388. In K. Wolf, K. D. Breunig, and G. Barth (ed.), Nonconventional yeasts in biotechnology, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 6.Beckman, M. 2006. Cell biology. Great balls of fat. Science 311:1232-1234. [DOI] [PubMed] [Google Scholar]
- 7.Browse, J., P. J. McCourt, and C. R. Somerville. 1986. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152:141-145. [DOI] [PubMed] [Google Scholar]
- 8.Czabany, T., K. Athenstaedt, and G. Daum. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771:299-309. [DOI] [PubMed] [Google Scholar]
- 8a.Dear, S., and R. Staden. 1991. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 19:3907-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreyra, R. G., N. I. Burgardt, D. Milikowski, G. Melen, A. R. Kornblihtt, E. C. Dell' Angelica, J. A. Santome, and M. R. Ermacora. 2006. A yeast sterol carrier protein with fatty-acid and fatty-acyl-CoA binding activity. Arch. Biochem. Biophys. 453:197-206. [DOI] [PubMed] [Google Scholar]
- 10.Fickers, P., P. H. Benetti, Y. Wache, A. Marty, S. Mauersberger, M. S. Smit, and J. M. Nicaud. 2005. Hydrophobic substrate utilization by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 5:527-543. [DOI] [PubMed] [Google Scholar]
- 11.Fickers, P., F. Fudalej, M. T. Le Dall, S. Casaregola, C. Gaillardin, P. Thonart, and J. M. Nicaud. 2005. Identification and characterization of LIP7 and LIP8 genes encoding two extracellular triacylglycerol lipases in the yeast Yarrowia lipolytica. Fungal Genet. Biol. 42:264-274. [DOI] [PubMed] [Google Scholar]
- 12.Fickers, P., M. T. Le Dall, C. Gaillardin, P. Thonart, and J. M. Nicaud. 2003. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods 55:727-737. [DOI] [PubMed] [Google Scholar]
- 13.Folch, J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 14.Holdsworth, J. E., M. Veenhuis, and C. Ratledge. 1988. Enzyme activities in oleaginous yeasts accumulating and utilizing exogenous or endogenous lipids. J. Gen. Microbiol. 134:2907-2915. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, E. P. 1962. The biosynthesis of phosphatides and triglycerides. Dtsch. Med. Wochenschr. 87:99-102. [DOI] [PubMed] [Google Scholar]
- 16.Kleijn, R. J., J. M. Geertman, B. K. Nfor, C. Ras, D. Schipper, J. T. Pronk, J. J. Heijnen, A. J. van Maris, and W. A. van Winden. 2007. Metabolic flux analysis of a glycerol-overproducing Saccharomyces cerevisiae strain based on GC-MS, LC-MS and NMR-derived C-labelling data. FEMS Yeast Res. 7:216-231. [DOI] [PubMed] [Google Scholar]
- 17.Kurat, C. F., K. Natter, J. Petschnigg, H. Wolinski, K. Scheuringer, H. Scholz, R. Zimmermann, R. Leber, R. Zechner, and S. D. Kohlwein. 2006. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281:491-500. [DOI] [PubMed] [Google Scholar]
- 18.Laffargue, A., A. de Kochko, and S. Dussert. 2007. Development of solid-phase extraction and methylation procedures to analyze free fatty acids in lipid-rich seeds. Plant Physiol. Biochem. 45:250-257. [DOI] [PubMed] [Google Scholar]
- 19.Le Dall, M. T., J. M. Nicaud, and C. Gaillardin. 1994. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 26:38-44. [DOI] [PubMed] [Google Scholar]
- 20.Meesters, P. A., and G. Eggink. 1996. Isolation and characterization of a delta-9 fatty acid desaturase gene from the oleaginous yeast Cryptococcus curvatus CBS 570. Yeast 12:723-730. [DOI] [PubMed] [Google Scholar]
- 21.Mlicková, K., E. Roux, K. Athenstaedt, S. d'Andrea, G. Daum, T. Chardot, and J. M. Nicaud. 2004. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 70:3918-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen, H. T., A. Dieterich, K. Athenstaedt, N. H. Truong, U. Stahl, and E. Nevoigt. 2004. Engineering of Saccharomyces cerevisiae for the production of l-glycerol 3-phosphate. Metab. Eng. 6:155-163. [DOI] [PubMed] [Google Scholar]
- 23.Papanikolaou, S., I. Chevalot, M. Komaitis, I. Marc, and G. Aggelis. 2002. Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures. Appl. Microbiol. Biotechnol. 58:308-312. [DOI] [PubMed] [Google Scholar]
- 24.Papanikolaou, S., L. Muniglia, I. Chevalot, G. Aggelis, and I. Marc. 2003. Accumulation of a cocoa-butter-like lipid by Yarrowia lipolytica cultivated on agro-industrial residues. Curr. Microbiol. 46:124-130. [DOI] [PubMed] [Google Scholar]
- 25.Querol, A., E. Barrio, T. Huerta, and D. Ramon. 1992. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 58:2948-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratledge, C. 2005. Single cell oils for the 21st century, p. 1-20. In Z. Cohen and C. Ratledge (ed.), Single cell oils. AOCS Press, Champaign, IL.
- 27.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Vigeolas, H., P. Waldeck, T. Zank, and P. Geigenberger. 2007. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol. J. 5:431-441. [DOI] [PubMed] [Google Scholar]
- 29.Wang, H. J., M.-T. Le Dall, Y. Waché, C. Laroche, J.-M. Belin, C. Gaillardin, and J.-M. Nicaud. 1999. Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J. Bacteriol. 181:5140-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]