Abstract
The composition of diatom-associated bacterial communities was studied with 14 different unialgal xenic diatom cultures isolated from freshwater epilithic biofilms of Lake Constance, Germany. A clear dominance of Alphaproteobacteria was observed, followed by Betaproteobacteria, Gammaproteobacteria, Bacteroidetes, and Verrucomicrobia. Pure cultures of the diatom Cymbella microcephala, which was found to be dominant in epilithic biofilms in Lake Constance, were cocultivated with six associated bacterial strains. All these bacterial strains were able to grow in C. microcephala cultures in the absence of organic cosubstrates. Diatom growth was generally enhanced in the presence of bacteria, and polysaccharide secretion was generally increased in the presence of Proteobacteria. The monomer composition of extracellular polysaccharides of C. microcephala changed in relation to the presence of different bacteria, but the dominant monomers were less affected. Our results indicate that these changes were caused by the diatom itself rather than by specific bacterial degradation. One Bacteroidetes strain strongly influenced carbohydrate secretion by the alga via extracellular soluble compounds. Biofilms were formed only in the presence of bacteria. Phylogenetic analysis and coculture studies indicate an adaptation of Proteobacteria and Bacteroidetes to the microenvironment created by the diatom biofilm.
Lake Constance is a large mesotrophic freshwater lake in Central Europe. The littoral zone of this lake is an area of high primary production. Rocks in these shallow-water zones are covered by greenish or brownish biofilms, sometimes several millimeters thick, consisting mainly of algae and bacteria. Interaction of algae and bacteria is assumed to be confined to the degradation and cycling of organic matter produced by the algae (22, 39). The role of specific classes of bacteria involved in such degradation of organic matter released by diatoms and other algae has been studied in marine (38) and in freshwater systems (11, 24, 32, 37, 40). Epilithic biofilms are complex communities based on interactions between the primary producers (algae and cyanobacteria) and bacteria, fungi, protozoa, insects, larvae, shellfish, etc. (32). Nonaxenic unialgal diatom cultures also harbor a distinct assemblage of associated bacteria, which have been termed satellite bacteria (8, 39). Usually such algal cultures are maintained for several years, and thus the associated bacteria undergo selection. The literature reports on Proteobacteria and Bacteroidetes as the major bacterial partners found in diatom mats and in diatom cultures from worldwide sampling places and different habitats (10, 11, 22, 30, 32, 38, 39, 49). As diatoms are the major primary producers and early colonizers of surfaces (1), studies on associated bacteria in unialgal cultures and cocultures of axenic diatoms and bacteria could help elucidate the role of these organisms in natural biofilm formation.
Biofilms are stabilized by extracellular polymeric substances (EPS). Parts of the EPS are soluble, whereas other parts are associated with the cell or the substratum, forming structure that range from jelly-like to solid, termed bound EPS. The extracellular polysaccharides of benthic diatoms are commonly composed of rhamnose, fucose, xylose, mannose, galactose, glucose, and other monomers; galactose and glucose often form the major part (10, 12, 43, 45). These sugar monomers are also found in natural biofilms dominated by diatoms (7, 41). Such extracellular polysaccharides may be used by heterotrophic organisms as a carbon source (19, 20, 21, 26). In fact, many diatom cultures are permanently contaminated with bacteria although most diatom culture media do not contain appreciable amounts of organic compounds. Therefore, cross-feeding between the auto- and heterotrophs is very likely, revealing a strong interdependency which is thought to be one of the major forces of microbial coevolution of mixed biofilm communities (46). Grossart et al. (22) reported on fluctuations of various bacteria during different growth phases of diatom cultures. Until now, diatoms and bacteria have rarely been cultivated in defined cocultures. The observed effects were often ambiguous. Growth of diatoms can be enhanced or can be suppressed by bacteria (6, 20, 21); the strength of adhesion to a substratum can also be increased (23) or reduced (51) or can even fluctuate, depending on the growth conditions of the associated bacteria (18). This variation is accompanied by changes within the diatom EPS (23), as indicated by lectin labeling (51). Thus, diatom-bacteria interactions may vary from being symbiotic to being antagonistic. For most diatoms it is still unknown whether they actually benefit from bacteria. Heterotrophic bacteria need organic matter for their energy metabolism, but it is still unclear what they feed on within the “habitat” created by diatom biofilm and whether they occupy specific niches. For instance Vibrio proteolytica is known to feed on exudates from Amphora coffaeformis (35).
In the present study we address the relation between diatom polysaccharide secretion and associated heterotrophic bacteria. We demonstrate that extracellular diatom polysaccharides can be substrates for these bacteria and that different bacterial strains utilize different parts of the polysaccharides. Interspecies relations were identified by mapping the community structure of diatom-associated bacteria in unialgal diatom cultures and by cocultivation of a representative diatom with the respective bacteria under defined conditions.
MATERIALS AND METHODS
Diatom cultures.
For isolation and cultivation of diatoms, 5 to 10 μl of biofilm material was scraped from stones collected in the littoral zone of Lake Constance, suspended in 1.0 to 1.5 ml of sterile diatom medium (DM) (47), and homogenized. Single diatom cells were isolated and cultured in DM. The cultivation parameters were a constant temperature of 16°C during a 16-h light period and 8 h of darkness. The light intensity varied from 30 to 60 μmol of photons m−2 s−1 using 58 W TLD Philips neon lamps. Cells were subcultured monthly and maintained in the same medium.
Isolation and cultivation of Cymbella microcephala and bacterial isolates.
C. microcephala was isolated and cultivated for more than 2 years as described above. This unialgal xenic culture was vortexed and diluted serially at 1:10 in DM. From dilutions of 105 to 107, 50 μl (each) was spread on 1.5% agar plates with different media: nutrient broth-soyotone-yeast extract (NSY) medium (modified from reference25) containing 0.1 g/liter peptone, yeast extract, and nutrient broth; maleic acid medium (modified from reference 13) without bromine thymol blue; and our medium B containing 14 mM KCl, 10 mM HEPES, 7.2 mM NaCl, 5 mM NH4Cl, 0.5 mM MgSO4, 0.15 mM phosphate (79% K2HPO4, 21% NaH2PO4 [wt/wt]), 0.01 mM CaCl2, 0.05% tryptone, 0.0005% yeast, and (per liter) 500 μl of trace element solution SL10 (50). The pH was 7.0. These agar plates were incubated under the conditions described above for diatom cultivation. Bacterial colonies that appeared different were picked with an inoculation loop and streaked at least three times on the same medium to obtain pure cultures. From these cultures, the 16S rRNA genes were amplified and sequenced as described below.
The diatom C. microcephala was purified from bacterial contaminants by spreading cells on 1.5% DM agar plates containing 5 μg/ml tetracycline and 5 μg/ml kanamycin. The absence of bacterial contaminants was verified by epifluorescence microscopy using the dyes 4′,6-diamidino-2-phenylindol (DAPI) or Sybr Green and by plating on the bacterial media described above.
C. microcephala was cocultivated with six different bacterial isolates in binary cultures or with all bacterial strains together. As a reference, the axenic diatom culture was used. All cultures were grown in 50 ml of DM in Erlenmeyer flasks. For every condition and for every harvesting point, three independent culture flasks were prepared. Diatom culture (1.5 ml) with a chlorophyll concentration of 0.31 μg/ml was used as an initial inoculum. Bacteria were added to C. microcephala cultures in similar amounts (optical density at 600 nm [OD600] × volume [in μl] = constant). As a further control, DM in test tubes was inoculated with bacteria as well. Cultures were grown as described above, and chlorophyll content (diatom growth), OD600, and carbohydrate concentration and composition were followed. Samples were taken at 4-day intervals over a cultivation period of 32 days.
Biofilms were removed from the glass surface with a rubber spatula, transferred to 50-ml centrifugation tubes, and suspended. Growth of diatoms and bacteria was measured via the OD600. At least five single measurements were done with every culture, and up to 20 were determined if strong biofilms and aggregates were formed. Growth of C. microcephala was quantified via chlorophyll content. From every culture, 1 ml was centrifuged at 16,100 × g for 20 min. The pellet was resuspended in 100 μl of methanol and vortexed for 20 min. After the addition of 900 μl of acetone, particles were spun down again, and the chlorophyll content was determined optically (28). For microscopy, 1 ml of every sample was fixed in 10% formaldehyde. For epifluorescence microscopy, samples dried on objective slides were incubated for 5 min with a 1.4 pM DAPI solution and then washed with water and subsequently with pure ethanol. After evaporation of the ethanol, the cells were embedded in the antifading agent AF1 (Citifluor, London).
Treatment of C. microcephala with spent medium of the bacterial strain 32.
Bacterial strain 32 belonging to the Bacteroidetes was grown in 50% LB medium as described above. Cultures were harvested in the stationary phase by centrifugation at 30,000 × g for 30 min at room temperature. The supernatant was either filter sterilized, autoclaved, or both and applied at a ratio of 0.2% to 50% (vol/vol) to freshly inoculated C. microcephala cultures. As a control, 50% LB medium was added to the diatom cultures. All cultures were grown in three replicates and checked daily by microscopy.
DNA extraction and 16S rRNA gene clone libraries.
Fourteen diatom species (5, 33) were used in xenic, unialgal cultures that were subcultured at least four to five times. For DNA extraction (modified from reference 34), cultures were centrifuged, and the cell pellet was frozen in liquid nitrogen and crushed with a pestle, mixed with 1 ml of cetyltrimethylammonium bromide buffer, and incubated at 65°C for 1 h. The sample was washed with a chloroform-isoamylalcohol mixture at 24:1 (vol/vol). DNA was precipitated with 0.7 volumes of isopropanol, washed with 80% ethanol, dried, and dissolved in 100 μl of 10 mM Tris-HCl, pH 8.0, with 1 mM EDTA. Aliquots (50 ng) of DNA were used to amplify 16S rRNA genes using the universal bacterial primers 27f 5′-AGA GTT TGA TCC TGG CTC AG-3′ (16) and 1492r 5′-TAC GGY TAC CTT ACG ACT T-3′ (48). PCR products were purified using a NucleoSpin kit (Macherey-Nagel, Germany), followed by insertion into the pGEM-T vector (Promega, Germany) and transformation into Escherichia coli XL1 Blue Excel (Stratagene, Heidelberg, Germany), according to the manufacturer's protocols. The inserted cloned 16S rRNA genes of 40 to 50 randomly selected colonies per clone library was amplified, digested with MspI (Fermentas) according to the manufacturer's instructions, and analyzed for restriction fragment length polymorphism by electrophoresis using 2% NuSieve agarose (NuSieve 3:1 Agarose; Cambrex Bio Science, Rockland, ME).
Sequencing and phylogenetic analysis.
At least 10% of the cloned 16S rRNA genes showing unique restriction patterns were sequenced. Sequence reactions were prepared with either the dye primer cycle sequencing ready reaction kit (SP6 and T7 primers) or the PRISM ready reaction dideoxy termination cycle sequencing kit (Perkin-Elmer). Sequences were obtained using an Applied Biosystems (model 3700) automated sequencer. BLAST searches were performed at NCBI (http://www.ncbi.nlm.nih.gov/) (4), and closely related sequences were retrieved. All sequences were checked for chimeras by dividing the sequence into two partial sequences and performing BLAST searches; sequences were phylogenetically analyzed using the ARB software package (version 2.5b; http://www.arb-home.de) (31). Sequences were added to the ARB database and aligned using the FAST Aligner tool as implemented in ARB. Only sequences larger than 1,400 nucleotides were used for alignment. Phylogenetic analysis was done using the maximum-likelihood, neighbor-joining, and maximum parsimony algorithms.
Chemical analyses.
Nitrate was assayed in cultures after high-performance liquid chromatography separation on an A06 column (Sykam) according to the manufacturer's description and was detected optically at a 254-nm wavelength. As a standard, 0.01 to 1 mM Ca(NO3)2 was used.
Polysaccharides were analyzed separately in the soluble and the cell-associated fractions. Cultures were centrifuged at 16°C at 5,250 × g for 10 min. The supernatant containing soluble EPS was separated from the pellet. To extract frustule-associated (bound) EPS, the pellet was resuspended in 5 ml of water and incubated for 1 h in a shaking water bath at 30°C. After centrifugation at 5,250 × g for 10 min (43), the obtained supernatant contained the bound EPS. Carbohydrate contents of soluble and bound EPS were measured optically using a phenol-sulfuric acid assay (15). As a standard, glucose was used at concentrations of 5 to 500 μg per ml. Polysaccharides were precipitated in 80% (vol/vol) ethanol at −20°C for at least 12 h (43), centrifuged at 5,252 × g and 4°C for 20 min, and dried in a laminar airflow cabinet. Polymers were hydrolyzed at 123°C for 20 min in 2 M trifluoroacetic acid (modified from reference 2). Then the trifluoroacetic acid was evaporated, and the remaining sugars were dissolved in 1 ml of water and analyzed via high-performance anion-exchange chromatography with pulsed amperometric detection (27) using equipment from Dionex. Mixtures of the d-isomers of arabinose, fructose, fucose, galactose, glucose, mannose, ribose, and xylose were used as reference compounds.
RESULTS
Analysis, isolation, and cultivation of diatom-associated bacteria.
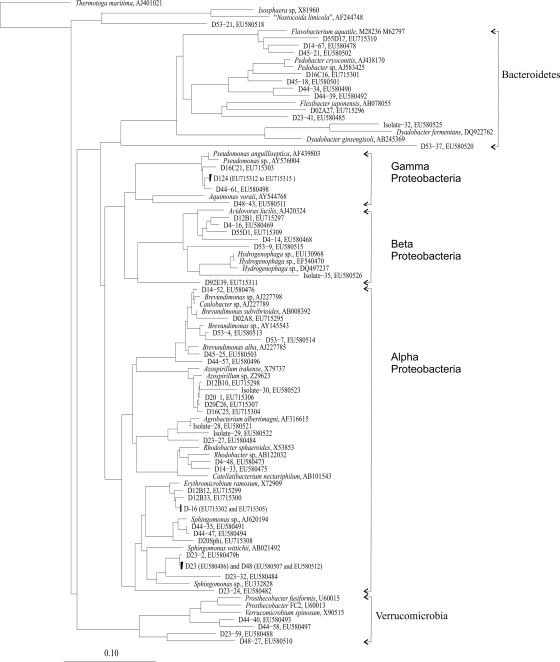
Single diatom cells were isolated from rocks of the littoral zone of Lake Constance by micromanipulation and were grown and maintained for 2 years together with the associated bacteria. Forty percent of the 16S rRNA genes cloned from diatom cultures were derived from heterotrophic bacteria, and 60% were from plastids. Among the bacteria, Alphaproteobacteria were dominant (59.2% of all bacterial sequences). Beta- and Gammaproteobacteria contributed 13% each, the Bacteroidetes group comprised 11%, and Verrucomicrobia spp. comprised 3%. Among the Alphaproteobacteria, sequences were related to five different clades (Fig. 1). One clade belonged to Erythromicrobium and Porphyrobacter, two clades belonged to Sphingomonas, one clade belonged to Brevundimonas, and one belonged to Azospirillum. Some sequences were related to Rhodobacter. Betaproteobacteria were mainly related to Acidovorax sp. or Hydrogenophaga sp. (Fig. 1), while most Gammaproteobacteria grouped with Pseudomonas sp. One clone grouped with Aquimonas voraii.
FIG. 1.
Phylogenetic tree of 16S rRNA gene sequences obtained from prokaryotic biomass associated with diatom cultures. Clones obtained in our study are identified by a clone number prefixed by a D. Representative 16S rRNA gene sequences of cultured and uncultured bacteria were used for the analysis, and only sequences of >1,400 nucleotides were considered. The tree was calculated by the neighbor-joining method showing 16S rRNA gene sequences recovered from the clone libraries of diatom-associated bacteria. NCBI accession numbers of clones and cultures are given; the bar represents 10% divergence. The tree was rooted with Thermotoga maritima as the outgroup.
Within the Bacteroidetes group, bacteria were related to Flavobacterium or Sphingobacterium (Fig. 1). Some sequences belonged to Verrucomicrobia and Planctomycetes (Fig. 1).
According to monthly counts of frustules from biofilms throughout the years 2004 and 2005, C. microcephala was found to be one of the dominant benthic diatoms in Lake Constance (data not shown). We cultivated this diatom in unialgal and in axenic cultures. Six strains of heterotrophic bacteria associated with C. microcephala were isolated from the nonaxenic culture in dilution series. Only strains abundant in 105 to 107 dilutions were studied further. Strains 28 and 29 were isolated in Doebereiner's medium, strains 30 and 32 were isolated in medium B, and strains 31 and 35 were isolated NSY medium. Strains 28, 29, 30, and 31 belonged to the Alphaproteobacteria, strain 35 belonged to the Betaproteobacteria, and strain 32 belonged to the Bacteroidetes.
Cocultivation of C. microcephala with isolated bacteria.
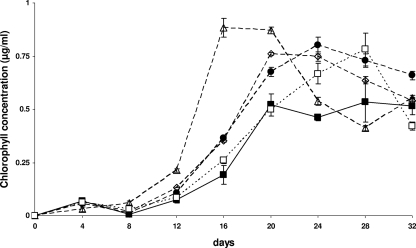
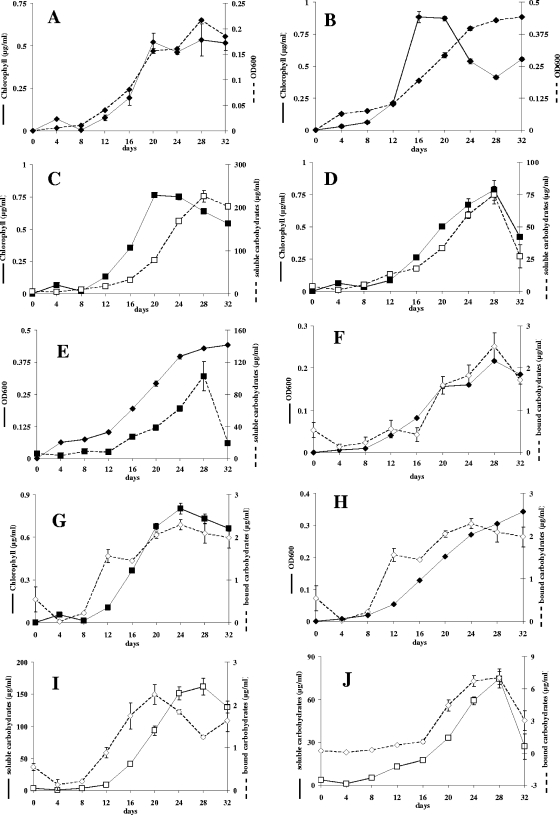
Cocultures of C. microcephala grown with the isolated associated bacterial strains yielded chlorophyll contents 11% to 66% higher than those of the axenic culture. Within 1 month, the cells reached chlorophyll concentrations up to 0.9 ± 0.001 μg ml−1 in liquid DM while the axenic cultures yielded a maximal chlorophyll content of 0.5 ± 0.09 μg/ml (400,000 cells ml−1 ± 10%). All cocultures with bacteria, except those containing strain 32, grew faster than the pure diatom culture. The axenic diatom culture showed maximal growth after 20 days. In the stationary phase, the chlorophyll content remained stable until the end of the cultivation period. In cocultures with bacteria, the chlorophyll content generally decreased toward the end of the cultivation period (Fig. 2). This phenomenon was most distinct in cultures inoculated with all bacterial strains together: the cultures reached maximal cell density and the highest chlorophyll content of all cultures after only 12 days, followed by a stationary phase lasting for 4 days before the chlorophyll content decreased (Fig. 2 and 3B). Cocultures with Alphaproteobacteria strains 28, 29, and 30 and with the Betaproteobacteria strain 35 reached their maximal chlorophyll concentrations at the same time as the axenic culture, the coculture with the Alphaproteobacteria strain 31 reached a peak 4 days later, and the coculture with the Bacteroidetes strain 32 reached a maximum 8 days later (Fig. 2). In axenic cultures, the OD600 values correlated with the chlorophyll content (Fig. 3A), and the same was true for the cocultures with strain 32. The cocultures with strains 31 and 35 and with all bacterial strains together showed an increasing OD600 at a time when the chlorophyll content declined (Fig. 3B). Similar phenomena were observed with the cocultures with Alphaproteobacteria strains 28, 29, and 30 but to a lesser extent.
FIG. 2.
Growth of C. microcephala in pure culture (solid line) or in coculture with bacterial isolates (broken lines). ▪, axenic C. microcephala; •, coculture with strain 31; □, coculture with strain 32; ⋄, coculture with strain 35; ▵, coculture with all strains combined.
FIG. 3.
Growth and product formation by C. microcephala in pure culture and in cocultures. Growth of axenic C. microcephala cultures (A) and cocultures with mixed bacterial strains (B) was measured as the OD600 (dashed line) and chlorophyll content (solid line). Chlorophyll content (solid line) and concentrations of soluble carbohydrates (dashed line) of C. microcephala cocultures with strain 35 (C) or 32 (D) were determined. (E) OD600 (solid line) and concentration of soluble carbohydrates (dashed line) of C. microcephala cocultivated with all bacterial isolates. (F) OD600 (solid line) and concentrations of bound carbohydrates (dashed lines) of axenic C. microcephala. In a coculture of C. microcephala and strain 31, chlorophyll content (solid line; G), OD600 (solid line; H) and concentrations of bound carbohydrates (dashed line in both panels) were determined. Concentrations are shown of bound (dashed line) and soluble (solid line) carbohydrates from cocultures with C. microcephala and strain 29 (I) or strain 32 (J).
The initial concentration of free nitrate in DM was 0.34 mM. Until day 28 of cultivation, the axenic diatom used 50% of the nitrate, and the cocultures used between 56% and 90%. There was always at least 30 μM nitrate left in all cultures.
Quantification of carbohydrate formation.
All Proteobacteria enhanced polysaccharide secretion by the diatom. The axenic culture reached concentrations up to 121 μg·ml−1 of soluble carbohydrates (up to 284 pg per diatom cell), whereas in all cocultures with Proteobacteria, the respective amount increased up to 226 μg·ml−1 or up to 444 pg per diatom cell (coculture with strain 35) (Table 1). The coculture with the Bacteroidetes strain showed a decreased polysaccharide concentration during the cultivation period, and lower polysaccharide contents were observed also in the cocultures with all bacterial isolates combined (Table 1).
TABLE 1.
Carbohydrate contents of growing C. microcephala cultures on days 20 and 28 after inoculation
| Cocultured strain | Amt of soluble carbohydrate ata:
|
Amt of bound carbohydrate ata:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Day 20
|
Day 28
|
Day 20
|
Day 28
|
|||||
| μg/ml | pg/cell | μg/ml | pg/cell | μg/ml | pg/cell | μg/ml | pg/cell | |
| None | 46.3 ± 2.8 | 111 | 121 ± 18.2 | 284 | 1.6 ± 0.2 | 3.8 | 2.5 ± 0.3 | 5.9 |
| Alphaproteobacteria strain 28 | 113 ± 4.5 | 209 | 171 ± 1.6 | 408 | 2.7 ± 0.6 | 5 | 1.6 ± 0.1 | 3.9 |
| Alphaproteobacteria strain 29 | 93.1 ± 7.3 | 199 | 162 ± 13 | 407 | 2.2 ± 0.2 | 4.8 | 1.2 | 3.1 |
| Alphaproteobacteria strain 30 | 79.2 ± 10.1 | 170 | 167 ± 9.5 | 310 | 2.1 ± 0.2 | 4.5 | 1.8 ± 0.1 | 3.3 |
| Alphaproteobacteria strain 31 | 74.9 ± 2.7 | 139 | 192 ± 12 | 328 | 2.1 ± 0.1 | 3.8 | 2.1 ± 0.2 | 3.6 |
| Bacteroidetes strain 32 | 33.2 ± 0.4 | 83 | 74.5 ± 6.7 | 119 | 4.4 ± 0.5 | 12.2 | 7 ± 0.6 | 11.2 |
| Betaproteobacteria strain 35 | 78.2 ± 3 | 129 | 226 ± 13.7 | 444 | 4.1 ± 0.6 | 6.8 | 2.7 | 5.2 |
| All | 38.4 ± 2.5 | 55.1 | 103 ± 18 | 312 | 2.8 ± 1 | 4 | 14 | 42.5 |
Amounts are given as micrograms of carbohydrate per milliliter of culture and as picograms of carbohydrates per diatom cell.
Bound carbohydrates were formed by diatoms in axenic culture to a maximal concentration of 2.5 μg·ml−1 (∼6 pg per diatom cell), whereas cocultures with the Betaproteobacteria strain 35 reached 41 μg·ml−1 (∼7 pg per diatom cell). The maximal concentration of bound carbohydrates varied from 2.1 μg·ml−1 to 3.0 μg·ml−1 for cocultures with Alphaproteobacteria. All cocultures with Alphaproteobacteria showed a decreasing ratio of bound carbohydrates to diatom cell number toward the end of the cultivation period. Maximum formation of bound carbohydrates was observed in cultures with the Bacteroidetes strain 32 and in that with all bacteria together (Table 1). In both cultures, the ratio of bound carbohydrates to diatom cell number increased strongly toward the end of the cultivation period.
Chlorophyll and soluble carbohydrate concentrations in axenic C. microcephala cultures were correlated, but the carbohydrate content increased with a delay of 8 days compared to the chlorophyll content (Fig. 3C). This was true for all cultures except the cocultures with strain 32, where both graphs nearly coincided (Fig. 3D). In the cocultures with the Betaproteobacteria strain 35 and those with all bacterial strains, a stagnating OD was followed by a decline in the concentration of soluble carbohydrates by 81% within the last 4 days (Fig. 3E).
The concentrations of bound carbohydrates within axenic C. microcephala cultures correlated with the chlorophyll concentrations and with the OD600 (Fig. 3A and F). This was also true for cocultures with strains 32 and 35. With all cocultures incubated with Alphaproteobacteria strains (strains 28, 29, 30, and 31), a parallel increase in the chlorophyll concentrations and the content of bound carbohydrates was observed, but at the same time the level of bound EPS decreased when the OD600 stagnated or increased (Fig. 3G and H).
The situation was entirely different with the cocultures inoculated with all bacteria together. Here, the bound carbohydrate contents correlated with the OD600 but not with the chlorophyll content. While the latter decreased, more cell-associated carbohydrates were found (data not shown).
The amounts of bound and soluble carbohydrates were strictly correlated. An increase of bound carbohydrates was followed approximately 8 days later by an increase in soluble carbohydrates (Fig. 3I). This was true for the axenic culture and for nearly all cocultures, with the exception of the cocultures with strain 32 and cultures with all bacterial strains together. Here, both carbohydrate fractions increased and decreased simultaneously (Fig. 3J).
Analysis of carbohydrate composition.
Soluble carbohydrates isolated from axenic C. microcephala cultures contained 40 to 50% galactose and 30 to 40% mannose/xylose monomers. Furthermore, about 7.2% rhamnose and 3.5% fucose were detected throughout the cultivation period. The level of mannose/xylose decreased slightly, whereas the galactose content increased toward the middle of the cultivation period. After 16 days, the proportion of galactose decreased and that of mannose/xylose increased. The glucose content was constantly reduced until complete absence after 20 days of cultivation. Similar results were observed for N-acetyl-d-glucosamine (GlcNAc), which decreased until day 16 of cultivation. Arabinose and fructose proportions were below one percent and fluctuated randomly.
All cocultures with the different bacteria showed similar proportions of monomers, whereas the described fluctuations of mannose/xylose and of galactose were always larger, as with the axenic culture. The strongest decrease of mannose/xylose was found in the cocultures with all bacterial strains together, reaching a minimum of 13.2% on day 16 of cultivation; the galactose proportion increased simultaneously to a maximum of 74.1%. The proportions of glucose, GlcNAc, rhamnose, and fucose were generally similar to that in the axenic culture in all situations. All other monomers fluctuated randomly within a proportion of 1.5%.
Cell-associated polysaccharides of the axenic C. microcephala cultures contained a similar percentage of mannose/xylose (30 to 40%) and of galactose (50 to 60%) as did the soluble carbohydrates. Different from the soluble carbohydrates, relevant proportions of glucose were found also toward the end of the cultivation period in the bound polysaccharides of the axenic cultures (4.7% at day 32). Arabinose and fructose were absent; fucose, rhamnose, and GlcNAc fluctuated at low proportions. In cocultures with strain 32 or those with all bacterial strains together, GlcNAc was missing. Cocultures either showed a decrease in glucose content (strains 29 and 30) or complete absence of this monomer (cocultures with all strains), or the proportion fluctuated randomly. In all cocultures, the proportions of fucose (up to 7%) did not change throughout the cultivation period, different from the axenic culture. All other monomers showed fluctuations at a lower level; fructose was never detected.
Cocultures of C. microcephala with the Bacteroidetes strain 32.
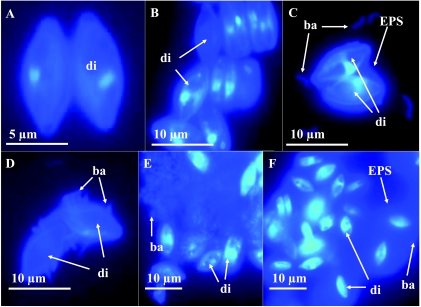
C. microcephala cocultures with Bacteroidetes strain 32 formed a polysaccharide capsule surrounding the diatom cells with an estimated volume of up to 10 times that of the algal cell. This capsule was stained by DAPI (Fig. 4F) and was also visible in phase-contrast microscopy. Axenic C. microcephala cells did not form these capsules. The concentration of soluble carbohydrates was the lowest of all cocultures containing single bacteria, whereas the concentration of bound carbohydrates was highest. The soluble fraction showed a higher level of fucose and rhamnose, the bound fraction had the lowest fucose content, and GlcNAc was completely absent.
FIG. 4.
Cell-cell aggregates formed by C. microcephala (di) under different culture conditions. Epifluorescence photomicrographs of DAPI preparations of C. microcephala cells grown either axenically (A), or cocultivated with strain 28 (B), 29 (C), 30 (D), 31 (E) or 32 (F). In all cocultures, bacteria (ba) show a strain-specific assembly in relation to the diatom. In cocultures with strain 32 (F), C. microcephala secretes an EPS capsule.
Capsule formation was induced also after the addition of bacterial culture supernatant. After 3 to 4 days of incubation, the first algal cells were surrounded by this gel-like matrix. Also filter-sterilized or autoclaved culture supernatant added at a ratio of 0.2% to 10% (vol/vol) caused this effect. No difference was observed between frozen and fresh bacterial supernatants. This effect was caused neither by the medium (50% LB) itself nor by the pH of the culture supernatant (6.9 to 7.3).
Cocultures and biofilm structure.
Epifluorescence microscopy of DAPI preparations revealed that, depending on the bacterium in each defined coculture combination, different cell aggregates and biofilm structures developed (Fig. 4). Exclusively, Betaproteobacteria strain 35 grew suspended, showing visible turbidity during cocultivation. The characteristic visual patterns regarding cell aggregation, such as turbidity and biofilm or capsule formation in cocultures of C. microcephala with single bacterial strains, were observed also in the mixed cultures.
DISCUSSION
Phylogenetic analysis of diatom-associated bacteria.
Although the diatoms used in this study represented different genera, the associated bacteria often exhibited striking similarities in their 16S rRNA gene sequences. They were dominated by Alphaproteobacteria as also reported earlier (22, 38). Bacterial communities associated with aggregates from planktonic diatom blooms in Lake Constance were dominated by Alpha- and Betaproteobacteria and by Bacteroidetes as well (40). The dominant 16S rRNA gene sequences grouped with Sphingomonas, Caulobacter, and Rhizomonas or with Brevundimonas and Mycoplasma and formed a clade with data bank sequences obtained from lake snow microaggregates (42). Other sequences were similar to sequences of Roseobacter, including Azospirillum-related sequences that were described earlier to be associated with marine diatom assemblages (3). Sequences of Betaproteobacteria in this study were related to Hydrogenophaga and Acidovorax that had also been found in diatom-derived microaggregates (11) and in lake snow of Lake Constance (40). Most of the sequences of Gammaproteobacteria were related to the eel pathogen Pseudomonas anguilliseptica (14). 16S rRNA gene sequences related to Bacteroidetes were often amplified from diatom cultures. These sequences were found in epilithic biofilms in Lake Constance (data not shown) and appear to be associated frequently with diatoms (22, 30, 38). Interestingly, the abundant types of 16S rRNA gene sequences derived from our samples have recent common ancestors although the tested diatoms were phylogenetically highly diverse including raphid and araphid species. Since other 16S rRNA gene sequences derived from diatom-associated prokaryotes (10, 38, 39) confirm this observation, diatoms might be regarded generally as a microhabitat to which especially Proteobacteria and Bacteroidetes have adapted and evolved separately, independent of whether the diatoms are planktonic or benthic, raphid or araphid, freshwater- or saltwater-adapted, or terrestrial or found in polar ice.
Cocultivation of C. microcephala with bacterial isolates.
In our cocultures with the ubiquitous freshwater diatom C. microcephala, we showed that the diatoms produced the organic carbon source for these bacteria. Further, we confirm studies in which heterotrophic bacteria supported diatom growth (17, 23) although opposite observations were reported as well (6). Apparently, the bacteria release substances that support growth of C. microcephala, or they consume substances that might otherwise inhibit diatom growth. Since nitrate availability was not a limiting factor, bacterial N2 fixation can be ruled out as a possible means of support.
The measured OD600 values can be regarded as a rough estimate of the total cell numbers including diatoms and bacteria. Graphs of chlorophyll concentrations and OD600 values coincide with axenic cultures, thus confirming the reliability of both methods. An increasing OD600 and simultaneous decreasing chlorophyll content should be due to increased bacterial growth. Increasing bacterial growth while diatom growth stagnates can be explained either as bacterial exploitation of a substrate derived from the diatom, e.g., glycolate, or secreted polymeric organic matter (20, 21), or phosphate released from dead diatoms. In cocultures with the Bacteroidetes strain 32, the OD600 and the chlorophyll content increased and decreased in parallel. Obviously, bacteria and diatoms grew simultaneously, either due to the production of an unknown growth-supporting factor or by a direct bacterial influence on the growth of C. microcephala.
Formation of carbohydrates.
The fraction of soluble carbohydrates contained glucose that derived from soil extract that was added to the culture medium. During cocultivation glucose disappeared completely from the soluble fraction, probably due to bacterial consumption. However, glucose disappeared also in the axenic culture and was not detectable after 16 days. Obviously, it was consumed or converted also by the diatom. A similar phenomenon was observed with GlcNAc.
All Proteobacteria in this study enhanced secretion of soluble polysaccharides by the diatom, probably caused by an unknown bacterial factor influencing the diatom. In the axenic cultures and in most cocultures, diatom growth was followed by the accumulation of secreted carbohydrates in the medium. This was not the case in cocultures with strain 35 or in those with all strains together. Here, a decrease of soluble carbohydrates, an increasing OD600, and rising concentrations of frustule-associated carbohydrates indicate that soluble carbohydrates were preferentially used by the Betaproteobacteria strain 35. Interestingly, the coculture with strain 35 produced the highest amounts of soluble carbohydrates. Betaproteobacteria are typically found attached to lake snow often rich in dead or dying diatoms (11, 40) in the water column of Lake Constance. Therefore, these bacteria are likely to utilize dissolved polymers and to degrade dead algal cells. Strain 35 grew freely suspended in the culture flask, indicating that it might have been found only accidentally in the biofilm.
The Alphaproteobacteria appear to utilize cell-bound polysaccharides as their carbon source. In these cocultures, the content of cell-associated carbohydrates decreased simultaneously with the chlorophyll content, while at the same time the OD600 increased, and soluble sugars started to accumulate. Alphaproteobacteria are known to be associated ubiquitously with diatoms, independent of the habitat of the algae (10, 11, 22, 30, 32, 38, 39, 49). Adaptation of Alphaproteobacteria to this habitat appears likely. They might feed on frustule-associated organic matter because these carbohydrates are permanently produced by the alga and do not diffuse into the surrounding water column. This hypothesis is supported by the observation that these Alphaproteobacteria were found to be mainly embedded in the diatom-bacteria biofilms.
Bacteroidetes strain 32 influences the secretion of diatom carbohydrates via soluble molecules.
The Bacteroidetes strain 32 apparently strongly influenced the diatom carbohydrates by drastically decreasing the content of soluble polysaccharides and increasing the level of bound polysaccharides. Microscopic observation showed the formation of capsules around the diatoms. Low concentrations of soluble EPS and high amounts of bound EPS were observed also in cocultures with all bacteria. Bacteroidetes are often found on diatom-rich detritus (30), e.g., in diatom cultures in the late stationary phase (22, 38). They degrade complex polymers (29, 41) and colonize solid substrates rapidly (36). In microscopic observations, we often found these bacteria on the surfaces of capsules surrounding the diatoms. The observed decrease of soluble sugars could be caused by immediate bacterial consumption or by reduced secretion of soluble EPS to form preferentially bound carbohydrates. Since this effect can also be induced by autoclaved bacterial culture supernatant, we assume that the bacterium produces a thermostable molecule which induces capsule secretion, possibly as a protection against any kind of threat (e.g., predation and toxins).
Single bacterial strains and mixed bacteria in coculture with C. microcephala.
Microscopic observation of the cocultures showed the formation of specific structures of cell aggregates of C. microcephala and single bacterial strains (Fig. 4). In the cocultures with mixed bacteria, all types of such structures were found. Specific effects of isolate 32 on diatom carbohydrates, namely, high amounts of frustule-associated carbohydrates (capsule formation) and low concentrations of soluble carbohydrates, were also measured in the mixed cocultures. Every single strain appears to use a substrate deriving directly from the diatoms, thereby forming characteristic aggregates together with the diatom, whether or not other bacteria are present. Obviously, the diatom provides various niches for different bacteria and benefits from their presence. In natural biofilms, such niche formation might, besides other factors, explain the success and distribution pattern of certain diatoms and associated bacteria. The microorganisms may adapt to each other and create a kind of microenvironment optimized for interacting partners. Diatoms and bacteria might support each other by an equilibrium of cross-feeding, possibly optimized by exchange of chemical factors. Such associations can be specific or random. It is likely that cross-feeding partners may change due to various factors such as cell density, presence of other microorganisms and their secretions, availability of nutrients, or abiotic factors such as light, temperature, water currents, etc. Further, such interactions appear to initialize formation of diatom biofilms and aggregates as this has been shown so far with marine microbial communities (20, 21).
Acknowledgments
We thank Anja Dullius (Department of Biology, University of Konstanz) for help in carbohydrate analysis. We thank Linda Medlin (AWI Bremerhaven) for identification of the isolates. We also thank Elke Hespeler, Axel Meyer, and Walter Salzburger (Department of Biology, University of Konstanz) for help in sequencing.
We are grateful for support by the University of Konstanz and for a grant from the Deutsche Forschungsgemeinschaft, grant SFB454 Bodensee-Littoral (TP B1 to B.S. and TP B11 to P.G.K.).
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Ács, É. 1998. Short-term fluctuations in the benthic algal compositions on artificial substratum in a large river (river Danube, near Budapest). Verh. Int. Verein. Limnol. 26:1653-1656. [Google Scholar]
- 2.Albersheim, P., D. J. Nevis, P. D. English, and A. Karr. 1967. A method for the analysis of sugars in plant cell-wall polysaccharides by gas-liquid chromatography. Carbohydr. Res. 5:340. [Google Scholar]
- 3.Allgaier, M., H. Uphoff, A. Felske, and I. Wagner-Döbler. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Bahulikar, R. A., and P. G. Kroth. 2007. Localization of EPS components secreted by freshwater diatoms using differential staining with fluorophore-conjugated lectins and other fluorochromes. Eur. J. Phycol. 42:199-208. [Google Scholar]
- 6.Baker, K. H., and D. S. Herson. 1978. Interactions between the diatom Thalassiosira pseudonana and an associated pseudomonad in a mariculture system. Appl. Environ. Microbiol. 35:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battin, T. J., L. A. Kaplan, J. D. Newbold, X. Cheng, and C. Hansen. 2003. Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl. Environ. Microbiol. 69:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, W. H. 1984. Bacterial adaptation to low-nutrient conditions as studied with algal extracellular products. Microb. Ecol. 10:217-230. [DOI] [PubMed] [Google Scholar]
- 9.Bhaskar, P. V., H.-P. Grossart, N. B. Bhosle, and M. Simon. 2005. Production of macroaggregates from dissolved exopolymeric substances (EPS) of bacterial and diatom origin. FEMS Microb. Ecol. 53:255-264. [DOI] [PubMed] [Google Scholar]
- 10.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A. McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brachvogel, T., B. Schweitzer, and M. Simon. 2001. Dynamics and bacterial colonization of microaggregates in a large mesotrophic lake. Aquat. Microb. Ecol. 26:23-35. [Google Scholar]
- 12.Chiovitti, A., A. Bacic, J. Burke, and R. Wetherbee. 2003. Heterogeneous xylose-rich glycans are associated with extracellular glycoproteins from the biofouling diatom Craspedosauros australis (Bacillariophyceae). Eur. J. Phycol. 38:351-360. [Google Scholar]
- 13.Doebereiner, J., and F. O. Pedrosa. 1992. Nitrogen-fixing bacteria in nonleguminous crop plants. Springer-Verlag, Madison, WI.
- 14.Doménech, A., J. F. Fernández-Garayzábal, J. A. García, M. T. Cutuli, M. Blanco, A. Gibello, M. A. Moreno, and L. Domínguez. 1999. Association of Pseudomonas anguilliseptica infection with “winter disease” in sea bream, Sparus aurata L. J. Fish Dis. 22:69-71. [Google Scholar]
- 15.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-3556. [Google Scholar]
- 16.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukami, K., T. Nishijima, and Y. Ishida. 1997. Stimulative and inhibitory effects of bacteria on the growth of microalgae. Hydrobiologia 358:185-191. [Google Scholar]
- 18.Gawne, B., Y. Wang, K. Hoagland, and M. R. Gretz. 1998. Role of bacteria and bacterial exopolymer in the attachment of Achnanthes longipes. Biofouling 13:137-156. [Google Scholar]
- 19.Giroldo, D., A. A. H. Vieira, and B. S. Paulsen. 2003. Relative increase of deoxy sugars during microbial degradation of an extracellular polysaccharide released by a tropical freshwater Thalassiosira sp. (Bacillariophyceae). J. Phycol. 39:1109-1115. [Google Scholar]
- 20.Grossart, H.-P., and M. Simon. 2007. Interactions of planktonic algae and bacteria: effects on algal growth and organic matter dynamics. Aquat. Microb. Ecol. 47:163-176. [Google Scholar]
- 21.Grossart, H.-P., G. Czub, and M. Simon. 2006. Algae-bacteria interactions and their effects on aggregation and organic matter flux in the sea. Environ. Microbiol. 8:1074-1084. [DOI] [PubMed] [Google Scholar]
- 22.Grossart, H.-P., F. Levold, M. Allgaier, M. Simon, and T. Brinkhoff. 2005. Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7:860-873. [DOI] [PubMed] [Google Scholar]
- 23.Grossart, H.-P. 1999. Interactions between marine bacteria and axenic diatoms (Cylindrotheca fusiformis, Nitschia laevis and Thalassiosira weissflogii) incubated under various conditions in the lab. Aquat. Microb. Ecol. 19:1-11. [Google Scholar]
- 24.Grossart, H.-P., M. Simon, and B. E. Logan. 1997. Formation of macroscopic organic aggregates (lake snow) in a large lake: the significance of transparent exopolymer particles, phytoplankton, and zooplankton. Limnol. Oceanogr. 42:1651-1659. [Google Scholar]
- 25.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haynes, K., T. A. Hofman, C. J. Smith, A. S. Ball, G. J. C. Underwood, and A. M. Osborn. 2007. Diatom-derived carbohydrates as factors affecting bacterial community composition in estuarine sediments. Appl. Environ. Microbiol. 73:6112-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahnel, J. B., P. Ilieva, and F. H. Frimmel. 1998. HPAE-PAD—a sensitive method for the determination of carbohydrates. Fresenius J. Anal. Chem. 360:827-829. [Google Scholar]
- 28.Jeffrey, S. W., and G. F. Humphrey. 1975. New spectrophotometric equation for determining chlorophyll a, b, c1 and c2. Biochem. Physiol. Pflanzen. 167:194-204. [Google Scholar]
- 29.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microb. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 30.Knoll, S., W. Zwisler, and M. Simon. 2001. Bacterial colonization of early stages of limnetic diatom aggregates. Aquat. Microb. Ecol. 25:141-150. [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makk, J., E. Acs, K. Marialigeti, and G. Kovacs. 2003. Investigations on the Danube gravel-biofilm diatom-associated bacterial communities. Biologia (Bratisl.) 58:729-742. [Google Scholar]
- 33.Medlin, L., I. Jung, R. A. Bahulikar, K. Mendgen, P. G. Kroth, and W. H. C. F. Kooistra. 2008. Evolution of the diatoms. VI. Assessment of the new genera in the araphids using molecular data. Nova Hedwig. Beih. 133:81-100. [Google Scholar]
- 34.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, R. E., K. E. Cooksey, and J. C. Priscu. 1986. Stimulation of bacterial DNA synthesis by algal exudates in attached algal-bacterial consortia. Appl. Environ. Microbiol. 52:1177-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinhassi, J., F. Azam, J. Hempälä, R. A. Long, J. Martinez, U. L. Zweifel, and A. Hagström. 1999. Coupling between bacterioplankton species composition, population dynamics and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 37.Riemann, L., and A. Winding. 2001. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 42:274-285. [DOI] [PubMed] [Google Scholar]
- 38.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schäfer, H., B. Abbas, H. Witte, and G. Muyzer. 2002. Genetic diversity of “satellite” bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42:25-35. [DOI] [PubMed] [Google Scholar]
- 40.Schweitzer, B., I. Huber, R. Amann, W. Ludwig, and M. Simon. 2001. α- and β-Proteobacteria control the consumption and release of amino acids on lake snow aggregates. Appl. Environ. Microbiol. 67:632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shewan, J. M., and T. A. Mc Meekin. 1983. Taxonomy (and ecology) of Flavobacterium and related genera. Annu. Rev. Microbiol. 37:233-252. [DOI] [PubMed] [Google Scholar]
- 42.Simon, M., H.-P. Grossart, B. Schweitzer, and H. Ploug. 2002. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28:175-211. [Google Scholar]
- 43.Staats, N., B. De Winder, L. J. Stal, and L. R. Mur. 1999. Isolation and characterisation of extracellular polysaccharides from the epipelic diatoms Cylindrotheca closterium and Navicula salinarum. Eur. J. Phycol. 34:161-169. [Google Scholar]
- 44.Taylor, I. S., D. M. Paterson, and A. Mehlert. 1999. The quantitative variability and monosaccharide composition of sediment carbohydrates associated with intertidal diatom assemblages. Biogeochemistry 45:303-327. [Google Scholar]
- 45.Underwood, G. J. C., M. Boulcott, C. A. Raines, and K. Waldron. 2004. Environmental effects on exopolymer production by marine benthic diatoms: dynamics, changes in composition and pathways of production. J. Phycol. 40:293-304. [Google Scholar]
- 46.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe, M. M. 2005. Freshwater culture media, p. 429-538. In R. A. Anderson (ed.), Algal culturing techniques. Elsevier Academic Press, Philadelphia, PA.
- 48.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss, P., B. Schweitzer, R. Amann, and M. Simon. 1996. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl. Environ. Microbiol. 62:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies in dissimilatory sulfate-reducing bacteria that decompose fatty acids. Characterisation of filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:167-172. [Google Scholar]
- 51.Wigglesworth-Cooksey, B., and K. Cooksey. 2005. Use of fluorophore-conjugated lectins to study cell-cell interactions in model marine biofilms. Appl. Environ. Microbiol. 71:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]