Abstract
Here we present the first oligonucleotide DNA microarray analysis of global gene expression changes in the obligate intracytoplasmic pathogen Rickettsia prowazekii using temperature upshift as a model stress condition, and we describe a methodology for isolating highly purified rickettsial RNA. In toto, 23 transcripts were significantly increased by temperature upshift (≥2.0-fold; P < 0.05), and no transcripts demonstrated reproducible decreases. Array results for three heat shock-inducible mRNAs were confirmed using quantitative reverse transcription-PCR.
Rickettsia prowazekii is the etiological agent of epidemic typhus fever in humans and a Centers for Disease Control-designated select agent. R. prowazekii resides exclusively within the cytosol of eukaryotic cells unbounded by a host-derived membrane vesicle (for a review, see reference 13). In vivo, R. prowazekii infects gut epithelial cells of the louse vector, Pediculus humanus corporis, and (primarily) endothelial cells of the human host. Flying squirrels have been identified as a nonhuman reservoir for R. prowazekii, and individuals with recrudescent Brill-Zinsser disease serve as human reservoirs for this pathogen (6, 26). Circumstances in which humans are forced to live in confined, filthy conditions hold the potential for reemergent outbreaks of this pathogen (22, 24). Despite its small genome size (only 835 putative open reading frames [ORFs]) (1) and growth in a metabolite-replete, stable environment, R. prowazekii has maintained regulation of both enzyme function and gene expression (7-10, 17, 18), indicating that rickettsiae sense changes in their environment and can respond accordingly.
To date, R. prowazekii has proven recalcitrant to classical bacterial genetics techniques (4, 15, 19-21, 23, 25); thus, the use of gene knockouts and reporter fusions is not a viable strategy for studying global regulation. Further, the use of microarrays to assess global changes in rickettsial gene expression has been limited to a single, recent study comparing the Shelia Smith and Iowa strains of R. rickettsii (11). This study reported that only four genes significantly differed in expression levels between the two strains. In the present study, we have employed DNA oligonucleotide microarrays to perform the first high-throughput analyses of global R. prowazekii gene expression in response to temperature upshift as a model environmental stress. We present a method of isolating highly enriched R. prowazekii total RNA away from contaminating host cell nucleic acids for DNA microarray analysis of all 835 putative R. prowazekii ORFs and demonstrate that R. prowazekii reprograms gene expression in response to temperature upshift.
R. prowazekii infection of L929 mouse fibroblast cells and RNA isolation.
Analyses of obligate intracellular organisms are complicated by the need to isolate the bacteria away from contaminating host cells and their constituents. In a total RNA extraction of rickettsia-infected host cells, the rickettsial RNA makes up less than 10% of the total based on rRNA (see Fig. S1A in the supplemental material). We reasoned that removing host cell contaminating RNA would reduce background during array hybridization and analysis in addition to allowing the use of minimal amounts of RNA to keep cDNA synthesis and labeling both efficient and cost effective. Therefore, we have optimized a technique using differential centrifugation to produce high-quality rickettsial RNA suitable for microarray analysis. It is likely that this technique will be easily adapted for use on other obligate intracellular organisms.
L929 mouse fibroblast cells were infected with R. prowazekii (Madrid E strain) at a multiplicity of infection of 50 (to give 5 to 10 rickettsiae per cell and >95% of the total cells infected), as previously described (3). After 48 h of growth, a condition that routinely yielded approximately 200 to 300 rickettsiae per infected cell, one-half of the flasks were transferred to 42°C for 30 min. Infected L cells (1,850 cm2) from both the control (34°C) and heat shock (42°C) conditions were harvested by trypsin treatment and collected by centrifugation. Trypsinization and all subsequent steps were performed in the presence of a 20% (vol/vol) concentration of DNA/RNA Protect (Sierra Diagnostics, California) to preserve nucleic acid integrity. We tested various reagents that are used to preserve RNA integrity and determined that the DNA/RNA Protect reagent from Sierra Diagnostics proved optimal for use with rickettsiae, presumably due to a lower level of viscosity allowing for efficient recovery of bacteria during the centrifugation steps of the protocol. The rickettsia-infected L-cell pellets from each condition were suspended in 1 ml of SPGMg-Sierra (sucrose, 0.281 M; KH2PO4, 3.76 mM; K2HPO4, 7.1 mM; glutamic acid, 5 mM; MgCl2, 10 mM; pH 7.0) (5, 27), which contains 20% (vol/vol) Sierra DNA/RNA Protect. Rickettsiae were released by ballistic shearing using a Mini-Beadbeater blender (BioSpec Products, Oklahoma) to deliver a 5-s pulse, followed by incubation on ice for 20 s (repeated three times). The lysate was removed, and the beads were washed three times with SPGMg-Sierra, resulting in approximately 4 ml of lysate. L-cell debris was removed by centrifugation (5 min, 1,150 × g, 24°C), the supernatant transferred to a 30-ml Corex tube, and rickettsiae collected by centrifugation (15 min, 9,600 × g, 4°C). The supernatant was discarded, and 0.1 ml of SPGMg-Sierra was added to the rickettsial pellet, which was mulled with a pestle. A 1-ml volume of RNA Wiz (Ambion) was added and the suspension mixed. Total RNA was extracted as per the manufacturer's directions, but in lieu of the final precipitation step, a 0.56 volume of 100% ethanol was added slowly with thorough mixing prior to application onto a Qiagen RNeasy mini-spin column. Following an on-column DNase I (Qiagen) digestion step (as per the manufacturer's directions), total RNA was eluted from the column twice (final volume of 60 μl) in RNase-free water containing 0.1 mM EDTA (pH 8.0). After release of the rickettsiae by ballistic shearing and differential centrifugation, the rickettsial rRNA constituted ∼38% of the total RNA in the sample (see Fig. S1B in the supplemental material).
We have successfully employed two methods to substantially reduce the levels of contaminating host RNA from the preparation prior to microarray analyses. The first involved the use of Ambion's MICROBEnrich technology. Host RNA levels were quantified, and the sample was prepared and processed as per the manufacturer's directions, resulting in the removal of ∼90% of the remaining eukaryotic rRNA (see Fig. S1C in the supplemental material). Alternatively, rickettsiae can be further purified after isolation by ballistic shearing and differential centrifugation, described above. Instead of the RNA isolation procedure and MICROBEnrich steps, the rickettsial pellet collected from the high-speed spin step was suspended (mulled and homogenized) in 3 ml of SPG-Sierra (containing no MgCl2). The sample was layered over 30 ml of SPG-Sierra containing 25% Renograffin (14) in an Oakridge centrifuge tube. Centrifugation was carried out at 26,000 × g, 60 min, 4°C. The resulting cell pellet was washed once in SPG-Sierra and RNA isolated as described above. This step yields rickettsial RNA of comparable purity to that of the MICROBEnrich-purified sample (compare Fig. S1C and D in the supplemental material). For the array experiments presented here, only total rickettsial RNA prepared using the MICROBEnrich method was used.
R. prowazekii ORF DNA microarray analyses.
One of the hallmarks of a typhus infection is the presentation of fever in the infected individual. Furthermore, analysis of the R. prowazekii genome suggested the presence of two sigma factor subunits of the RNA polymerase (1). The first is a verified homologue of the vegetative sigma factor, σD (2, 16), and the second is annotated as a homologue of the heat shock sigma factor, σH. Finally, there is evidence that R. prowazekii is able to adapt and grow at 40°C in cell culture model systems (12). Based on these observations, we rationalized that R. prowazekii might modulate gene expression in response to temperature upshift, making this an ideal condition to standardize analyses of global transcriptional control in this obligate intracellular pathogen. Purified rickettsial RNA (2 μg) from 34°C and 42°C heat shock samples was analyzed on an oligonucleotide DNA microarray as described in the text of the supplemental material. Details on array construction and validation are also described in the text of the supplemental material (also see Fig S2 in the supplemental material).
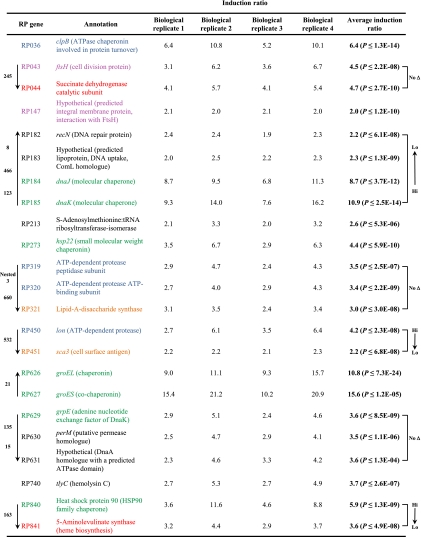
The data presented in Table 1 represent a compilation of the technical replicates for each biological replicate. Only genes that displayed a ≥2.0-fold (P < 0.05) change for all oligonucleotide probes in all four biological replicates are reported. In addition, an overall average induction ratio was calculated using all data from all biological replicates and the P value from the t test reported. In toto, the data illustrate that 23 rickettsial genes were induced ≥2.0-fold after 30 min of incubation at 42°C. In one preliminary experiment, we examined gene regulation at 15 min (one biological replicate only; data not shown) but ultimately determined that 30 min of incubation at 42°C was the shortest time tested that gave consistent, reproducible results. It is entirely possible that extended incubation at elevated temperatures could reveal additional genes or temporally regulated subsets of extreme heat shock genes. Finally, it is interesting to note that no genes with significant downregulation are reported. Our analyses revealed that there were some genes in some experiments that showed downregulation, but overall, there was no reproducibility between all four biological replicates (data not shown). Again, it is possible that extending the challenge past 30 min might reveal transcripts that are degraded/downregulated.
TABLE 1.
Heat shock induction ratios from four independent biological replicatesa
Only genes that displayed a ≥2.0-fold heat shock induction in all four experiments are shown (P ≤ 0.05). With the exception of RP627, every gene upregulated by heat shock was probed with two 70-mer oligonucleotides (in triplicate on the array). Data from all four biological replicates (two oligonucleotide probes from replicate, dye swap arrays) were combined and the overall induction ratio calculated and analyzed by a two-tailed t test. The overall average induction from these four experiments is shown to the right along with the P value. Arrows to the left of the table denote polarity, with a downward-facing arrow representing the positive-sense strand. The intergenic spacing in base pairs is given. Arrows to the right of the table denote groupings of interest, where “Hi → Lo” indicates that the induction ratios decrease in a polar fashion. “No Δ” denotes no change in the induction ratio for any gene in the given cluster. The color scheme groups genes with a (putative) common function: blue, protein turnover; light purple, cell division; green, molecular chaperonins; red, metabolism; orange, structural proteins; black, no apparent grouping.
Overall, 57% of the genes reported as significantly upregulated in this study are annotated homologues of known heat-shock-inducible genes. As interpolated from Table 1, 26% of the heat-shock-induced genes were putative proteases and 30% were putative chaperones. In addition, the two R. prowazekii ORFs annotated as heat shock genes hsp22 (RP273) and hsp90 (RP840) were induced by temperature upshift. A protective role for the remaining 43% of the heat-shock-induced R. prowazekii genes is not intuitively obvious based on their annotated functions. This subset of genes may play a heretofore-unknown role in the R. prowazekii response to elevated temperatures. Alternatively, these genes may show an inadvertent induction based upon their proximity to known heat shock genes. Thirty-five percent of the R. prowazekii heat shock genes have no known role in heat stress responses but lie immediately downstream of heat shock genes expected to play a protective role. As shown in Table 1 (right column), there are two distinct patterns evident. The first pattern consists of three loci (i.e., groups of successive genes that are all heat shock induced) where the genes with predicted roles in heat stress demonstrate the highest levels of induction and the genes with unknown roles lie distal and show lower induction ratios (reminiscent of a polar effect if these genes are indeed transcribed as an operon). The second pattern consists of two loci where genes with predicted heat stress roles and those without predicted roles display very similar induction ratios. Nine percent of the R. prowazekii heat-shock-induced genes reported here have no known role in heat stress response and are not proximal to genes with predicted heat stress function.
Real-time qRT-PCR.
To validate the results generated by microarray analysis, we assayed the levels of mRNA corresponding to the R. prowazekii ORFs that demonstrated low, medium, and high induction (RP044, RP626, and RP629) using the Cepheid Smart Cycler real-time RT-PCR platform as previously described (3). The RNA used in these experiments came from the same control and heat shock samples that were used for microarray analysis. Separate reverse transcription (RT) reaction mixtures containing rickettsial RNA and reverse primers specific for the RP044, RP626, and RP629 ORFs were prepared (see Table S1 in the supplemental material). In all RT reaction mixtures, RNA loading was normalized such that 250 ng of 16S rRNA (based on the Agilent bioanalysis) was added. In all cases, control RT reactions containing no Superscript II reverse transcriptase were run to confirm the absence of contaminating DNA. These samples showed reaction profiles similar to primer controls where no template was added. The quantitative RT-PCR (qRT-PCR) results (using RNA from biological replicates 3 and 4) confirmed the overall induction trends determined by microarray analyses, although the absolute induction values differed slightly (see Fig. S3 in the supplemental material).
In conclusion, we have presented an efficient methodology for the isolation of highly purified RNA from the obligate intracytoplasmic pathogen R. prowazekii. This methodology was used to assay rickettsial gene regulation in response to temperature upshift by microarray hybridization. We have determined that rickettsiae alter the expression of a regulon of 23 genes presumably involved in repair of protein damage elicited by the temperature upshift. This report is the first microarray analysis of R. prowazekii using a model stress response, and the RNA isolation method described here should be useful for studies of other obligate intracellular organisms.
Microarray data accession number.
The complete microarray data set generated in this study is deposited for public access in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE12377.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant R01AI-45533 from the National Institute of Allergy and Infectious Diseases to H.H.W.
The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIAID or the NIH.
All microarray design, construction, and hybridization were performed on a fee-for-services basis at the Washington University Microarray Core Genome Sequencing Center (St. Louis, MO). We thank Seth D. Crosby and Michael Heinz for their contributions. In addition, we thank Rosemary Roberts and Andrew Woodard for technical assistance during the early stages of this project.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowdki, A. K. Naslund, A.-S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-143. [DOI] [PubMed] [Google Scholar]
- 2.Aniskovitch, L. P., and H. H. Winkler. 1996. Rickettsia prowazekii sigma factor σ73 can be overexpressed in Escherichia coli and promotes RNA polymerase binding and transcription. Microbiology 142:901-906. [DOI] [PubMed] [Google Scholar]
- 3.Audia, J. P., and H. H. Winkler. 2006. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 188:6261-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldridge, G. D., N. Burkhardt, M. J. Herron, T. J. Kurtti, and U. G. Munderloh. 2005. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl. Environ. Microbiol. 71:2095-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bovarnick, M. R., J. C. Miller, and J. C. Snyder. 1950. The influence of certain salts, amino acids, sugars, and proteins on the stability of rickettsiae. J. Bacteriol. 59:509-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozeman, F. M., S. A. Masiello, M. S. Williams, and B. L. Elisberg. 1975. Epidemic typhus rickettsiae isolated from flying squirrels. Nature 255:545-547. [DOI] [PubMed] [Google Scholar]
- 7.Cai, J., H. Pang, D. O. Wood, and H. H. Winkler. 1995. The citrate synthase-encoding gene of Rickettsia prowazekii is controlled by two promoters. Gene 163:115-119. [DOI] [PubMed] [Google Scholar]
- 8.Cai, J., and H. H. Winkler. 1993. Identification of tlc and gltA mRNAs and determination of in situ RNA half-life in Rickettsia prowazekii. J. Bacteriol. 175:5725-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, J., and H. H. Winkler. 1996. Transcriptional regulation in the obligate intracytoplasmic bacterium Rickettsia prowazekii. J. Bacteriol. 178:5543-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, J., and H. H. Winkler. 1997. Transcriptional regulation of the GLTA and TLC genes in Rickettsia prowazekii growing in a respiration-deficient host cell. Acta Virol. 41:285-288. [PubMed] [Google Scholar]
- 11.Ellison, D. W., T. R. Clark, D. E. Sturdevant, K. Virtaneva, S. F. Porcella, and T. Hackstadt. 2008. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 76:542-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudima, O. S. 1982. Reproduction of vaccine and virulent Rickettsia prowazekii strains in continuous cell lines at different temperatures. Acta Virol. 26:390-394. [PubMed] [Google Scholar]
- 13.Hackstadt, T. 1996. The biology of rickettsiae. Infect. Agents Dis. 5:127-143. [PubMed] [Google Scholar]
- 14.Hanson, B. A., C. L. Wisseman, Jr., A. Waddell, and D. J. Silverman. 1981. Some characteristics of heavy and light bands of Rickettsia prowazekii on Renografin gradients. Infect. Immun. 34:596-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, Z. M., A. M. Tucker, L. O. Driskell, and D. O. Wood. 2007. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 73:6644-6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marks, G. L., H. H. Winkler, and D. O. Wood. 1992. Isolation and characterization of the gene coding for the major sigma factor of Rickettsia prowazekii DNA-dependent RNA polymerase. Gene 121:155-160. [DOI] [PubMed] [Google Scholar]
- 17.Phibbs, P. V., Jr., and H. H. Winkler. 1982. Regulatory properties of citrate synthase from Rickettsia prowazekii. J. Bacteriol. 149:718-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phibbs, P. V., Jr., and H. H. Winkler. 1981. Regulatory properties of partially purified enzymes of the tricarboxylic acid cycle of Rickettsia prowazekii, p. 421-430. In W. Burgdorfer and R. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, NY.
- 19.Qin, A., A. M. Tucker, A. Hines, and D. O. Wood. 2004. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 70:2816-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rachek, L. I., A. Hines, A. M. Tucker, H. H. Winkler, and D. O. Wood. 2000. Transformation of Rickettsia prowazekii to erythromycin resistance encoded by the Escherichia coli ereB gene. J. Bacteriol. 182:3289-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachek, L. I., A. M. Tucker, H. H. Winkler, and D. O. Wood. 1998. Transformation of Rickettsia prowazekii to rifampin resistance. J. Bacteriol. 180:2118-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raoult, D., J. B. Ndihokubwayo, H. Tissot-Dupont, V. Roux, B. Faugere, R. Abegbinni, and R. J. Birtles. 1998. Outbreak of epidemic typhus associated with trench fever in Burundi. Lancet 352:353-358. [DOI] [PubMed] [Google Scholar]
- 23.Renesto, P., E. Gouin, and D. Raoult. 2002. Expression of green fluorescent protein in Rickettsia conorii. Microb. Pathog. 33:17-21. [DOI] [PubMed] [Google Scholar]
- 24.Tarasevich, I., E. Rydkina, and D. Raoult. 1998. Outbreak of epidemic typhus in Russia. Lancet 352:1151. [DOI] [PubMed] [Google Scholar]
- 25.Troyer, J. M., S. Radulovic, and A. F. Azad. 1999. Green fluorescent protein as a marker in Rickettsia typhi transformation. Infect. Immun. 67:3308-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss, E., B. L. Elisberg, F. M. Bozeman, R. A. Ormsbee, C. B. Philip, C. L. Wisseman, Jr., P. Fiset, D. Paretsky, C. M. Downs, and W. J. Vinson. 1968. Rickettsiae and rickettsial diseases. Science 159:553-556. [DOI] [PubMed] [Google Scholar]
- 27.Winkler, H. H. 1976. Rickettsial permeability: an ADP-ATP transport system. J. Biol. Chem. 251:389-396. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.