Abstract
Antimicrobial peptides were isolated from a phage display peptide library using bacterial magnetic particles (BacMPs) as a solid support. The BacMPs obtained from “Magnetospirillum magneticum” strain AMB-1 consist of pure magnetite (50 to 100 nm in size) and are covered with a lipid bilayer membrane derived from the invagination of the inner membrane. BacMPs are easily purified from a culture of magnetotactic bacteria by magnetic separation. Approximately 4 × 1010 PFU of the library phage (complexity, 2.7 × 109) was reacted with BacMPs. The elution of bound phages from BacMPs was performed by disrupting its membrane with phospholipase D treatment. Six candidate peptides, which were highly cationic and could bind onto the BacMP membrane, were obtained. They exhibited antimicrobial activity against Bacillus subtilis but not against Escherichia coli and Saccharomyces cerevisiae. The amino acid substitution of the selected peptide, KPQQHNRPLRHK (peptide 6-7), to enhance the hydrophobicity resulted in obvious antimicrobial activity against all test microorganisms. The present study shows for the first time that a magnetic selection of antimicrobial peptides from the phage display peptide library was successfully achieved by targeting the actual bacterial inner membrane. This BacMP-based method could be a promising approach for a high-throughput screening of antimicrobial peptides targeting a wide range of species.
Cationic antimicrobial peptides are part of the innate immunity of many species, including humans. The antimicrobial peptides were expected to be novel sources of antibiotics to treat multidrug-resistant bacteria due to the different antimicrobial mechanisms from nontraditional antibiotics. They show low toxicity for the host but rapid effects against gram-negative and gram-positive bacteria. Most cationic antimicrobial peptides consist of 12 to 50 amino acids and have a net positive charge of >2 due to excess arginine and lysine residues and ∼50% hydrophobic amino acids. These peptides can interact with the outer leaflet of the bacterial membrane, which has negatively charged phospholipids, and can cross or penetrate the membrane, resulting in cell lysis subsequent to the formation of pores (37).
Recently, the screening of antimicrobial peptides from a phage display peptide library has been conducted for its specific binding properties, using whole Escherichia coli cells and Eimeria acervulina sporozoites as targets (7, 24). However, these methods contained troublesome and tedious operations, such as washing the cell membrane and loosely bound phages with detergent, collecting microbial cells with bound phages by centrifugation, and phage eluting by a detergent or acidic buffer at pH 2.2. The use of detergents and acidic solutions may cause damage to the microbial cell membrane, resulting in a lower recovery of microbial cells and/or bound phages. Furthermore, antimicrobial peptides integrated into the membrane cannot be efficiently eluted by the elution conditions described above. Until now, little information on the efficient screening of antimicrobial peptides targeting the actual membrane has been reported. Recently, a bead-based process to overcome these disadvantages has been proposed, using liposome-conjugated magnetic beads (34) and lipopolysaccharide (LPS)-conjugated epoxy beads (11). The benefit of using the bead-based process was realized by more efficient reactions and washes. The use of a bead-based process could simplify the bound/free separation. Furthermore, because beads have a high surface area-to-volume ratio, this enables us to enhance the efficiency of peptide binding in biopanning.
Our research group has demonstrated the application of bacterial magnetic particles (BacMPs) obtained from “Magnetospirillum magneticum” AMB-1 as magnetic supports for biomolecules (1, 12, 16, 19, 21, 22, 35, 36). BacMPs are easily purified from a culture of magnetotactic bacteria by magnetic separation using a permanent magnet. BacMPs are 50 to 100 nm in size and enveloped by a lipid bilayer membrane. The lipid bilayer membrane of BacMPs was shown to be derived from a cytoplasmic membrane in magnetotactic bacteria (27). Recently, a specific integration of an antimicrobial peptide, temporin L, into the BacMP membrane has been observed (31). The peptide temporin L was originally isolated from the red frog Rana temporaria as a cationic peptide with antimicrobial activity (26). The basic amino acids in temporin L played an important role in its integration into the BacMPs. In contrast, other cationic peptides, such as the ribotoxin L3 loop (14) and the arginine chain peptides (8), which have translocation activity against a eukaryotic cell membrane, were not integrated into the BacMP membrane. These results suggest that the use of BacMPs as a solid support will be useful for the selection of antimicrobial peptides targeting the bacterial membrane.
In this study, efficient selection of membrane-bound peptides from the phage display peptide library was demonstrated by using BacMPs. Prior to peptide screening, a recovery method for bound phages from the BacMP membrane was established by disrupting its membrane with phospholipase D (PLD) treatment. Six peptides, which were highly cationic and could bind onto the BacMP membrane, were obtained as candidates. The antimicrobial activity of selected peptides was then evaluated. Furthermore, the effect of the substitution of hydrophilic residues in a selected peptide on antimicrobial activity was investigated.
MATERIALS AND METHODS
Preparation of BacMPs.
The magnetotactic bacterium M. magneticum AMB-1 was cultured microaerobically in magnetic spirillum growth medium (3) at room temperature for 6 to 7 days. The stationary-phase cells were centrifuged at 11,344 × g for 10 min at 4°C, resuspended in phosphate-buffered saline (10 mM, pH 7.4), and disrupted by three passes through a French press at 1,500 kg/cm2 (Ohtake Works Co., Ltd., Tokyo, Japan). BacMPs were magnetically isolated from disrupted cell fractions using a neodymium-boron (Nd-B) magnet. The BacMPs were washed 10 times with HEPES (2-[4-(2-hydroxyethyl)-1-iperazinyl]-ethanesulfonic acid) buffer (10 mM, pH 7.4) and 3 times with HEPES buffer containing 1 M sodium chloride to electrostatically remove membrane surface proteins attached to BacMPs. The BacMPs (1 mg) were suspended with 1 U proteinase K (Wako Pure Chemical Industries, Ltd., Osaka, Japan) in HEPES buffer for 30 min at room temperature with pulsed sonication (1-min sonication at 10-min intervals for 30 min) to further remove membrane proteins (31). The BacMPs were separated magnetically from the reaction mixture using an Nd-B magnet and washed five times with HEPES buffer (10 mM, pH 7.4). To determine the surface protein content on the BacMP membranes, purified BacMPs were treated with 7 M urea, 2 M thiourea, 4% (wt/vol) CHAPS (3-[(3-chloramidopropyl)-dimethylammonio]-1-propanesulfonate), and 40 mM Tris-based solution overnight at 4°C. The proteins in the supernatant were measured by the Bradford method (4).
Lipid extraction from BacMP membrane by PLD.
In order to recover phage bound to BacMPs, BacMPs were suspended in 10 mM Tris buffer (pH 8.0) containing 100 U/ml PLD (Sigma Chemical Co., St. Louis, MO), 150 mM sodium chloride, and 100 μM calcium chloride and incubated for 1.5 h at room temperature with agitation. To confirm membrane removal, the remaining membrane on BacMPs was extracted by a mixture of chloroform-methanol (2:1) as described by Kates (10) and finally dissolved with chloroform. The membrane fraction was analyzed by thin-layer chromatography (TLC) using a silica gel plate (Silica Gel 60F254; Merck, Tokyo, Japan) with chloroform-methanol-water (65:25:4, vol/vol/vol) as the development solvent and detected by Dittmer reagent (25).
Selection of BacMPs binding phage.
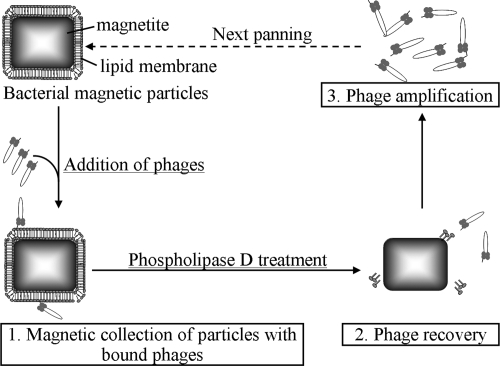
The experimental procedure for the biopanning using BacMPs is presented in Fig. 1. The Ph.D.-12 phage display peptide library kit with a complexity of 2.7 × 109 (New England BioLabs, Beverly, MA) was used for six rounds of affinity selection. Selection against the BacMP membrane was performed by incubating approximately 4 × 1010 PFU of the library phage (complexity, 2.7 × 109) with BacMPs (300 μg) in 50 mM Tris buffer containing 150 mM sodium chloride and 0.1% Tween 20 (pH 7.5) for 1 h at room temperature with agitation. The BacMPs were washed five times with HEPES containing 1 M sodium chloride and magnetically separated from nonspecific phages. Then, the BacMP membrane was disrupted by PLD treatment as described above, and the phages associated with the BacMP membrane were recovered from the supernatant after the magnetic separation of BacMPs. To propagate phages for the second round, an aliquot of the supernatant (220 μl) containing the phage was added to 20 ml of exponentially growing E. coli ER2738 culture and incubated for 4.5 h at 37°C. After bacterial cells were removed by centrifugation at 9,100 × g for 10 min twice, 1/6 (vol/vol) of PEG solution (20% polyethylene glycol 8000, 2.5 M sodium chloride in distilled water) was then added to the supernatant containing the phages and incubated overnight at 4°C to precipitate the phages. The phages were precipitated by centrifugation at 9,100 × g. The pellets were suspended in Tris-buffered saline (TBS) containing 0.02% sodium azide and used for the next round of biopanning. Phage titering was performed according to the manufacturer's instructions (Ph.D.-12 instruction manual, New England BioLabs, Beverly, MA).
FIG. 1.
Schematic procedure for biopanning using BacMPs.
Sequencing of phage DNA.
Phages obtained after the final round of selection were plated on agar plates, and clones were randomly selected for DNA isolation. The recovered phages (0.05 μl) were mixed with 200 μl of exponentially growing E. coli ER2738 cells, and the mixture was suspended in the top agarose layer (3 ml, 45°C) and incubated on an LB (1% Bacto-Tryptone, 0.5% yeast extract, 0.5% sodium chloride) agar plate containing X-Gal (5-bromo-4-chloro-3-inodyl-β-d-glucuronic acid) and IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 37°C. Plaque-forming phages (positive phages) were randomly selected at each round (14 to 48 clones) and amplified in E. coli ER2738 cells. After purification of phages by precipitation using PEG solution, the DNA sequences of individual phages were analyzed.
Peptide synthesis.
Nonlabeled peptides were synthesized by a standard 9-fluorenylmethoxycarbonyl-based solid-phase method with a peptide synthesizer (PSSM-8; Shimadzu Corp., Kyoto, Japan), purified by high-performance liquid chromatography and analyzed by mass spectrometry (LCQ Deca XP; Thermo Electron Corp., Chicago, IL). Deprotection of the N terminus protecting group (9-fluorenylmethoxycarbonyl) was achieved by 30% piperidine in dimethylformamide. The side chain amine of the lysine, arginine, and glutamine molecules is protected by a Boc group, a Pbf group, and a Trt group, respectively, all of which were deprotected by trifluoroacetic acid including 2.5% (vol/vol) m-cresol, 7.5% (vol/vol) ethanedithiol, and 7.5% (vol/vol) thioanisole. Labeled peptides with fluorescein isothiocyanate (FITC) at the C terminus were synthesized by Invitrogen Japan K.K. (Tokyo, Japan). Protean (Lasergene; DNAStar Inc., Madison, WI) was used to predict the pI. The total hydropathy index (THI) of peptides was calculated based on the hydropathy index of each amino acid (13).
Evaluation of BacMP-peptide binding.
FITC-labeled peptides (final concentration, 30 μM) were mixed with BacMPs (350 μg/ml) in HEPES buffer for 30 min at room temperature with pulsed sonication. After washing five times with HEPES containing 1 M NaCl, the fluorescence intensity of FITC-labeled peptides on BacMPs was measured using a spectrofluorometer (excitation, 495 nm; emission, 520 nm). The fluorescence intensity was expressed in arbitrary units.
Evaluation of antimicrobial activity.
The antimicrobial activity of the peptides was investigated against Escherichia coli K-12 as a model gram-negative bacterium and against Bacillus subtilis (BGSC 1A1) as a model gram-positive bacterium. Antimicrobial peptides showing antimicrobial activities frequently have toxicity to eukaryotic cells, including human cells. To evaluate the effect of selected peptides on eukaryotic cells, Saccharomyces cerevisiae (NRBC 0224) was also investigated as a model eukaryote. Each final concentration of peptides (0.1, 1, 10, or 100 μM) was added to 105 CFU/ml of cells diluted with TBS. After incubation for 1 to 8 h at 37°C, the samples were diluted with TBS, spread on LB or YPG (1% Bacto-Tryptone, 0.5% yeast extract, 2% glucose) agar plates, and incubated overnight at 37°C. The antimicrobial activity of each peptide was evaluated via CFU. The number of CFU per milliliter was calculated from the average colony number of three plates.
Assessment of hemolytic activity.
Cationic peptides showing antimicrobial activities frequently cause hemolysis; i.e., they are toxic to human cells. To evaluate the possibility that the selected peptides can cause the hemolysis of red blood cells, hemolytic activity was examined as follows: 2% goat erythrocytes (Nippon Biotest Laboratories Inc., Tokyo, Japan) were washed once and resuspended in TBS. The erythrocyte solution was incubated with 100 μM of peptides for 1 to 8 h at 37°C. The samples were then centrifuged at 800 × g for 5 min. The increase in absorbance upon hemoglobin release in the supernatant was evaluated by measuring the absorbance at 550 nm (A550) using a spectrophotometer (ALOKA Co., Ltd., Tokyo, Japan). The increase of A550 when 1% Triton X-100 was added is defined as 100%.
Hemolytic activity.
Hemolytic activity was determined using the following equation: (A550 in the presence of peptide − initial A550)/(A550 in the presence of 1% Triton X-100 − initial A550) × 100.
RESULTS
Preparation of BacMPs for extracting phages.
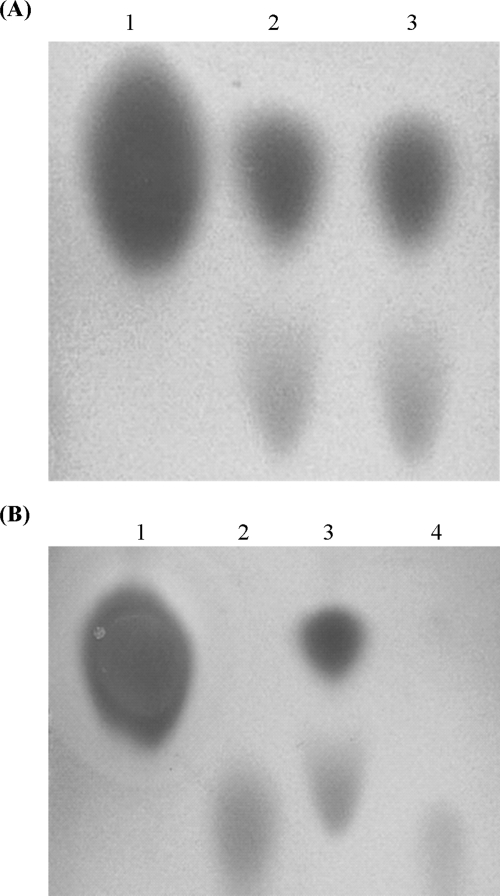
The BacMPs were suspended with proteinase K to remove membrane proteins. To clarify the protein and lipid contents of BacMPs before and after proteinase K treatment, proteins and lipids on the surfaces of BacMPs were quantified. The BacMP membrane contained 32 μg proteins and 188 μg phospholipids per mg particles. Among them, 31 μg of the membrane proteins was removed by proteinase K treatment (Table 1). On the other hand, TLC analysis showed that the phospholipids on BacMPs remained unchanged after proteinase K treatment (Fig. 2A; Table 1), in which phosphatidylethanolamine (PE) and an unidentified phospholipid were detected. These results suggest that the surface proteins on the BacMP membrane were efficiently removed by proteinase K treatment. The amount of protein that remained on BacMP was expected to be negligible for the following peptide screening.
TABLE 1.
Amounts of protein and phospholipid after enzymatic treatment of BacMPs
| Type of treatment | Protein (μg/mg particles) | Phospholipid (μg/mg particles) |
|---|---|---|
| Nontreatment | 32 | 188 |
| Proteinase K | 1 | 168 |
| Proteinase K + PLD | 1 | 13 |
FIG. 2.
TLC analysis of phospholipids. (A) Proteinase K (1 U/ml; 1 ml) was added to the BacMP suspension (final concentration, 1 mg/ml). After being washed, BacMPs were suspended in chloroform-methanol (2:1) to extract the lipid membrane. The lipid fraction was finally dissolved with chloroform and subjected to TLC analysis (lane 3). The sample obtained from nontreated BacMPs was also subjected as a control (lane 2). PE was used as a standard (lane 1). (B) BacMPs treated with proteinase K were further treated with PLD (lane 4). Nontreated BacMPs (lane 3), PE (lane 1), and phosphatidic acid (lane 2) were used as a control and standards. All samples were applied to a silica gel plate with chloroform-methanol-water (65:25:4, vol/vol/vol) and detected by Dittmer reagent.
Furthermore, the lipid content on the BacMP membrane was evaluated before and after PLD treatment.
After the PLD-treated BacMPs were magnetically separated, the remaining lipid on the BacMP surface was further extracted with chloroform-methanol (2:1). The sample solution was applied to a TLC plate. TLC bands of phospholipids extracted from the BacMP membrane were almost undetectable after PLD treatment (Fig. 2B). Phosphatidic acid, which was the product generated by the hydrolysis of phospholipids by PLD, was slightly detected. These results indicated that the BacMP membrane was efficiently hydrolyzed and released into the supernatant. Therefore, PLD treatment was used for membrane extraction and phage elution in the following experiments.
Selection and characterization of BacMP membrane-bound peptides.
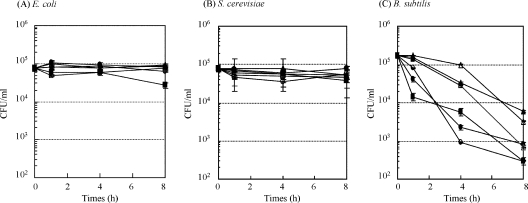
To select specific peptide sequences against the BacMP membrane, DNA sequences of 180 positive phages were analyzed from the first through sixth rounds (Fig. 1). However, a high frequency of a specific motif was not observed in the peptides. On the other hand, DNA sequencing indicates that the proportion of cationic peptides from randomly selected phage clones, which had pI values above 10, increased along with the panning process (Table 2). These results indicated that affinity selection was successfully accomplished. After 6 rounds of panning, six different peptides, including 5-24, 6-2, 6-7, 6-8, 6-17, and 6-45, were obtained as candidate peptides (Table 3). All of the peptides had complete matched peptide sequences in multiple clones. The THI of peptides was calculated based on the hydropathy index of each amino acid according to the method of Kyte and Doolittle (13). The amino acid sequences of candidate peptides (5-24, 6-2, 6-7, 6-8, 6-17, and 6-45) showed hydrophilic properties, with 8 to 25% hydrophobic amino acids. All of the candidate peptides showed antimicrobial activity against only B. subtilis, but not against E. coli and S. cerevisiae (Fig. 3). In contrast, no antimicrobial activity against all test microorganisms was observed when peptides 2-2, 2-20, 3-1, 3-4, 3-15, and 3-22 were used. These results suggest that an efficient selection of antimicrobial peptides was successfully achieved by using BacMP-based screening.
TABLE 2.
Number of eluted phages and the proportion of cationic peptides at each round of biopanning
| Round | Input phage (PFU) | Elute phage (PFU) | Ratio of elute to input phage | % Cationic peptides (no. of sequenced phage clones)a |
|---|---|---|---|---|
| 1 | 1.5 × 1011 | 9.5 × 106 | 6.3 × 10−5 | 13.6 (24) |
| 2 | 2.9 × 1010 | 2.7 × 108 | 9.3 × 10−3 | 18.2 (46) |
| 3 | 1.4 × 1011 | 3.3 × 107 | 2.4 × 10−4 | 31.8 (14) |
| 4 | 1.1 × 1011 | 1.3 × 107 | 1.2 × 10−4 | 63.2 (24) |
| 5 | 5.4 × 1010 | 1.4 × 107 | 2.6 × 10−4 | 76.2 (24) |
| 6 | 1.3 × 1010 | 7.5 × 107 | 5.8 × 10−3 | 65.0 (46) |
Cationic peptides were defined as having pI values above 10, calculated from the sequence analysis software Protean.
TABLE 3.
Sequences of peptides bound onto BacMPs obtained from phage display peptide library after 4 to 6 rounds of panning
| Peptide (round) | Sequencea | pI | THIb | Frequencyc |
|---|---|---|---|---|
| 2-2 (2) | ATETLARSLRLF | 11.0 | 3.1 | 1/46 |
| 2-20 (2) | YKHGMVTVGSTP | 9.5 | −2.7 | 1/46 |
| 3-1 (3) | KSLSRHDHIHHH | 9.8 | −21.2 | 2/14 |
| 3-4 (3) | HQTVVRPIPLFR | 12.5 | −0.1 | 1/14 |
| 3-15 (3) | ASHMSWLGPGLR | 11.2 | −1.3 | 1/14 |
| 3-22 (3) | SSLYPARLQGMS | 9.7 | −2.4 | 1/14 |
| 5-24 (5) | QFNVQKVPKSKP | 10.8 | −15.0 | 2/24 |
| 6-2 (6) | KPIHHHPHLPLK | 10.5 | −13.3 | 2/48 |
| 6-7 (4&6) | KPQQHNRPLRHK | 12.5 | −33.1 | 2/48 |
| 6-8 (4&6) | KIPHPEHPTKFR | 10.5 | −15.1 | 2/48 |
| 6-17 (5&6) | GPVHKHLPKAHK | 10.9 | −20.4 | 3/48 |
| 6-45 (4&6) | VFAGKPSHKPPH | 10.6 | −11.4 | 2/48 |
The amino acids are given in the one-letter code. Basic amino acids are bold.
THI of peptides was calculated based on the hydropathy index of each amino acid.
Number of clones with identical sequences in randomly sequenced clones.
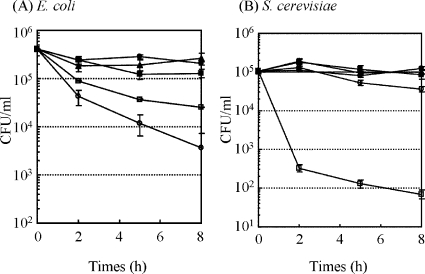
FIG. 3.
CFU of E. coli (A), B. subtilis (B), and S. cerevisiae (C) after inoculation with selected peptides. Synthesized peptides (final concentration, 100 μM) were added to approximately 105 CFU/ml of cells. After incubation for 1 to 8 h at 37°C, the samples were spread on an agar plate and incubated overnight at 37°C. •, peptide 5-14; ▪, peptide 6-2; ▴, peptide 6-7; ○, peptide 6-8; □, peptide 6-17; ▵, peptide 6-45.
To provide additional support for the identified peptides binding to the BacMPs, the binding of peptides onto BacMPs was examined using synthesized peptides labeled with FITC. All of the peptides, which showed antimicrobial activity against B. subtilis (i.e., peptides 5-24, 6-2, 6-7, 6-8, 6-17, and 6-45), bound onto the BacMP surface (results not shown). Among them, peptide 6-7 showed the highest binding capacity against the BacMP membrane.
Effect of amino acid substitutions on antimicrobial activity.
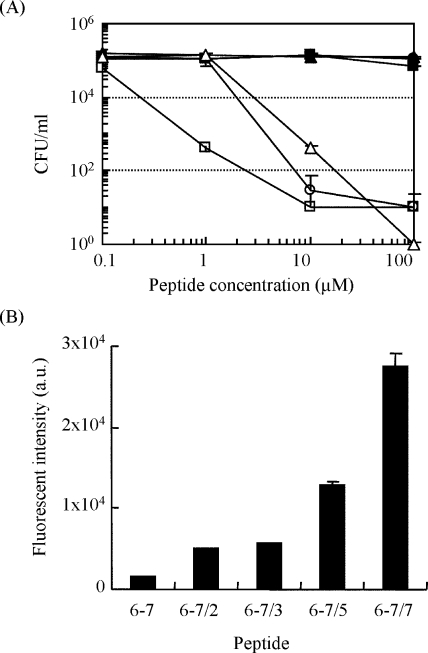
In order to improve the antimicrobial activity of a selected peptide, 6-7, the substitution at specific sites of the peptide was performed using peptide 6-7 as a template, which showed the highest binding capacity against the BacMP membrane. Proline, glutamine, and asparagine or histidine were substituted with phenylalanine, valine, or tryptophan, respectively (peptides 6-7/2 to 6-7/7) (Table 4). The THI by Kyte and Doolittle (13) showed that hydrophobicity increased with the substitutions. The antimicrobial activity of peptides 6-7/5 and 6-7/7 at 100 μM against B. subtilis was 1.1 × 104 times higher than that of the unmodified peptide 6-7 (Fig. 4A). The obvious activity was observed by incubating 1 or 10 μM of the modified peptides for 1 h, while no significant activity was obtained by incubating 100 μM of peptide 6-7 for 4 h (Fig. 3C). The modified peptide 6-7/5, where proline was substituted with phenylalanine and glutamine/asparagine was substituted with valine, showed a much higher level of activity than peptides 6-7/2 and 6-7/3. This phenomenon indicated that specific amino acids did not affect the antimicrobial activity. A more remarkable increase of the antimicrobial activity was observed in peptide 6-7/7 by further substituting histidine into tryptophan, resulting in slightly greater hydrophobicity. The antimicrobial activities of peptide 6-7/7 at 1 μM were 2.6 × 102 times higher than those of the unmodified peptide 6-7 (Fig. 4A). These results suggest that an amphiphilic feature of modified peptides was considered a possible reason for antimicrobial activity. Furthermore, the binding assay of FITC-labeled peptide with the BacMP membrane indicated that higher binding onto the BacMP membrane may be related to the antimicrobial activity (Fig. 4B).
TABLE 4.
Sequences of synthesized peptides with amino acid substitutions
| Peptide | Peptide sequencea | pI | THIb |
|---|---|---|---|
| 6-7 | KPQQHNRPLRHK | 12.5 | −33.1 |
| 6-7/2 | KFQQHNRFLRHK | 12.5 | −24.3 |
| 6-7/3 | KPVVHVRPLRHK | 12.5 | −10.0 |
| 6-7/5 | KFVVHVRFLRHK | 12.5 | −1.2 |
| 6-7/7 | KFVVWVRFLRWK | 12.5 | 3.4 |
Amino acids are given in the one-letter code. Basic amino acids and substituted amino acids are bold and underlined, respectively.
THI of peptides was calculated based on the hydropathy index of each amino acid.
FIG. 4.
(A) CFU of B. subtilis incubated on agar culture medium after the addition of modified peptide under the different concentrations. The modified peptides were added to approximately 105 CFU/ml of cells. After incubation for 1 h at 37°C, the samples were spread on an agar plate and incubated overnight at 37°C. •, peptide 6-7 (original); ▪, peptide 6-7/2; ▴, peptide 6-7/3; ○, peptide 6-7/5; □, peptide 6-7/7; ▵, temporin L (positive control). (B) Fluorescence intensity of FITC-labeled peptides bound onto BacMPs. FITC-labeled peptides (final concentration, 30 μM) were mixed with BacMPs (350 μg/ml) in HEPES buffer for 30 min at room temperature with pulsed sonication. After washing the FITC-labeled peptides five times with HEPES containing 1 M NaCl, the fluorescence intensity of FITC-labeled peptide on BacMPs was measured using a spectrofluorometer (excitation, 495 nm; emission, 520 nm).
A similar tendency was observed when E. coli and S. cerevisiae were used as targets (Fig. 5). The CFU of S. cerevisiae was reduced 103-fold by incubating with peptide 6-7/7 at 100 μM for 8 h (Fig. 5B). In contrast, lower activity was observed against E. coli (Fig. 5A) than against B. subtilis and S. cerevisiae. The lower activity was expected to be due to the inhibitory effect of LPS on the E. coli cell surface. To confirm the effect of LPS on the antimicrobial activity against E. coli, bacterial cells were treated with a chelating agent, EDTA, prior to the peptide addition to disrupt the stabilizing effect of the outer membrane. A remarkable increase in antimicrobial activity was observed after the 1 mM EDTA treatment. These results indicated that the lower level of activity against E. coli was due to the inhibitory effect of LPS. All modified peptides scarcely exhibited hemolytic activity against goat erythrocytes by incubating for 4 h at 100 μM (Table 5). These activities were much less than those of antimicrobial peptide LL-37, which indicated 10% hemolytic activity by incubating for 1 h at 60 μM (5).
FIG. 5.
CFU of E. coli (A) and S. cerevisiae (B) on agar culture medium after addition of modified peptides. The modified peptides (final concentration, 100 μM) were added to approximately 105 to 106 CFU/ml cells. After incubation for 1 to 8 h at 37°C, the samples were spread on an agar plate and incubated overnight at 37°C. •, peptide 6-7 (original); ▪, peptide 6-7/2; ▴, peptide 6-7/3; ○, peptide 6-7/5; □, peptide 6-7/7.
TABLE 5.
Hemolytic activity of each peptide against goat erythrocytesa
| Peptide | % Hemolytic activity at:
|
|
|---|---|---|
| 1 h | 4 h | |
| 6-7/2 | 0.0 ± 1.0 | 1.5 ± 1.3 |
| 6-7/3 | 0.0 ± 0.6 | 1.5 ± 1.9 |
| 6-7/5 | 0.0 ± 0.5 | 2.0 ± 2.0 |
| 6-7/7 | 0.0 ± 0.5 | 0.0 ± 1.4 |
Goat erythrocytes (2%) were incubated with 100 μM peptides at 37°C. The absorbance increase by hemoglobin release in the supernatant was evaluated. The increase of A550 by the addition of 1% Triton X-100 was defined as 100%. Triplicate measurements for each experiment were performed.
DISCUSSION
Until now, little information on the efficient screening of antimicrobial peptides targeting bacterial cells has been reported, although high-throughput screening using liposome-conjugated magnetic beads was recently reported (34). In this study, a magnetic selection of antimicrobial peptides using bacterial cell membrane on BacMPs obtained from a magnetotactic bacterium was demonstrated. The BacMPs consist of pure magnetite (Fe3O4) enveloped with a stable lipid membrane, which is 5- to 6-nm thick, indicating formed unilamellar vesicles. It was suggested that the BacMP membrane was derived from the invagination of the inner membrane in the gram-negative bacterium M. magneticum AMB-1 (21). The lipid bilayer membrane of BacMPs was shown to be derived from the cytoplasmic membrane in magnetotactic bacteria (27). The major lipids are phospholipids, fatty acid chains within the range of 12 to 18 carbons (20). PE accounted for approximately 50% of the total phospholipids present. Because negatively charged phospholipid membranes were the main target for antimicrobial peptides, the BacMP membrane is regarded as an ideal model for screening antimicrobial peptides.
On the other hand, phage elution from the membrane is a fundamental process for peptide screening. An acid treatment is commonly used for phage elution. However, acid treatment was not suitable for this study because antimicrobial peptides probably bound onto the membrane by both electrostatic and hydrophobic interactions. Therefore, lipid extraction was required to recover membrane-associated phages. Several methods for the extraction of lipids from the BacMP membrane, such as chloroform-methanol (2:1) (20) or 1% sodium dodecyl sulfate treatment with boiling (1), have been established; however, these methods were also not suitable for phage elution because of fatal damage to the bound phages by these detergents or organic solvents. Therefore, PLD treatment was employed as a mild method of extracting lipid membranes and eluting phages from BacMPs in this study.
Six cationic peptides (5-24, 6-2, 6-7, 6-8, 6-17, and 6- 45) showed antimicrobial activity against only B. subtilis, not against E. coli and S. cerevisiae (Fig. 3). Furthermore, no hemolytic activity was observed under the same conditions. This phenomenon agreed with the relatively higher level of activity of cationic antimicrobial peptides against prokaryotes (17, 37), especially gram-positive bacteria that lack LPS (23). On the other hand, BacMP membrane originating from the inner (cytoplasmic) membrane in gram-negative bacteria (21), which is composed mainly of negatively charged phospholipids, was used for peptide screening. Therefore, selected peptides could tightly bind to the microbial membrane by electrostatic interaction but not by penetrating the outer membrane or inserting into the cell membrane, resulting in no antimicrobial activity against E. coli and S. cerevisiae.
In general, the position of the basic amino acids in the sequence and the number of hydrophobic amino acids between the charged residues affect antimicrobial activity (2). Furthermore, aromatic amino acids, such as phenylalanine or tryptophan, have been found at the membrane interface to stabilize peptide in the membrane (33). Recently, analysis of antimicrobial peptides by a complete substitution library revealed that alanine and proline, in addition to acidic amino acids, do not affect their activity (9). Based on the knowledge described above, the modification of peptide 6-7 was performed by substituting hydrophilic amino acids with hydrophobic amino acids or aromatic amino acids. The basic amino acids, lysine and arginine, which can easily bind to negatively charged membranes, were localized on the N or C terminus of the peptides (Table 3). Furthermore, another basic amino acid, histidine, was present in many copies, although the expected frequency of histidine was calculated as 3.1% in the Ph.D. library. Therefore, the histidine residues may contribute to the electrostatic interaction between the peptides and the BacMP membrane. However, a high frequency of a specific motif was not observed in the peptides.
There are two hypothesized reasons to justify the high binding of the selected peptides to the bacterial cell membrane. Firstly, the obtained peptides bound onto the BacMP membrane under the conditions when the electrostatic interactions between the membrane and highly cationic peptides were cancelled. Previous reports indicated that highly cationic peptides, the ribotoxin L3 loop (pI, 11.4), and the arginine chain peptide (pI, 13.1) could bind the BacMP membrane by electrostatic interactions but were completely removed from the membrane by being washed with HEPES buffer containing 1 M NaCl. The peptides obtained in this experiment were still bound onto the BacMP membrane under the same conditions. These results suggest that the peptides were tightly bound onto the BacMP membrane.
The second reason was due to the antimicrobial characteristics of the peptides. The obtained peptides showed higher activity against the gram-positive bacterium Bacillus subtilis. In general, a lower level of activity of antimicrobial peptides against eukaryotes is observed because the outer leaflet of the eukaryotic cell membrane is stabilized by cholesterol, weakening the net negative charge (17, 37). Furthermore, the antimicrobial peptides bind the negatively charged LPS of gram-negative bacteria which serves as a barrier to prevent the arrival of peptides in the inner membrane (23). These tendencies were well suited to the results shown in Fig. 3. In order to confirm the barrier effect of LPS in E. coli, the effect of disrupting the outer membrane by EDTA on antimicrobial activity against E. coli was investigated. EDTA is thought to chelate magnesium ions from the LPS of the outer membrane of gram-negative bacteria, causing the outer cell surface to become more permeable (6). The antimicrobial activity was increased 10-fold when 100 μM of peptides was incubated at 37°C (data not shown). This result also supports the hypothesis described above that the selected peptides showed high levels of binding activity against the microbial cell membrane.
The antimicrobial activity of selected peptides was improved by substituting amino acids and increasing the hydrophobicity. The modified peptides exhibited little hemolytic activity. On the other hand, amphiphilic (i.e., cationic and hydrophobic) peptides, which potentially showed broad spectra against microorganisms, were not obtained by the first screening from the random peptide library using BacMPs. One of the possible reasons was due to the association of these peptides with hydrolyzed lipids, resulting in the lower infection efficiency of the phages to E. coli cells. However, our proposed method, i.e., the first screening of membrane-bound peptides from the random peptide library using BacMPs and the subsequent substitution of amino acids in the peptides, will be one powerful approach to obtain the antimicrobial peptide efficiently.
The lipid bilayer imparts BacMPs with better dispersion qualities than those of artificial magnetic particles, and superior dispersion permits various applications of BacMPs. Antibody and DNA have been fixed on BacMPs and applied to automated immunoassays (29, 30, 32) and DNA detection systems (15, 18, 28) as magnetic supports. The fully automated system with BacMPs allows simultaneous execution and will be applied to the peptide screening using BacMPs in future work.
Acknowledgments
This work was funded in part by Grants-in-Aid for Specially Promoted Research, no. 13002005, and for Young Scientists (A), no. 17686073, from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 24 October 2008.
REFERENCES
- 1.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 2.Bender, V., M. Ali, M. Amon, E. Diefenbach, and N. Manolios. 2004. T cell antigen receptor peptide-lipid membrane interactions using surface plasmon resonance. J. Biol. Chem. 279:54002-54007. [DOI] [PubMed] [Google Scholar]
- 3.Blakemore, R. P., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Ciornei, C. D., T. Sigurdardottir, A. Schmidtchen, and M. Bodelsson. 2005. Antimicrobial and chemoattractant activity, lipopolysaccharide neutralization, cytotoxicity, and inhibition by serum of analogs of human cathelicidin LL-37. Antimicrob. Agents Chemother. 49:2845-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutter, C. N., and G. R. Siragusa. 1995. Population reductions of gram-negative pathogens following treatments with nisin and chelators under various conditions. J. Food Prot. 58:977-983. [DOI] [PubMed] [Google Scholar]
- 7.da Silva, A., Jr., U. Kawazoe, F. F. Freitas, M. S. Gatti, H. Dolder, R. I. Schumacher, M. A. Juliano, M. J. da Silva, and A. Leite. 2002. Avian anticoccidial activity of a novel membrane-interactive peptide selected from phage display libraries. Mol. Biochem. Parasitol. 120:53-60. [DOI] [PubMed] [Google Scholar]
- 8.Futaki, S., T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda, and Y. Sugiura. 2001. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 276:5836-5840. [DOI] [PubMed] [Google Scholar]
- 9.Hilpert, K., R. Volkmer-Engert, T. Walter, and R. E. Hancock. 2005. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23:1008-1012. [DOI] [PubMed] [Google Scholar]
- 10.Kates, M. 1972. Lipid extraction procedures. North-Holland Publishing Co., Amsterdam, The Netherlands.
- 11.Kim, Y. G., C. S. Lee, W. J. Chung, E. M. Kim, D. S. Shin, J. H. Rhim, Y. S. Lee, B. G. Kim, and J. Chung. 2005. Screening of LPS-specific peptides from a phage display library using epoxy beads. Biochem. Biophys. Res. Commun. 329:312-317. [DOI] [PubMed] [Google Scholar]
- 12.Kuhara, M., H. Takeyama, T. Tanaka, and T. Matsunaga. 2004. Magnetic cell separation using antibody binding with protein A expressed on bacterial magnetic particles. Anal. Chem. 76:6207-6213. [DOI] [PubMed] [Google Scholar]
- 13.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 14.Langedijk, J. P. 2002. Translocation activity of C-terminal domain of pestivirus Erns and ribotoxin L3 loop. J. Biol. Chem. 277:5308-5314. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama, K., H. Takeyama, E. Nemoto, T. Tanaka, K. Yoda, and T. Matsunaga. 2004. Single nucleotide polymorphism detection in aldehyde dehydrogenase 2 (ALDH2) gene using bacterial magnetic particles based on dissociation curve analysis. Biotechnol. Bioeng. 87:687-694. [DOI] [PubMed] [Google Scholar]
- 16.Matsunaga, T., Y. Okamura, and T. Tanaka. 2004. Biotechnological application of nano-scale engineered bacterial magnetic particles. J. Mater. Chem. 14:2099-2105. [Google Scholar]
- 17.Matsuzaki, K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1462:1-10. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa, T., R. Hashimoto, K. Maruyama, T. Tanaka, H. Takeyama, and T. Matsunaga. 2006. Capture and release of DNA using aminosilane-modified bacterial magnetic particles for automated detection system of single nucleotide polymorphisms. Biotechnol. Bioeng. 94:862-868. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura, C., J. G. Burgess, K. Sode, and T. Matsunaga. 1995. An iron-regulated gene, magA, encoding an iron transport protein of Magnetospirillum sp. strain AMB-1. J. Biol. Chem. 270:28392-28396. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura, N., K. Hashimoto, and T. Matsunaga. 1991. Immunoassay method for the determination of immunoglobulin G using bacterial magnetic particles. Anal. Chem. 63:268-272. [DOI] [PubMed] [Google Scholar]
- 21.Okamura, Y., H. Takeyama, and T. Matsunaga. 2001. A magnetosome-specific GTPase from the magnetic bacterium Magnetospirillum magneticum AMB-1. J. Biol. Chem. 276:48183-48188. [DOI] [PubMed] [Google Scholar]
- 22.Okamura, Y., H. Takeyama, and T. Matsunaga. 2000. Two-dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillum sp. AMB-1. Appl. Biochem. Biotechnol. 84-86:441-446. [DOI] [PubMed] [Google Scholar]
- 23.Papo, N., and Y. Shai. 2005. A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 280:10378-10387. [DOI] [PubMed] [Google Scholar]
- 24.Pini, A., A. Giuliani, C. Falciani, Y. Runci, C. Ricci, B. Lelli, M. Malossi, P. Neri, G. M. Rossolini, and L. Bracci. 2005. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob. Agents Chemother. 49:2665-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw, B. R., J. L. Corden, C. G. Sahasrabuddhe, and K. E. Van Holde. 1974. Chromatographic separation of chromatin subunits. Biochem. Biophys. Res. Commun. 61:1193-1198. [DOI] [PubMed] [Google Scholar]
- 26.Simmaco, M., G. Mignogna, S. Canofeni, R. Miele, M. L. Mangoni, and D. Barra. 1996. Temporins, antimicrobial peptides from the European red frog Rana temporaria. Eur. J. Biochem. 242:788-792. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka, M., Y. Okamura, A. Arakaki, T. Tanaka, H. Takeyama, and T. Matsunaga. 2006. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics 6:5234-5247. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, T., K. Maruyama, K. Yoda, E. Nemoto, Y. Udagawa, H. Nakayama, H. Takeyama, and T. Matsunaga. 2003. Development and evaluation of an automated workstation for single nucleotide polymorphism discrimination using bacterial magnetic particles. Biosens. Bioelectron. 19:325-330. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, T., and T. Matsunaga. 2001. Detection of HbA(1c) by boronate affinity immunoassay using bacterial magnetic particles. Biosens. Bioelectron. 16:1089-1094. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, T., and T. Matsunaga. 2000. Fully automated chemiluminescence immunoassay of insulin using antibody-protein A-bacterial magnetic particle complexes. Anal. Chem. 72:3518-3522. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, T., H. Takeda, Y. Kokuryu, and T. Matsunaga. 2004. Spontaneous integration of transmembrane peptides into a bacterial magnetic particle membrane and its application to display of useful proteins. Anal. Chem. 76:3764-3769. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, T., H. Takeda, F. Ueki, K. Obata, H. Tajima, H. Takeyama, Y. Goda, S. Fujimoto, and T. Matsunaga. 2004. Rapid and sensitive detection of 17beta-estradiol in environmental water using automated immunoassay system with bacterial magnetic particles. J. Biotechnol. 108:153-159. [DOI] [PubMed] [Google Scholar]
- 33.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 34.Xie, Q., S. Matsunaga, Z. Wen, S. Niimi, M. Kumano, Y. Sakakibara, and S. Machida. 2006. In vitro system for high-throughput screening of random peptide libraries for antimicrobial peptides that recognize bacterial membranes. J. Pept. Sci. 12:643-652. [DOI] [PubMed] [Google Scholar]
- 35.Yoshino, T., F. Kato, H. Takeyama, M. Nakai, Y. Yakabe, and T. Matsunaga. 2005. Development of a novel method for screening of estrogenic compounds using nano-sized bacterial magnetic particles displaying estrogen receptor. Anal. Chim. Acta 532:105-111. [Google Scholar]
- 36.Yoshino, T., M. Takahashi, H. Takeyama, Y. Okamura, F. Kato, and T. Matsunaga. 2004. Assembly of G protein-coupled receptors onto nanosized bacterial magnetic particles using Mms16 as an anchor molecule. Appl. Environ. Microbiol. 70:2880-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]