Abstract
The crenarchaeal order Sulfolobales collectively contain at least five major terminal oxidase complexes. Based on genome sequence information, all five complexes are found only in Metallosphaera sedula and Sulfolobus tokodaii, the two sequenced Sulfolobales capable of iron oxidization. While specific respiratory complexes in certain Sulfolobales have been characterized previously as proton pumps for maintaining intracellular pH and generating proton motive force, their contribution to sulfur and iron biooxidation has not been considered. For M. sedula growing in the presence of ferrous iron and reduced inorganic sulfur compounds (RISCs), global transcriptional analysis was used to track the response of specific genes associated with these complexes, as well as other known and putative respiratory electron transport chain elements. Open reading frames from all five terminal oxidase or bc1-like complexes were stimulated on one or more conditions tested. Components of the fox (Msed0467 to Msed0489) and soxNL-cbsABA (Msed0500 to Msed0505) terminal/quinol oxidase clusters were triggered by ferrous iron, while the soxABCDD′ terminal oxidase cluster (Msed0285 to Msed0291) were induced by tetrathionate and S0. Chemolithotrophic electron transport elements, including a putative tetrathionate hydrolase (Msed0804), a novel polysulfide/sulfur/dimethyl sulfoxide reductase-like complex (Msed0812 to Msed0818), and a novel heterodisulfide reductase-like complex (Msed1542 to Msed1550), were also stimulated by RISCs. Furthermore, several hypothetical proteins were found to have strong responses to ferrous iron or RISCs, suggesting additional candidates in iron or sulfur oxidation-related pathways. From this analysis, a comprehensive model for electron transport in M. sedula could be proposed as the basis for examining specific details of iron and sulfur oxidation in this bioleaching archaeon.
Certain extremely thermoacidophilic archaea are promising candidates for biomining operations targeting the recovery of base and precious metals (44). These microorganisms grow at elevated temperatures where abiotic ferrous oxidation rates are accelerated and where passivation of mineral surfaces from reduced inorganic sulfur compounds (RISCs) is minimal (44). To tap into the biotechnological potential of these microorganisms, their physiological characteristics that relate to metal mobilization need to be better understood, especially mechanisms underlying biooxidation of iron (Fe2+) and RISCs. Biooxidation implicates membrane-associated protein complexes that mediate electron transport, which seemingly determines the capability and capacity of extreme thermoacidophiles for metal and sulfur mobilization. However, the complexes required for bioleaching have not been established. Models that address the specific and collective function of electron transport complexes in the Sulfolobales are needed to provide a physiological framework for exploring the intricacies of biological metal and sulfur oxidation.
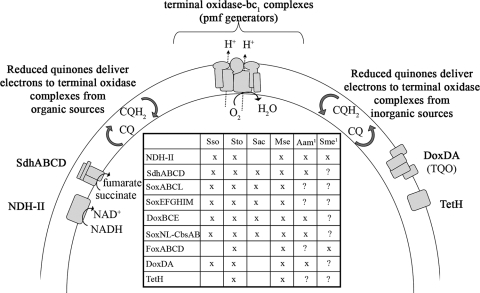
Current knowledge of respiratory electron transport chains (ETC) in the Sulfolobales is based primarily on separate biochemical characterization studies performed in various members of this order (Fig. 1 and Table 1). Based on studies in Acidianus ambivalens, Sulfolobus metallicus, and Sulfolobus tokodaii (3, 17, 24, 32, 40), electrons from NADH or succinate oxidation presumably enter ETC via NADH:quinone oxidoreductases (NDH-II enzymes) or succinate:quinone oxidoreductases (SdhABCD; type E). Reduced quinones (typically caldariella or Sulfolobus-type quinones) then deliver electrons to one of several terminal oxidase complexes, composed of cytochrome oxidase and/or bc1 complex-like subunits. These subunits are related to aa3 terminal oxidase complexes in A. ambivalens (DoxBCE) (14, 37, 42) and Sulfolobus acidocaldarius (SoxABCD-SoxL) (16, 34, 35), the bb3 terminal oxidase complex in S. acidocaldarius (SoxEFGHIM) (11, 29, 33), the putative FoxAB terminal oxidase in S. metallicus (the recent discovery of which coincided with the first report of ferrous iron oxidation in S. tokodaii) (5), and bc1-complex in S. acidocaldarius (SoxLN) (22). Both FoxAB and SoxNL are associated with cytochrome b558/566-like proteins (FoxCD and CbsAB), which exhibit characteristics found in other terminal oxidase subunits (i.e., glycosylated like SoxBC and conformational flexibility like Rieske proteins) and are probably involved in ETC in a similar manner (5, 21, 22, 37, 47). Terminal oxidase complexes are important in extreme thermoacidophiles as proton pumps for maintaining intracellular pH and generating proton motive force. Other proteins may contribute to proton motive force generation by using electrons from Fe2+ and RISCs to reduce quinones. Leading candidates for this function include the thiosulfate:quinone oxidoreductase (TQO) in A. ambivalens (37), as well as the tetrathionate hydrolase (TetH) in the mesoacidophilic bioleaching bacteria Acidithiobacillus caldus and Acidithiobacillus ferrooxidans (9, 26, 46). Recent work with A. ferrooxidans has shown that the respiratory ETC encoded in the rus and petI operons are involved in uphill electron transport from Fe2+ oxidation, with electrons being donated via the blue copper protein, rusticyanin, to a bc1 complex operating in reverse, possibly to NDH-II (8, 15, 23). However, the recently completed genome sequence of the bioleacher Metallosphaera sedula revealed that these operons are not present (2, 12). Although some individual components are similar (i.e., blue copper protein and PetAB-like subunit sequences), no capacity for uphill electron transfer has been demonstrated to date in the Sulfolobales.
FIG. 1.
Composite model of respiratory electron transport chain components in the Sulfolobales. Inset table notes components identified in selected Sulfolobales. Sso, S. solfataricus; Sto, S. tokodaii; Sac, S. acidocaldarius; Mse, M. sedula; Aam, A. ambivalens; Sme, S. metallicus. Genome sequences are not yet available for organisms with a superscript 1. pmf, protein motive force.
TABLE 1.
Components of respiratory ETC in the order Sulfolobales
| Component | ORF(s) | Homolog present in: | Reference | Proposed function |
|---|---|---|---|---|
| NDH-II | Msed2059 | A. ambivalens | 17 | Transfer of electron from NADH to quinones |
| S. metallicus | 3 | |||
| SdhABCD | Msed0674-Msed0677 | A. ambivalens | 32 | Transfer of electron from succinate to quinones |
| S. tokodaii | 24 | |||
| Terminal oxidases | ||||
| SoxABCDL | Msed0285-Msed0291 | S. acidocaldarius | 16 | aa3-type transferring electrons to O2 (proton pump) |
| SoxEFGHIM | Msed0319-Msed0324 | S. acidocaldarius | 29 | bb3-type transferring electrons to O2 (proton pump) |
| DoxBCE | Msed2030-Msed2032 | A. ambivalens | 14 | aa3-type transferring electrons to O2 (proton pump) |
| SoxNL-CbsAB | Msed0500-Msed0504 | S. acidocaldarius | 22 | Electron transfer, direction unknown |
| FoxABCD | Msed0477-Msed0478, Msed0480, Msed0484-Msed0485 | S. metallicus/S. tokodaii | 5 | Electron transfer, direction unknown |
| DoxDA | Msed0363-Msed0364 | A. ambivalens | 37 | Transfer of electron from thiosulfate to quinones |
| TetH | Msed0804 | A. ferrooxidans | 26 | Disproportionation of tetrathionate |
| A. caldus | 9 |
Functional genomics approaches are potentially useful for gleaning the detailed features of respiratory ETC chains with respect to biological iron and sulfur oxidation, since genes encoding ETC components in both mesoacidophilic and extremely thermoacidophilic bioleachers apparently are differentially transcribed in the presence of RISC- or Fe2+-based substrates (5, 8, 27, 43, 51). Utilizing this observation, the global transcriptional response of bioleacher M. sedula to the presence and/or absence of inorganic electron donors was investigated during growth on yeast extract. Transcriptome response was used to support the role of previously characterized proteins in ferrous iron and RISC-induced ETC and to propose new candidates for members of ETC. The model resulting from this analysis provides the basis for probing metabolic features of iron and sulfur oxidation in extreme thermoacidophiles.
MATERIALS AND METHODS
Growth of M. sedula in the presence of inorganic iron and sulfur compounds.
M. sedula (DSMZ 5348) was grown aerobically at 70°C in an orbital shaking oil bath at 70 rpm on DSMZ 88 medium (pH 2), supplemented with 0.1% yeast extract (Y medium). Growth medium was supplemented with 10 g/liter of FeSO4·7H2O (1% [wt/vol]; 36 mM) (YFS medium), 20 g/liter of elemental sulfur (S0) (2% [wt/vol]) (YS medium), 10 g/liter K2S4O6 (1% [wt/vol]; 33 mM) (YKT medium), or 6 g/liter K2SO4 (29 mM) (YKS medium). Cells were acclimated to supplemented medium through three passages before harvesting. On the fourth passage, an initial density 5 × 106 ± 1 × 106 cells/ml was used in two 1-liter bottles containing a 300-ml volume (four 1-liter bottles were used for the FeSO4-supplemented condition). Cultures in mid-exponential phase (∼24 h postinoculation; 4 × 107 to 7 × 107 cells/ml) were harvested by quickly chilling and then centrifuging at 9,510 × g for 15 min at 4°C.
M. sedula oligonucleotide microarray construction and transcriptional response analysis.
A spotted whole-genome oligonucleotide microarray based on at least 2,256 protein-coding open reading frames (ORFs) was constructed (2,328 probes with 5 replicates of each, yielding 11,640 spots per array), as described previously (2). Probes (60-mers) were designed using OligoArray, version 2.1, software (45) and synthesized by Integrated DNA Technologies (Coralville, IA).
RNA was extracted and purified (RNAqueous; Ambion), reverse transcribed (Superscript III; Invitrogen), repurified, labeled with either Cy3 or Cy5 dye (GE Healthcare), and hybridized to one of five microarray slides (Corning). Slides were scanned on a Perkin-Elmer scanner, and raw intensities were quantitated using ScanArray Express, version 2.1.8, software. Normalization of data and statistical analysis were performed using JMP Genomics, version 3.1, software (SAS, Cary, NC). In general, significant differential transcription, or “response,” was defined to be relative changes at or above 2 (where a log2 value of ±1 equals a twofold change) with significance values at or above the Bonferroni correction, which was 5.4 (equivalent to a P value of 4.0 × 10− 6) for this data set.
Comparative genomics analysis.
For comparative genomics, M. sedula gene sequences and annotations were downloaded from the Joint Genome Institute microbial genome website (http://genome.ornl.gov/microbial/msed/28feb07/). Other sequences referenced were obtained from the GenBank, with the exception of A. ferrooxidans ATCC 23270 sequences, which were obtained from The Institute for Genome Research's Comprehensive Microbial Resource. The Basic Local Alignment Search Tool (BLAST) searches were conducted using a BLOSUM62 matrix with either the NCBI protein-protein blast program (BLASTP) against the Swissprot database, the nonredundant database, or against the genomes of Metallosphaera, three Sulfolobus species, and Ferroplasma; the Oak Ridge National Library Microbial BLAST server's BLASTP program against the Msed database; or the Comprehensive Microbial Resource tblastn program against the A. ferrooxidans genome.
Microarray data accession number.
Raw data, as well as final log2 relative changes, have been deposited in the NCBI Gene Expression Omnibus (GEO) database under series accession number GSE12044.
RESULTS
Genome sequence analysis was used in conjunction with transcriptomics to examine the presence and potential function of ETC in M. sedula. Previous reports on the biochemical function of specific proteins implicated in electron transfer in other extremely thermoacidophilic archaea (and mesoacidophiles as well) were incorporated into the analysis. From this, a model could be proposed for ETC that included new annotations for putative proteins in the in M. sedula genome sequence.
Quinone reduction from organic electron donors.
Other than the efforts with NDH-II mentioned above, little work has been done with type I NADH dehydrogenases (NDH-I) in extremely thermoacidophilic archaea. In fact, the lack of ORFs with similarity to NADH and flavin mononucleotide binding regions in NDH-I enzymes suggests that the actual electron donor may not be NADH (40). Type III NDHs have not been found in extremely thermoacidophilic archaea, and type IV NDHs (NDH-IV) have just recently been discovered in archaea (39). The best candidates for NDH-I subunits (NuoACDHIJKLN; Msed1895 to Msed1903), NDH-II (Msed2059), and NDH-IV (Msed0428) in M. sedula were not differentially transcribed during growth in the presence of Fe2+ or RISCs. In addition to succinate dehydrogenase (SdhABCD), encoded by Msed0674 to Msed0677, another appears to be encoded by Msed1112 to Msed1116 (SdhACD-FumAB), which has two iron-sulfur subunits more similar to class I fumarase than SdhB. Neither locus responded to Fe2+ or RISCs. Because ORFs encoding quinones in members of the Sulfolobales have yet to be annotated, transcriptional response information is not readily available.
Quinone reduction via inorganic electron donors.
The M. sedula genome encodes a putative TetH (Msed0804) (2); in fact, Msed0804 was upregulated under both RISC-supplemented conditions, i.e., growth on YKT (at 32-fold this was the largest change noted in this data set) and YS (Table 2; see also Table S1 in the supplemental material) media. Genes encoding mesoacidophile TetH proteins (A. ferrooxidans TetH [10, 26] and A. caldus TetH [9, 46]) were also upregulated on S4O62− and S0, although A. caldus TetH induction was less conclusive since S0 concentrations were fourfold lower than those used for A. ferrooxidans and M. sedula studies (5g/liter versus 20 g/liter). A. caldus TetH contains a pyrrolo-quinoline quinone (PQQ) binding motif, shown to interact with a quinone (46). Msed0804 also contains a PQQ-binding motif, presenting the possibility that it donates electrons to quinol oxidases (bc1 complexes). TQO (DoxDA), which oxidizes S2O32− to S4O62−, connects to ETC in a similar manner (37). M. sedula encodes a putative TQO (Msed0363 and Msed0364) with a separate DoxD-like sequence (Msed0374). Because of TQO's RISC oxidation function, it was hypothesized that transcription of these subunits would be higher in the presence of RISCs. However, these ORFs in M. sedula did not respond to any conditions tested (see Table S1 in the supplemental material).
TABLE 2.
M. sedula genome loci responding to ferrous iron and RISC
| Cluster | Msed ORF | Compared media | Fold change | Significancea (−log10P value) | Supplemental documentation |
|---|---|---|---|---|---|
| soxABCDD′Lb | 0289 | YKT vs YFS | 13.9 | 16.4 | Table S7 |
| 0290 | 2.4 | 4.2 | |||
| 0291 | 3.3 | 4.3 | |||
| 0285 | 14.8 | 17.9 | |||
| 0286 | 4.9 | 12.1 | |||
| 0288 | 2.7 | 3.1 | |||
| soxEFGHIM | 0323 | YFS vs YS | 1.2 | 0.7 | Table S8 |
| 0322 | 1.5 | 1.4 | |||
| 0321 | 3.0 | 10.5 | |||
| 0320 | 1.6 | 9.1 | |||
| 0319 | 1.1 | 0.6 | |||
| 0324 | 1.8↓ | 2.2 | |||
| foxAA′BCDEFGHIJc | 0484 | Y vs YFS | 2.0↓ | 3.7 | Table S8 |
| 0485 | 14.9 | 10.6 | |||
| 0480 | 2.1 | 3.5 | |||
| 0478 | 1.5 | 1.1 | |||
| 0477 | 1.0 | 0.0 | |||
| 0475 | 1.2↓ | 0.8 | |||
| 0474 | 1.4↓ | 1.6 | |||
| 0469 | 1.3↓ | 3.0 | |||
| 0468 | 1.5↓ | 8.8 | |||
| 0473 | 1.6↓ | 7.4 | |||
| soxNL-cbsABA | 0500 | YFS vs YKT | 5.1 | 13.3 | Table S8 |
| 0501 | 1.2 | 0.6 | |||
| 0502 | 1.7 | 6.7 | |||
| (1147) | |||||
| 0502 | 2.6 | 7.1 | |||
| (1148) | |||||
| 0503 | 2.0 | 7.8 | |||
| 0504 | 6.4 | 11.3 | |||
| tetH, sre reductase-like/dms reductase-like | 0804 | YKT vs Y | 31.9 | 20.0 | Tables S1, S3 |
| 0812 | 13.5 | 13.3 | |||
| 0814 | 8.2 | 12.1 | |||
| 0815 | 9.9 | 16.3 | |||
| 0816 | 3.4 | 12.8 | |||
| 0817 | 2.6 | 8.4 | |||
| 0818 | 1.6 | 2.9 | |||
| hdrABC-like | 1542 | YKT vs YFS | 5.5 | 7.0 | Table S5 |
| 1543 | 12.6 | 11.1 | |||
| 1544 | 5.5 | 6.9 | |||
| 1545 | 6.5 | 5.1 | |||
| 1546 | 5.0 | 5.9 | |||
| 1547 | 1.8 | 4.4 | |||
| 1548 | 10.1 | 12.1 | |||
| 1549 | 19.0 | 10.9 | |||
| 1550 | 12.3 | 10.0 | |||
| doxBCE | 2030 | YS vs YFS | 1.5↓ | 2.0 | Tables S7, S8 |
| 2031 | 2.4 | 3.8 | |||
| 2032 | 1.8↓ | 3.4 |
The Bonferroni correction for this data set was 5.4.
soxD′ (Msed0286) is the homolog of a gene encoding a small, previously unnamed polypeptide that purified with SoxABCD (see reference 35).
foxA′ (Msed0485) is the second copy of foxA (Msed0484); only one copy was originally described in reference 5.
New components of Fe(II)-induced electron transport chains.
A blue copper protein, rusticyanin, is implicated in electron transport from iron oxidation, and one version of this protein has been extensively characterized in A. ferrooxidans (4, 51, 53). The M. sedula genome encodes four blue copper protein-like sequences: two sulfocyanins (Msed0323 and Msed0826) and two rusticyanins (2). However, none of these ORFs responded to any significant extent under any of the conditions tested (see Table S2 in the supplemental material). In most cases, levels for these ORFs were slightly below average (least-square means [LSM] of <0) compared to all transcripts, with Msed0323 > Msed1206 > Msed0966 > Msed0826 under all conditions tested (see Fig. S1 in the supplemental material).
New components of RISC-induced electron transport chains.
The Msed0810 to Msed0818 locus was stimulated on YS and YKT media (Fig. 2 and Table 2; see also Table S3 in the supplemental material). With the exceptions of Msed0811, Msed0813, and Msed0818, all ORFs in this locus were upregulated at least twofold on the RISCs, with maximum induction of Msed0812 on YKT versus YFS (18-fold). Homologous putative proteins, found similarly clustered in S. solfataricus, S. tokodaii, and S. acidocaldarius (see Table S4 in the supplemental material), belong to the dimethyl sulfoxide (DMSO) reductase family, which includes thiosulfate reductase (PhsABC) from Salmonella enterica serovar Typhimurium (1, 20), polysulfide reductase (PsrABC) from Wolinella succinogenes (30), chlorate reductase (ClrABC) from Ideonella dechloratans (50), dimethyl sulfide dehydrogenase (DdhABC) from Rhodovulum sulfidophilum (36), and sulfur reductase (SreABC) from A. ambivalens (31). This family of proteins is mostly found in anaerobic bacteria although anoxic conditions appear to be required for transcriptional regulation but not activity (1, 20). Members of this family are heterotrimers in which the catalytic α and β subunits are both hydrophilic and contain 4Fe-4S and/or 3Fe-4S coordination sites (as recognized by characteristic Cys-rich motifs). The α subunit also binds a Mo cofactor, specifically molybdopterin guanine dinucleotide. A chaperone protein, often encoded between the β and γ subunits, helps in proper folding/assembly of the α and β subunits. The hydrophobic γ subunit functions as the membrane anchor. Msed0814 and Msed0815 encode the equivalents of the α (the only bacterial N-terminal domain [NTD] conserved) and β subunits, respectively, while Msed0816 is related to TorD/DmsD-like chaperone/assembly proteins, specifically A. ambivalens SreE, I. dechloratans ClrD, and R. sulfidophilum DdhD. Msed0812, a predicted transmembrane protein on the opposite strand from the rest of the Msed0810 to Msed0818 cluster, is related to the membrane anchors W. succinogenes PsrC and A. ferrooxidans SreC (Afe_0918) (membrane anchor sequences have been reported to be less conserved than the catalytic subunits) (50) and also appears to be similar to formate-dependent nitrate reductase membrane anchor proteins (NrfD like; COG3301). In some cases the γ subunit is thought to be a cytochrome b involved with electron transfer (36). However, Msed0812 has no similarity to any cytochrome b encoded elsewhere in its genome. Msed0817 is homologous to A. ambivalens SreD, thought to be an FeS-binding electron transfer protein involved with SreABC. The remaining three members of the cluster are hypothetical proteins: one with weak homology to a transcriptional regulator domain possibly involved with the regulation of this gene cluster's transcription (Msed0810; COG1414), the second with weak homology to a GTP-binding domain and ATP-dependent helicase/nuclease that may be associated with molybdopterin guanine dinucleotide cofactor synthesis (Msed0811), and the third with a tetratricopeptide repeat domain that has been associated with assembly of multisubunit proteins (Msed0813; cd00189).
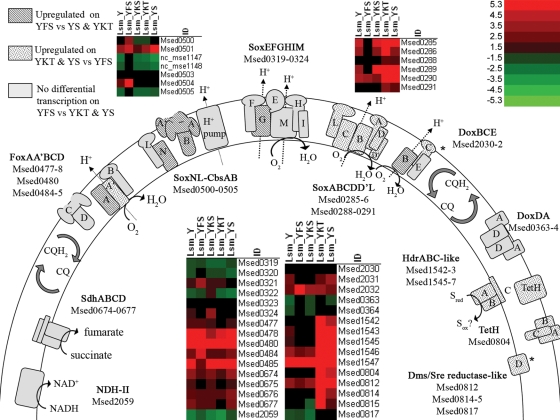
FIG. 2.
Proposed model of M. sedula respiratory electron transport chain(s) and transcriptional response to tested conditions. Model proposed is based on previous work done in other Sulfolobales. Media are as described in Materials and Methods. YKS, Y medium plus potassium sulfate. Grayscale patterns represent differential transcription [minimum log2 relative change of 0.9 and −log10(P value) of ≥2.5] between specified test conditions, while heat plots depict normalized transcription levels for each test condition compared to an average LSM of 0. Heat plots were constructed using the software Array File Maker, version 4.0 (7). For Msed2031 and Msed2032, specified differential transcription was observed for YS versus YFS medium but not for YKT versus YFS medium; for Msed0817, specified differential transcription was observed for YKT versus YFS medium but not for YS versus YFS medium (sites marked by asterisks). Differential transcription log2 relative changes and significance values for each ORF under all test conditions can be found under GEO accession number GSE12044.
The Msed1540s region (Msed1542 to Msed1550) was found to be differentially transcribed on YS and YKT medium compared to YFS medium (Fig. 2 and Table 2; see Table S5 in the supplemental material). With the exception of Msed1547, all ORFs in this locus were upregulated at least 2.0-fold, with an overall stronger transcriptional response for YKT than for YS medium. Maximum induction for this cluster was achieved by Msed1549 on YKT versus YFS (19-fold) medium. Although not differentially transcribed, Msed1547 was highly transcribed under all conditions tested, with an LSM ranging from 3.4 to 4.4 compared to the experimental loop's normalized average gene expression level of 0.0. As opposed to most of the region of Msed0810 to Msed0818, the Msed1540s region appears to be transcribed at higher than average levels on Y medium alone. RISCs boosted transcription further, while Fe2+ led to a slight decrease for the four ORFs compared to Y medium alone (data not shown).
ORFs related to the Msed1540s region are all found similarly clustered in S. solfataricus, S. tokodaii, and S. acidocaldarius (see Table S6 in the supplemental material). The mesoacidophilic bioleaching bacterium A. ferrooxidans and the hyperthermophilic bacterium Aquifex aeolicus also contain a similar cluster. The most-studied similar sequences, however, are the heterodisulfide reductases (HdrABC) found in the methanogenic archaea (18, 19).
ORFs Msed1542 to Msed1544 overlap with one another (by 4 and 14 bp, respectively). Although Msed1544 is not similar to previously characterized proteins, Msed1542 (306 amino acids [aa]) and the central region of Msed1543 (100 aa out of 230 aa) are related to the HdrB and HdrC subunits of several mesophilic and hyperthermophilic methanogens. Msed1546 and Msed1547 appear to encode additional HdrBC subunits (444 and 280 aa, respectively) but have slightly less homology to the same methanogen HdrBC subunits than Msed1542 and Msed1543. Interestingly, both M. sedula HdrC-like sequences (Msed1547 and Msed1543) share only 27% identity in their NTDs, which contain Cys-rich motifs typically corresponding to Fe-S binding regions, while the two HdrB-like sequences (Msed1546 and Msed1542) are not similar. Msed1545 (365 aa) is similar to the NTD and central regions of HdrA sequences of the same methanogens. None of these methanogens contains hdrA in the same neighborhood as hdrBC although Methanothermobacter marburgensis reportedly contains an hdrACB gene organization comparable to that found in M. sedula (18). Msed1548 (80 aa) is related to a SirA family protein, part of a two-component response regulation system in bacteria (IPR001455), and to COG0425, a predicted redox protein regulator in disulfide bond formation. This implicates Msed1548 in regulation of the Msed1540s Hdr-like region. Msed1549 and Msed1550 both lie upstream of this possible regulator, with Msed1550 on the opposite strand from the rest of the Msed1540s region. Msed1549 has no homology to any characterized protein. While Msed1550 is annotated as a DsrE family protein involved with intracellular sulfur reduction (IPR003787), it is not similar to DsrE of the dsr locus in Allochromatium vinosum (41).
Quinone oxidation via terminal oxidase complexes.
The SoxABCL terminal oxidase cluster is based on both quinol oxidase (SoxCL) and cytochrome oxidase (SoxAB) components (2, 16, 35). As expected (2, 27), soxABCL, Msed0288 to Msed0291, was significantly upregulated on both YS and YKT (15-fold for Msed0285) media compared to YFS medium (Fig. 2 and Table 2; see Table S7 in the supplemental material). This is consistent with previous reports of differential expression of terminal oxidase subunits as a function of substrate (27). Several ORFs upstream of soxABCL (Msed0283 to Msed0286; each gene encodes a putative protein of ∼100 to 150 aa) showed transcription patterns similar to soxABCL. Homologs to Msed0283 and Msed0284 are present in S. solfataricus (Sso2640 and Sso2641) and S. tokodaii (ST1697 and ST1698) although they are not colocated with the SoxABCL cluster. Msed0285 and Msed0286 are predicted membrane proteins whose homologs are colocated with the soxABCL clusters present in S. solfataricus (Sso2656 to Sso2662), S. tokodaii (ST0132 to ST0137), and S. acidocaldarius (Saci_2087 to Saci_2092). This suggests that they are associated with the SoxABCL terminal oxidase complex and possibly the two small polypeptides (one named SoxD) that purified with SoxABC in S. acidocaldarius (35). These polypeptides comigrated on a gel at 14 kDa (35); both Msed0285 and Msed0286 have predicted product masses of ∼14.5 kDa. SoxD was reported to be transcribed at the end of the soxABC operon, while the other peptide had a partial translation product that did not match known S. acidocaldarius sequences (35). Using the S. acidocaldarius genome, the unknown translation product can now be aligned with a segment of the NTD of Saci_2091, whose top hit in the M. sedula genome is Msed0286 (SoxD′). Msed0285 is similar to Saci_2092, likely encoding the originally described SoxD.
The DoxBCE terminal oxidase complex was originally isolated from A. ambivalens grown in the presence of S0 (42). Table S8 in the supplemental material shows that M. sedula doxBCE (Msed2030 to Msed2032) was not significantly induced by RISC substrates although normalized transcription levels (LSMs) indicate that these subunits were constitutively transcribed under all conditions tested. Note that a separate doxB-like gene (Msed0570) was upregulated on both RISC substrates. A separate doxB locus was also identified in A. ambivalens although its function has not been confirmed (42).
The SoxEFGHIM terminal oxidase complex was originally isolated and studied in S. acidocaldarius (11, 29, 33). More recent studies in M. sedula showed soxM to be upregulated on yeast extract (YE) medium compared to a reduced YE concentration medium supplemented with Fe(II) or S0. However, it was not clear whether this differential transcription was triggered by the presence of Fe(II) and RISCs or the decrease in Y medium from 0.1% to 0.02% (27). Here, when the concentration of Y was held constant for all conditions tested, significant upregulation of soxM was seen only on Y versus YFS medium (see Table S8 in the supplemental material). Interestingly, the cytochrome b subunit transcript, soxG (Msed0321), was upregulated on YFS medium (two- to threefold) compared to Y, YKT, and YS media (see Table S8 in the supplemental material). Examination of the gene neighborhoods of sequenced Sulfolobales shows that, unlike soxABC and doxBCE, soxHGM are not colocated on the same strand; soxM is located on the strand opposite soxHG in S. acidocaldarius, S. solfataricus, S. tokodaii, and M. sedula.
The SoxNL-CbsAB and FoxABCD clusters were originally described in S. acidocaldarius (22) and S. metallicus (5), respectively. Work in M. sedula showed that cbsA is upregulated on pyrite compared to YE medium or reduced YE concentration medium supplemented with S0 (27), while work in S. metallicus showed that foxABCD is upregulated on Fe(II) versus S0 (5). Preliminary transcriptional response analysis of M. sedula's soxLN-cbsAB and foxABCD clusters on Y medium plus Fe(II) versus Y medium showed that soxLN-cbsAB and the first copy of foxA (Msed0484) were upregulated on Y medium plus Fe(II) versus Y medium, but foxBCD was downregulated under the same conditions. The second copy of foxA (foxA′; Msed0485) showed no significant differential transcription (2). Here, when M. sedula was exposed to higher amounts of Fe(II) for longer periods of time, soxN-cbsAB and the first copy of foxA were significantly upregulated on YFS versus Y medium, while foxB and foxA′ were significantly downregulated (2-fold and 15-fold, respectively) under the same conditions (see Table S8 in the supplemental material). Transcripts from soxL and foxCD (CbsAB-like sequences) showed no significant differential transcription under these conditions. Normalized transcription levels for foxAA′BCD were all above average, with 70% of foxAA′BC LSMs ranging from 2.0 to 5.2, indicating that these genes are highly transcribed under all conditions tested. Interestingly, foxA maximum transcription levels occurred on YFS medium, the same conditions under which foxA′ minimum transcription levels occurred (see Fig. S2 in the supplemental material). Also of note, Msed0502 is listed as a pseudogene in GenBank; however, microarray probes for this ORF (nc_mse1147 and nc_mse1148) showed significant upregulation on YFS versus Y medium and RISCs (Fig. 2; see Table S8 in the supplemental material) and on Y medium plus Fe(II) versus Y medium (from reference 2) (see GEO accession no. GSE11296). The translated Msed0502 draft sequence fragments share similarities with CbsA cytochromes b, like the CbsA encoded by Msed0504. Although microarray probe design aims to avoid cross-hybridization, it was verified via an independent nucleotide BLAST search of the M. sedula genome that probes for Msed0502 hybridize fully with sections of Msed0502 only and not Msed0504.
Fe(II)- and RISC-induced hypothetical proteins.
In some cases, ORFs encoding hypothetical or conserved hypothetical proteins responded to the tested conditions by as much as 13-fold for Msed1905 on YS versus YFS medium (see Table S9 in the supplemental material). In some instances the differentially transcribed ORFs may be involved with the respiratory electron transport chains (i.e., Msed0509, a subunit of a pyruvate flavodoxin/ferredoxin oxidoreductase complex), but in most cases there is insufficient annotation to suggest a respiration-related function. These ORFs merit further investigation.
DISCUSSION
With the sequencing of the first genome from an extremely thermoacidophilic bioleacher, it has become evident that M. sedula does not contain the same iron oxidation pathways as those previously studied in the mesoacidophile A. ferrooxidans. Little or no previous work has been done on the iron oxidation pathways in extremely thermophilic bioleachers, but potential candidates in these pathways can be proposed from transcriptome and bioinformatic analysis, including novel components with no similarity to other proteins with known iron oxidation functions. A similar situation exists with some aspects of sulfur oxidation although an arguably larger body of literature exists on bacterial sulfur metabolism.
The complexity of sulfur chemistry presents a significant challenge to understanding archaeal inorganic sulfur metabolism (28). For example, both S0 and tetrathionate (S4O62−) contain S-S bonds with S in the zero oxidation state. However, S4O62− also contains S in the +5 oxidation state, which potentially impacts the S-responsive genes in the M. sedula transcriptome. Both RISCs are subject to abiotic disproportionation reactions although these are unfavorable at a pH of <4 (28). However, there is a possibility that small amounts of thiosulfate (S2O32−) and sulfide (S2−) in the case of S0, may also be formed. Furthermore, S2O32− at a pH of <4 forms sulfite (SO32−) and S0. Thus, despite the relatively high purity of RISC supplements, it is possible that M. sedula experienced sulfur as S0, SO32−, S2−, and S2O32−. In addition to the abiotic production of S0, SO32−, and S2O32−, TetH may catalyze a disproportionation reaction, splitting S4O62− into other sulfur compounds, namely, S0, HSO3−, and SO42− (28). Experimental data suggest that the membrane-bound TetH in A. ferrooxidans produces S2O32− instead of HSO3− (26) and that the periplasmic TetH in A. caldus produces S2O32− and S5O62− instead of HSO3− (46). Reactions like these could potentially add significant amounts of S2O32− and S5O62− RISCs to the M. sedula medium, a possibility that needs to be considered when interpreting transcriptome information.
By utilizing the same amount of complex organic substrate in all conditions tested (Y at a concentration of 0.1%), the experimental loop used here was designed to eliminate differential transcription caused by the type or concentration of heterotrophic substrate present. This was done so that the focus would be on the response to ferrous iron or RISCs (inorganic electron donors). No significant differences in growth rate/yield were observed under any conditions tested. However, it must also be considered that the presence of a heterotrophic substrate may impact the response to Fe(II) and/or RISC differently than under autotrophic conditions. For example, the response of the fox gene cluster in S. metallicus (an autotroph) (5) may be different than in M. sedula growing under heterotrophic conditions (2; also this work). While more details of M. sedula autotrophy have recently become available (6), the influence of autotrophy, heterotrophy, or even mixotrophy on bioleaching associated functions such as iron and sulfur oxidation is not known.
The role of the blue copper protein rusticyanin as an intermediate Fe(II)-sourced electron transfer protein between cytochromes c (Cyc2 and Cyc1 or CycA1) has been studied in A. ferrooxidans (8, 51-53), but the significance of rus-like sequences in extremely thermoacidophilic archaeal bioleachers is unclear. The presence of the blue copper protein sulfocyanin in M. sedula is expected as it has already been studied in S. acidocaldarius (11, 29). Since the proposed role of both of these blue copper proteins involves electron transfer, it is somewhat surprising to see no differential response of these four ORFs in M. sedula transcriptional studies.
The putative DMSO reductase-like enzyme complex (Msed0810 to Msed0818) would not be expected to involve sulfur as a terminal electron acceptor since all M. sedula cultures involved were grown aerobically. A phylogram based on A. ambivalens SreA showed how different DMSO reductase-like molybdoenzymes cluster (i.e., DMSO reductases distinct from sulfur reductases and distinct from formate dehydrogenases, e.g.) (31). An expanded phylogram (see Fig. S3 in the supplemental material) shows that sequences encoding similar functions group together, with the exception of Aae1234 (DmsA), although this Aquifex aeolicus ORF annotated name is not based on experimental data showing DSMO as the most likely substrate. Msed0814 and its Sulfolobus Mo-oxidoreductase homologs appear to group into a new subclade separate from the polysulfide and sulfur reductases, sharing a deep, but common root with the sulfide-oxidizing R. sulfidophilum DdhA. Taken together with the transcriptional response from growth on S0 and S4O62−, a catalytic function other than reduction of the sulfur substrate seems likely (i.e., RISC oxidation).
It is interesting that Msed1542 to Msed1550 encode proteins with similarities to Hdr in methanogens. The role of Hdr in methanogens is to reduce the CoM-CoB heterodisulfide, regenerating both cofactors for further use in methanogenesis. M. sedula is not known either to contain cofactors CoM or CoB or to participate in methanogenesis; therefore, HdrABC subunits in the region of Msed1542 to Msed1550 are believed to target a different substrate. Phylogenetic grouping appears to support this hypothesis (Fig. S4 in the supplemental material shows a phylogram of HdrB-like sequences that are believed to contain the catalytic/active site for Hdr in M. marburgensis [18]). The Msed1540s region is not predicted to contain transmembrane proteins, but the methanogen Hdr complex activity has been found in the membrane following gentle disruption (49). Alternative considerations should include association of the Hdr complex with other transmembrane proteins outside the gene neighborhood or the possibility that some Hdr-associated subunits may be integral membrane proteins containing amphipathic helices not predicted with TMM software, as seen in the HdrB-like SucC sequence from A. ambivalens (32).
Comparative genomics has revealed that the sequenced Sulfolobales collectively contain almost all the terminal oxidase (or bc1 complex-quinol oxidase) complexes mentioned here. This raises the question as to why multiple terminal oxidases are needed (while SoxM is a bb3 type, both SoxB and DoxB are aa3 types [40]; FoxA is yet unknown). The results presented here suggest that the relative presence of some terminal oxidase complexes is indirectly regulated by the presence of Fe(II) or RISCs. Also in M. sedula it has been noted that near duplications of genes exist within some of these complex clusters (i.e., cbsA, Msed0502 and Msed0504; foxAA′, Msed0484 and Msed0485; and doxB, Msed0570 and Msed2032). The purpose for near duplication is not clear, although in the case of cbsA and foxAA′, the difference in transcriptional profile suggests some differences in regulation and, thus, the potential for functional distinction. It has been noted that while cytochromes b (a586), SoxC and SoxG, from S. acidocaldarius have significant sequence similarity, the redox potential of SoxG is considerably lower than that of SoxC, impacting electron donor/acceptor options (16, 29). However, in Rubrivivax gelatinosus, similar Rieske subunits of the bc1 complex are known to substitute for each other to retain complex functionality (38).
One notable exception to the multiple terminal oxidase observation is the fox cluster (containing soxHB-like and cbsAB-like sequences) present in S. metallicus, S. tokodaii, and M. sedula (all shown to be iron oxidizers) and absent in S. solfataricus and S. acidocaldarius (no demonstrated iron oxidation capabilities). Considering this grouping, along with the observation that some fox sequences have low similarities to NADH or IMP dehydrogenase sequences (data not shown), suggests that the fox cluster may be the best candidate for uphill electron transport should this pathway exist in extremely thermoacidophilic bioleachers. However, definitive work demonstrating the operation of this cluster in the reverse (uphill) direction would be required to support this hypothesis.
In addition to the A. ferrooxidans model for iron oxidation, other bacterial models are emerging. For mesoacidophilic Leptosprillum group II bacteria, Fe(II) electrons are hypothesized to be transferred to a cytochrome (cyt572) at the outer membrane and then be transported across the periplasm by a reddish cytochrome (cyt579) (48). In certain photosynthetic mesoneutrophiles, including Rhodobacter capsulatis, Rhodobacter strain SW2, and Rhodopseudomonas palustris, iron oxidation is thought to occur via a putative periplasmic c-type cytochrome (FoxE or PioA) which transfers electrons to a PQQ domain protein or a high-potential iron-sulfur protein (FoxY or PioC) (13, 25). Both operons also contain a putative iron transport component (FoxZ and PioB, respectively) that could facilitate access from the outer membrane to the periplasm although biochemical work confirming specific function remains to be completed. While the current bacterial models share the common theme of a cytochrome c serving as the initial/primary electron acceptor, the thermoacidophilic archaeal model is likely to be different as these organisms are not known to possess cytochromes c and also lack a true periplasmic space. Interestingly, M. sedula has been reported to possess a yellow cytochrome capable of direct reduction by Fe(II) which exhibits a reduced absorption spectra peak at 572 nm (7), the same wavelength reported for Leptospirillum group II bacteria's proposed initial Fe(II) electron acceptor. Recent studies in M. sedula and S. metallicus have hypothesized that the 572-nm absorption peak may be associated with either CbsA or the CbsA-like FoxC (5, 27), and previous characterization of CbsA in S. acidocaldarius suggested that it could serve an “ectoenzyme” function (21).
Summary.
The availability of several Sulfolobales genome sequences has facilitated examination of respiratory ETC components. M. sedula's capacity to oxidize iron and sulfur distinguish it from other sequenced Sulfolobales and currently limit the usefulness of comparative genomics in identifying previously unrecognized components of respiratory electron transport chains. However, for the first time, the transcriptional response of all the known components has been evaluated simultaneously and in some cases appears to be a function of potential chemolithotrophic substrate. This argues that these components (or their pretranscriptional regulation) are important for oxidation of iron (i.e., SoxNL-CbsAB and Fox cluster components) or RISCs (i.e., TetH and DMSO/sulfur reductase-like and Hdr reductase-like components). In addition, several hypothetical proteins with strong responses to Fe(II) and RISCs have been identified and will warrant further characterization to determine the nature of their involvement in Fe(II) and RISC oxidation. A key question going forward is how Fe(II) and RISC oxidation are impacted by autotrophic, heterotrophic, or mixotrophic modes of growth.
Supplementary Material
Acknowledgments
K.S.A. acknowledges an NIH T32 Biotechnology Traineeship for support. This work was also funded in part by a grant to R.M.K. from the U.S. National Science Foundation.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alami, N., and P. C. Hallenbeck. 1995. Cloning and characterization of a gene cluster, phsBCDEF, necessary for the production of hydrogen sulfide from thiosulfate by Salmonella typhimurium. Gene 156:53-57. [DOI] [PubMed] [Google Scholar]
- 2.Auernik, K. S., Y. Maezato, P. H. Blum, and R. M. Kelly. 2008. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74:682-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandeiras, T. M., C. A. Salgueiro, H. Huber, C. M. Gomes, and M. Teixeira. 2003. The respiratory chain of the thermophilic archaeon Sulfolobus metallicus: studies on the type-II NADH dehydrogenase. Biochim. Biophys. Acta 1557:13-19. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, M. L., I. Harvey, M. Sundararajan, R. Surendran, J. F. Hall, M. J. Ellis, M. A. Hough, R. W. Strange, I. H. Hillier, and S. S. Hasnain. 2006. Atomic resolution crystal structures, EXAFS, and quantum chemical studies of rusticyanin and its two mutants provide insight into its unusual properties. Biochemistry 45:2927-2939. [DOI] [PubMed] [Google Scholar]
- 5.Bathe, S., and P. R. Norris. 2007. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 73:2491-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, I. A., D. Kockelkorn, W. Buckel, and G. Fuchs. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782-1786. [DOI] [PubMed] [Google Scholar]
- 7.Blake, R. C., II, E. A. Shute, M. M. Greenwood, G. H. Spencer, and W. J. Ingledew. 1993. Enzymes of aerobic respiration on iron. FEMS Microbiol. Rev. 11:9-18. [DOI] [PubMed] [Google Scholar]
- 8.Bruscella, P., C. Appia-Ayme, G. Levican, J. Ratouchniak, E. Jedlicki, D. S. Holmes, and V. Bonnefoy. 2007. Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology 153:102-110. [DOI] [PubMed] [Google Scholar]
- 9.Bugaytsova, Z., and E. B. Lindstrom. 2004. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271:272-280. [DOI] [PubMed] [Google Scholar]
- 10.Buonfiglio, V., M. Polidoro, F. Soyer, P. Valenti, and J. Shively. 1999. A novel gene encoding a sulfur-regulated outer membrane protein in Thiobacillus ferrooxidans. J. Biotechnol. 72:85-93. [DOI] [PubMed] [Google Scholar]
- 11.Castresana, J., M. Lubben, and M. Saraste. 1995. New archaebacterial genes coding for redox proteins: implications for the evolution of aerobic metabolism. J. Mol. Biol. 250:202-210. [DOI] [PubMed] [Google Scholar]
- 12.Clark, T. R., F. Baldi, and G. J. Olson. 1993. Coal depyritization by the thermophilic archaeon Metallosphaera sedula. Appl. Environ. Microbiol. 59:2375-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croal, L. R., Y. Jiao, and D. K. Newman. 2007. The fox operon from Rhodobacter strain SW2 promotes phototrophic Fe(II) oxidation in Rhodobacter capsulatus SB1003. J. Bacteriol. 189:1774-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, T. K., C. M. Gomes, T. M. Bandeiras, M. M. Pereira, M. Teixeira, and D. L. Rousseau. 2004. Active site structure of the aa3 quinol oxidase of Acidianus ambivalens. Biochim. Biophys. Acta 1655:306-320. [DOI] [PubMed] [Google Scholar]
- 15.Elbehti, A., G. Brasseur, and D. Lemesle-Meunier. 2000. First evidence for existence of an uphill electron transfer through the bc1 and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J. Bacteriol. 182:3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleissner, M., U. Kaiser, E. Antonopoulos, and G. Schafer. 1997. The archaeal SoxABCD complex is a proton pump in Sulfolobus acidocaldarius. J. Biol. Chem. 272:8417-8426. [DOI] [PubMed] [Google Scholar]
- 17.Gomes, C. M., T. M. Bandeiras, and M. Teixeira. 2001. A new type-II NADH dehydrogenase from the archaeon Acidianus ambivalens: characterization and in vitro reconstitution of the respiratory chain. J. Bioenerg. Biomembr. 33:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Hamann, N., G. J. Mander, J. E. Shokes, R. A. Scott, M. Bennati, and R. Hedderich. 2007. A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis. Biochemistry 46:12875-12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedderich, R., J. Koch, D. Linder, and R. K. Thauer. 1994. The heterodisulfide reductase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic of pyridine-nucleotide-dependent thioredoxin reductases. Eur. J. Biochem. 225:253-261. [DOI] [PubMed] [Google Scholar]
- 20.Heinzinger, N. K., S. Y. Fujimoto, M. A. Clark, M. S. Moreno, and E. L. Barrett. 1995. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J. Bacteriol. 177:2813-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hettmann, T., C. L. Schmidt, S. Anemuller, U. Zahringer, H. Moll, A. Petersen, and G. Schafer. 1998. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bound b-type hemoprotein. J. Biol. Chem. 273:12032-12040. [DOI] [PubMed] [Google Scholar]
- 22.Hiller, A., T. Henninger, G. Schafer, and C. L. Schmidt. 2003. New genes encoding subunits of a cytochrome bc1-analogous complex in the respiratory chain of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bioenerg. Biomembr. 35:121-131. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, D. S., and V. Bonnefoy. 2007. Genetic and bioinformatic insights into iron and sulfur oxidation mechanisms of bioleaching organisms, p. 281-307. In D. E. Rawlings and D. B. Johnson (ed.), Biomining. Springer-Verlag, Berlin, Germany.
- 24.Iwasaki, T., A. Kounosu, M. Aoshima, D. Ohmori, T. Imai, A. Urushiyama, N. J. Cosper, and R. A. Scott. 2002. Novel [2Fe-2S]-type redox center C in SdhC of archaeal respiratory complex II from Sulfolobus tokodaii strain 7. J. Biol. Chem. 277:39642-39648. [DOI] [PubMed] [Google Scholar]
- 25.Jiao, Y., and D. K. Newman. 2007. The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189:1765-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanao, T., K. Kamimura, and T. Sugio. 2007. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 132:16-22. [DOI] [PubMed] [Google Scholar]
- 27.Kappler, U., L. I. Sly, and A. G. McEwan. 2005. Respiratory gene clusters of Metallosphaera sedula—differential expression and transcriptional organization. Microbiology 151:35-43. [DOI] [PubMed] [Google Scholar]
- 28.Kletzin, A. 2007. Metabolism of inorganic sulfur compounds in Archaea, p. 261-276. In R. A. Garrett and H. P. Klenk (ed.), Archaea: evolution, physiology, and molecular biology. Blackwell, Malden, MA.
- 29.Komorowski, L., W. Verheyen, and G. Schafer. 2002. The archaeal respiratory supercomplex SoxM from S. acidocaldarius combines features of quinole and cytochrome c oxidases. Biol. Chem. 383:1791-1799. [DOI] [PubMed] [Google Scholar]
- 30.Krafft, T., M. Bokranz, O. Klimmek, I. Schroder, F. Fahrenholz, E. Kojro, and A. Kroger. 1992. Cloning and nucleotide sequence of the psrA gene of Wolinella succinogenes polysulphide reductase. Eur. J. Biochem. 206:503-510. [DOI] [PubMed] [Google Scholar]
- 31.Laska, S., F. Lottspeich, and A. Kletzin. 2003. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149:2357-2371. [DOI] [PubMed] [Google Scholar]
- 32.Lemos, R. S., C. M. Gomes, and M. Teixeira. 2001. Acidianus ambivalens complex II typifies a novel family of succinate dehydrogenases. Biochem. Biophys. Res. Commun. 281:141-150. [DOI] [PubMed] [Google Scholar]
- 33.Lubben, M., S. Arnaud, J. Castresana, A. Warne, S. P. Albracht, and M. Saraste. 1994. A second terminal oxidase in Sulfolobus acidocaldarius. Eur. J. Biochem. 224:151-159. [DOI] [PubMed] [Google Scholar]
- 34.Lubben, M., B. Kolmerer, and M. Saraste. 1992. An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 11:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubben, M., A. Warne, S. P. Albracht, and M. Saraste. 1994. The purified SoxABCD quinol oxidase complex of Sulfolobus acidocaldarius contains a novel haem. Mol. Microbiol. 13:327-335. [DOI] [PubMed] [Google Scholar]
- 36.McDevitt, C. A., G. R. Hanson, C. J. Noble, M. R. Cheesman, and A. G. McEwan. 2002. Characterization of the redox centers in dimethyl sulfide dehydrogenase from Rhodovulum sulfidophilum. Biochemistry 41:15234-15244. [DOI] [PubMed] [Google Scholar]
- 37.Muller, F. H., T. M. Bandeiras, T. Urich, M. Teixeira, C. M. Gomes, and A. Kletzin. 2004. Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53:1147-1160. [DOI] [PubMed] [Google Scholar]
- 38.Ouchane, S., W. Nitschke, P. Bianco, A. Vermeglio, and C. Astier. 2005. Multiple Rieske genes in prokaryotes: exchangeable Rieske subunits in the cytochrome bc-complex of Rubrivivax gelatinosus. Mol. Microbiol. 57:261-275. [DOI] [PubMed] [Google Scholar]
- 39.Patridge, E. V., and J. G. Ferry. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188:3498-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira, M. M., T. M. Bandeiras, A. S. Fernandes, R. S. Lemos, A. M. Melo, and M. Teixeira. 2004. Respiratory chains from aerobic thermophilic prokaryotes. J. Bioenerg. Biomembr. 36:93-105. [DOI] [PubMed] [Google Scholar]
- 41.Pott, A. S., and C. Dahl. 1998. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144:1881-1894. [DOI] [PubMed] [Google Scholar]
- 42.Purschke, W. G., C. L. Schmidt, A. Petersen, and G. Schafer. 1997. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quatrini, R., C. Appia-Ayme, Y. Denis, J. Ratouchniak, F. A. Veloso, J. Valdes, C. Lefimil, S. Silver, F. Roberto, O. Orellana, F. Denizot, E. Jedlicki, D. S. Holmes, and V. Bonnefoy. 2006. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 83:263-272. [Google Scholar]
- 44.Rawlings, D. E., and D. B. Johnson. 2007. The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology 153:315-324. [DOI] [PubMed] [Google Scholar]
- 45.Rouillard, J. M., M. Zuker, and E. Gulari. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31:3057-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rzhepishevska, O. I., J. Valdes, L. Marcinkeviciene, C. A. Gallardo, R. Meskys, V. Bonnefoy, D. S. Holmes, and M. Dopson. 2007. Regulation of a novel Acidithiobacillus caldus gene cluster involved in reduced inorganic sulfur compound metabolism. Appl. Environ. Microbiol. 73:7367-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoepp-Cothenet, B., M. Schutz, F. Baymann, M. Brugna, W. Nitschke, H. Myllykallio, and C. Schmidt. 2001. The membrane-extrinsic domain of cytochrome b(558/566) from the archaeon Sulfolobus acidocaldarius performs pivoting movements with respect to the membrane surface. FEBS Lett. 487:372-376. [DOI] [PubMed] [Google Scholar]
- 48.Singer, S. W., C. S. Chan, A. Zemla, N. C. VerBerkmoes, M. Hwang, R. L. Hettich, J. F. Banfield, and M. P. Thelen. 2008. Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl. Environ. Microbiol. 74:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stojanowic, A., G. J. Mander, E. C. Duin, and R. Hedderich. 2003. Physiological role of the F420-non-reducing hydrogenase (Mvh) from Methanothermobacter marburgensis. Arch. Microbiol. 180:194-203. [DOI] [PubMed] [Google Scholar]
- 50.Thorell, H. D., K. Stenklo, J. Karlsson, and T. Nilsson. 2003. A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl. Environ. Microbiol. 69:5585-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarzabal, A., C. Appia-Ayme, J. Ratouchniak, and V. Bonnefoy. 2004. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology 150:2113-2123. [DOI] [PubMed] [Google Scholar]
- 52.Yarzabal, A., G. Brasseur, J. Ratouchniak, K. Lund, D. Lemesle-Meunier, J. A. DeMoss, and V. Bonnefoy. 2002. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng, J., M. Geng, Y. Liu, L. Xia, J. Liu, and G. Qiu. 2007. The sulfhydryl group of Cys138 of rusticyanin from Acidithiobacillus ferrooxidans is crucial for copper binding. Biochim. Biophys. Acta 1774:519-525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.