Abstract
In Escherichia coli, growth is limited at elevated temperatures mainly because of the instability of a single enzyme, homoserine o-succinyltransferase (MetA), the first enzyme in the methionine biosynthesis pathway. The metA gene from the thermophile Geobacillus kaustophilus cloned into the E. coli chromosome was found to enhance the growth of the host strain at elevated temperature (44°C), thus confirming the limited growth of E. coli due to MetA instability. In order to improve E. coli growth at higher temperatures, we used random mutagenesis to obtain a thermostable MetAE. coli protein. Sequencing of the thermotolerant mutant showed five amino acid substitutions: S61T, E213V, I229T, N267D, and N271K. An E. coli strain with the mutated metA gene chromosomally inserted showed accelerated growth over a temperature range of 34 to 44°C. We used the site-directed metA mutants to identify two amino acid residues responsible for the sensitivity of MetAE. coli to both heat and acids. Replacement of isoleucine 229 with threonine and asparagine 267 with aspartic acid stabilized the protein. The thermostable MetAE. coli enzymes showed less aggregation in vivo at higher temperature, as well as upon acetic acid treatment. The data presented here are the first to show improved E. coli growth at higher temperatures solely due to MetA stabilization and provide new knowledge for designing E. coli strains that grow at higher temperatures, thus reducing the cooling cost of bioprocesses.
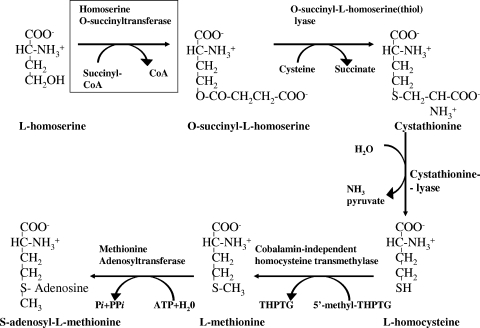
The growth of Escherichia coli, a mesophilic bacterium, is limited at elevated temperatures (13, 14). Quite unexpectedly, earlier investigations showed that E. coli did not grow at temperatures above 44°C because of the instability of a single protein, homoserine o-succinyltransferase (MetA) (13, 14, 15, 16). MetA (EC 2.1.3.46), the first enzyme in methionine biosynthesis (Fig. 1), catalyzes the transfer of succinate from succinyl-coenzyme A (succinyl-CoA) to l-homoserine (3, 22). Recent findings report that MetAE. coli tends to unfold even at 25°C in vitro; unfolding becomes maximal at 44°C and is followed by massive aggregation (5). In vivo, the soluble fraction of cytoplasmic proteins lacks MetAE. coli at temperatures higher than 44°C (5). MetA from Salmonella enterica is as sensitive to elevated temperature as to weak organic acids, including benzoate, propionate, and acetate (11). Moreover, hydrogen peroxide increases its sensitivity to both heat and acid and may oxidatively damage the destabilized MetA protein (11). Price-Carter and coworkers (11) suggested that an excess of MetAS. enterica synthesized at elevated temperatures and/or in the presence of weak organic acids leads to the accumulation of insoluble aggregates that are toxic to the cells and inhibit bacterial growth.
FIG. 1.
Biosynthesis of l-methionine and S-adenosyl-l-methionine in E. coli. Abbreviations: 5′-methyl-THPTG, 5′-methyltetrahydropteroyl-tri-l-glutamate; THPTG, tetrahydropteroyl-tri-l-glutamate.
In view of all of the foregoing data and the fact that MetA occupies the control point in methionine biosynthesis, it has been proposed that MetA plays a central role in the control of bacterial growth (2). MetA's high sensitivity to many stress factors suggests that it may serve as a sort of metabolic fuse, detecting unfavorable growth conditions (11). In this connection, it is notable that methionine relieves the inhibitory effect of high temperature and acetic acid on E. coli growth (7, 12, 13, 14). Thus, it would be quite interesting to try to widen the optimum growth temperatures of E. coli by increasing the stability of this enzyme.
In the present study, we used random mutagenesis to obtain a thermostable MetAE. coli mutant and to determine whether this alone can enhance the growth rate of E. coli at higher temperatures. Analysis of the evolved MetAE. coli protein revealed that the residues isoleucine 229 and asparagine 267 are independently responsible for the organism's sensitivity to heat and acid; replacing isoleucine 229 with threonine or asparagine 267 with aspartic acid stabilized the enzyme. An E. coli strain with thermostable MetAE. coli showed accelerated growth over a temperature range of 34 to 44°C. Quite interestingly, this thermostable MetAE. coli-producing strain was also resistant to acetic acid challenge.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| W3110 | Wild type | Laboratory stock |
| DH5α | F−supE44 hsdR17 recA1 gyrA96 endA1 thi-1 relA1 deoR λ− | 17 |
| JW3973 | ΔmetA Kanr | Keio collection (Japan) |
| WE | JW3973 carrying nonmutated metAE. coli on the chromosome, prototroph | This study |
| 333 | JW3973 carrying mutated metAE. coli-333 on the chromosome, prototroph | This study |
| T61 | JW3973 carrying metAE. coli with S61T substitution on the chromosome, prototroph | This study |
| V213 | JW3973 carrying metAE. coli with E213V substitution on the chromosome, prototroph | This study |
| T229 | JW3973 carrying metAE. coli with I229T substitution on the chromosome, prototroph | This study |
| D267 | JW3973 carrying metAE. coli with N267D substitution on the chromosome, prototroph | This study |
| K271 | JW3973 carrying metAE. coli with N271K substitution on the chromosome, prototroph | This study |
| WG | JW3973 carrying metAGeo on the chromosome, prototroph | This study |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm(DE3) | Novagen |
| G. kaustophilus | Wild type | KCTC 3397 |
| Plasmids | ||
| pACYC177 | Plasmid vector, Ampr Kanr | New England BioLabs |
| pET22b | Expression vector, Ampr | Novagen |
| pKD46 | λ red (gam bet exo) araC rep-101(Ts) Ampr | 4 |
| pPmetA | pACYC17 carrying metApE. coli, Ampr | This study |
| pMetA | pPmetA carrying metAE. coli, Ampr | This study |
| pMetA-333 | pPmetA carrying thermostable metAE. coli, Ampr | This study |
| pGeo-MetA | pPmetA carrying metAGeo, Ampr | This study |
Ampr, ampicillin resistance; Kanr, kanamycin resistance; metApE. coli, promoter of E. coli metA gene; metAE. coli, E. coli metA gene; metAGeo, G. kaustophilus metA gene.
Growth conditions.
E. coli strains were grown in minimal M9 medium (17) supplemented with glucose (0.2%), in LB (Difco), or in 2 × YT (17). Antibiotics were used in the following concentrations: ampicillin, 100 μg ml−1, kanamycin, 25 μg ml−1. l-Methionine was added to the medium to a final concentration of 50 μg ml−1. The strain Geobacillus kaustophilus KCTC 3397 was cultivated aerobically in nutrient broth (Difco) at 50°C.
Preparation of plasmid DNA.
Plasmid DNA was extracted from the cells with a plasmid mini-prep kit (SolGent Co., Ltd.). All of the restriction enzymes used in this study were purchased from New England BioLabs Inc.
Cloning of E. coli metA.
The natural promoter of metA was amplified from the genomic DNA of E. coli strain W3110 with primers metA1 (CGCCTACTCGAGATCGCAACGAGTTCCTCC) and metA2 (GCCTCAAAGCTTCATATGCTGATTACCTCACTACATACGC) and cloned into the XhoI and HindIII sites of plasmid vector pACYC177 to yield plasmid pPmetA. The structural metA gene amplified from the genomic DNA of E. coli W3110 with primers metA3 (CGCCTCCATATGCCGATTCGTGTGCCG) and metA4 (CGCCTCAAGCTTGGTGCCTGAGGTAAGGTGCTG) was cloned into the NdeI and HindIII sites of plasmid pPmetA to obtain plasmid pMetA.
Cloning of metA from thermophilic strain G. kaustophilus KCTC 3397.
The metAGeo gene from the genomic DNA was amplified with primers Geo1 (CGCCTCCATATGCCAATCAACATTCCAAAAG) and Geo2 (CAGGGCGATTGTCGAAACACG) and cloned into an NdeI restriction site of plasmid pPmetA. The resulting plasmid, pGeo-MetA, was transferred into strain E. coli JW3973(ΔmetA). The transformed cells were incubated on minimal M9 plates supplemented with glucose and ampicillin.
Library construction and selection of thermostable metA mutants.
The metA gene, together with its promoter region, was isolated from the pMetA plasmid, gel purified, and used as a template for error-prone PCR. Random mutagenesis was performed with a Diversify PCR random mutagenesis kit (Clontech Laboratories, Inc.) to obtain three, five, and eight nucleotide substitutions per kilobase. The PCR conditions were as described in the manual and included primers metA5 (GTAGTGAGGTAATCAGCATATG) and metA4. The PCR products were purified with a QIAquick PCR purification kit (Qiagen), amplified one more time with the same primers and TaKaRa Ex Taq polymerase, digested with the restriction enzymes NdeI and HindIII, and cloned into plasmid pPmetA. The resulting DNA mixture was transferred into freshly prepared E. coli JW3973(ΔmetA) cells by electroporation. The transformed cells were incubated at 37°C on M9 plates supplemented with glucose and ampicillin. The clones were then cultivated on solid M9 glucose medium at 44°C to select the growing ones. The fastest-growing mutant was identified during cultivation in liquid M9 glucose medium at 44°C.
Incorporation of metA mutants into the E. coli chromosome.
The metA mutants were transferred into the E. coli JW3973 (ΔmetA) chromosome with the lambda Red recombination system (4). The structural genes flanked by 50-bp nucleotide sequences up- and downstream were synthesized with primers metA8 (ATCTGGATGTCTAAACGTATAAGCGTATGTAGTGAGG) and metA9 (ATCGCTTAACGATCGACTATCACAGAAGATTAATCC), Vent polymerase (New England BioLabs Inc.), and the corresponding plasmids as templates. G. kaustophilus metA was amplified from the genomic DNA as the template with primers Geo4 (ATCTGGATGTCTAAACGTATAAGCGTATGTAGTGAGGTAATCAGGTTATGCCAATCAACATTCC) and Geo5 (ATCGCTTAACGATCGACTATCACAGAAGATTAATCCAGCGTTGGATTCATCAGGGCGATTGTCGAAACACG). Freshly prepared competent cells of strain E. coli JW3973 (ΔmetA) harboring helper plasmid pKD46 were transformed with 150 μg of PCR products by electroporation as described previously (4).
The transformed cells were incubated on M9 methionine-deficient medium plates supplemented with glucose at 37°C. The grown colonies were checked for loss of kanamycin resistance and cultivated on nonselective LB plates at 43°C to eliminate plasmid pKD46. The inserted metA mutants were amplified from the chromosome and sequenced.
Site-directed mutagenesis of metA.
To introduce multiple direct mutations into metA, a QuikChange Multi site-directed mutagenesis kit (Stratagene) was employed. The primers used were metT (GCCTGCTTTCAAACACACACCTTTGCAGGTCG), metD (CTATTTCCCGCACGATGATCCGCAAAATACACC), metK (GATCCGCAAAAGACACCGCGAGCGAGC), metT2 (GCCAGTAAAGATAAGCGCACTGCCTTTGTGACG), and metV (CTGGAAATTCTGGCAGTGACGGAAGAAGGG).
Cultivation of E. coli strains in a temperature gradient incubator.
Growth of E. coli strains in M9 glucose medium at different temperatures was studied with a temperature gradient incubator (TVS126MA; Adventec MSF Inc.). A single colony of each strain was cultivated in 5 ml of M9 medium overnight at 30°C. The overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in 300 ml of fresh M9 medium, dispensed into 15-ml tubes, and incubated at 30 to 47°C for 12 h with shaking. Growth was measured by monitoring the OD600 every 5 min.
Purification of aggregated and soluble proteins.
The metA chromosomal mutants of E. coli were grown in 75 ml of M9 glucose medium in flasks at 30°C to mid-exponential phase (OD600 of approximately 0.6). Twenty-five milliliters of each culture was shifted to 45°C for 40 min or treated with 30 mM acetic acid for 3 h at 30°C. The remaining 25 ml was used as a control. Aggregated and soluble protein fractions were purified as described previously (5, 19).
SDS-PAGE and immunoblotting.
Gel electrophoresis was performed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the proteins were electroblotted onto nitrocellulose membranes (Bio-Rad). MetA protein was detected with rabbit anti-MetA serum raised against a synthetic oligopeptide consisting of 74 to 94 amino acids (2) of MetA (Peptron Inc.) as the primary antibody and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Pierce) as the secondary antibody. The immunoblots were developed with a SuperSignal West Pico Chemiluminescent Substrate kit (Pierce), scanned with Image Reader Fujifilm LAS-3000, and analyzed with Lab Works software.
Cloning, expression, and protein purification.
The wild-type and mutated metA genes were cloned into the NdeI/HindIII restriction sites of plasmid pET22b in frame with a C-terminal six-histidine tag. The plasmid DNA was purified from ampicillin-resistant clones and sequenced to verify that the correct genes had been cloned. The resulting plasmids were transformed into competent E. coli BL21(DE3) cells. A single colony of strain E. coli BL21(DE3) harboring the corresponding plasmid was cultivated overnight at 30°C in 15 ml of LB medium with ampicillin. Half a liter of 2 × YT medium containing ampicillin was inoculated with an overnight culture, incubated at 30°C to an OD600 of 0.6, induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM final concentration), and then cooled to 22°C. After 4 h of induction, the cells were harvested by centrifugation and the pellets were resuspended in ice-cold buffer (50 mM Tris-HCl, 300 mM NaCl, 10 mM MgCl2, 5 mM imidazole, pH 7.5) at a ratio of 3 ml buffer/1 g of wet cells. The cells were lysed by incubation with 0.5 mg/ml lysozyme-1 mM phenylmethylsulfonyl fluoride-DNase I for 30 min with stirring at 4°C, followed by sonication for 10 × 20 s with 20-s intervals with a Branson Sonifier (model 450). The cell debris was removed by centrifugation at 12,000 × g for 40 min. The proteins were purified from the supernatants with Ni-nitrilotriacetic acid agarose (Qiagen). Two milliliters of agarose slurry was incubated with 6 ml of supernatant overnight at 4°C with rocking. The unbound proteins were removed by gravity filtration, and the agarose was washed with 4 ml of buffer (50 mM Tris-HCl, 300 mM NaCl, 100 mM imidazole, pH 7.5). The proteins were released from the agarose by elution with 6 ml of buffer (50 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole, pH 7.5), and the eluate was dialyzed against two changes of dialysis buffer (2 liters, 50 mM K-phosphate buffer, 150 mM NaCl, pH 7.6) and then against 50 mM K-phosphate buffer (pH 7.6) containing 150 mM NaCl and 50% glycerol for 6 h. The presence of pure protein in all of the samples was confirmed by SDS-PAGE.
Measurement of enzyme activity.
Reaction rates were determined by monitoring the decrease in absorbance at 232 nm caused by hydrolysis of the thioester bond of succinyl-CoA (3) in an ND1000 UV/Vis spectrophotometer (Nanodrop Technologies). Assay solutions containing 50 mM K-phosphate buffer (pH 7.5), 200 μM succinyl-CoA, 5 mM l-homoserine, and 1 μM protein in a final volume of 20 μl were incubated for 30 min at 40, 45, 50, 55, and 58°C or at 25°C in the presence of acetic acid. l-Homoserine was omitted from the control tubes. The reaction was started by adding the enzyme. The consumption of succinyl-CoA in the reaction was calculated as the difference between the values obtained in the presence and absence of l-homoserine. Three independent measurements were performed for each point.
Differential scanning calorimetry.
The thermal stabilities of the MetA proteins were measured calorimetrically over a temperature range of 15 to 90°C at a scan rate of 90°C/h. A VP-DSC calorimeter (MicroCal Inc.) was employed with 10 μM protein in 50 mM K-phosphate buffer (pH 7.5). Three scans were obtained with independent protein preparations.
RESULTS
l-Methionine stimulates E. coli growth.
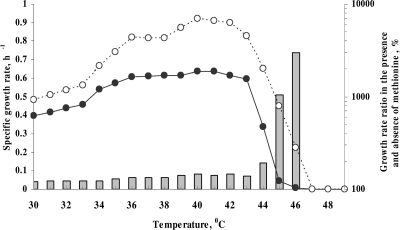
To confirm the stimulatory effect of methionine on E. coli growth at elevated temperatures, strain W3110 was cultivated in M9 glucose medium over a temperature range of 30 to 49°C with or without l-methionine (50 μg ml−1) in a temperature gradient incubator. Methionine accelerated E. coli growth at temperatures of greater than 30°C (Fig. 2). E. coli strain W3110 grew very slowly without methionine at 45°C and completely ceased to grow at 46°C or higher temperatures (Fig. 2), confirming previous findings (16). Supplementation of the culture medium with methionine allowed growth at 45 and 46°C, but no growth was detected at temperatures of greater than 47°C (Fig. 2). As expected, the methionine effect was more prominent at higher growth temperatures; the specific growth rate was increased 2-fold at 44°C and 10-fold at 45°C (Fig. 2).
FIG. 2.
Effect of methionine supplementation on the growth of E. coli strain W3110 at different temperatures. The strain was grown in M9 glucose minimal medium with (open circles) or without (filled circles) l-methionine supplementation in a temperature gradient incubator over a temperature range of 30 to 49°C. The columns indicate the ratios of specific growth rates in methionine-enriched (50 μg ml−1) and methionine-deficient media.
MetA from the thermophilic strain of G. kaustophilus KCTC 3397 accelerates E. coli growth at elevated temperature.
We checked whether MetA from the thermophilic strain would simply improve E. coli growth at higher temperatures. As a thermostable gene, we used metA from the strain G. kaustophilus KCTC 3397. This is an aerobic, gram-positive, endospore-forming bacterium that can grow at 37 to 75°C, with an optimum at 55 to 65°C (21). The metAGeo gene was cloned under the control of E. coli metAp on the pPmetA plasmid. The resulting plasmid, pGeo-metA, compensated the growth of the metA-null mutant on M9 medium. However, the specific growth rate of E. coli strain JW3973 (pGeo-MetA) was 20% less than that of E. coli JW3973(pMetA) at 37°C. We integrated metAGeo into the E. coli chromosome to yield strain WG. The metAGeo gene stimulated E. coli growth on solid M9 medium at 44°C (see Fig. 4).
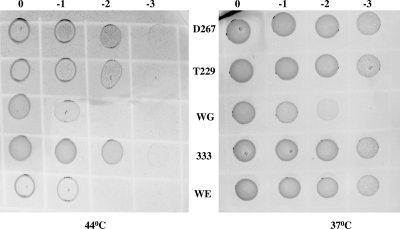
FIG. 4.
Thermostability of E. coli metA mutants. Samples were taken from logarithmically growing cultures at 30°C in M9 glucose medium, adjusted to equal density, and serially diluted in M9 medium (no glucose). Aliquots (5 μl) were spotted onto M9 glucose plates. Cells were incubated for 24 h at the indicated temperatures.
Directed evolution of MetA results in thermotolerant E. coli strains.
As methionine and thermostable MetAGeo relieve the impairment of E. coli growth at higher temperatures, we tried to obtain a more thermostable MetAE. coli protein by monitoring the enhancement of growth of E. coli cells carrying the metAE. coli mutation at higher temperatures. We supposed that thermostable MetAE. coli might accelerate E. coli growth not only at elevated but at mild temperatures, in contrast to MetAGeo, which lowers the specific growth rate of E. coli at 37°C. Random mutagenesis of metAE. coli was performed with error-prone PCR as described in Materials and Methods. We constructed a library of mutated metA genes in the pPmetA plasmid that completely restored the growth of the metA null mutant E. coli JW3973 on M9 glucose medium. The library, consisting of 490 clones, was incubated on M9 minimal plates at 44°C to select the growing clones. Twenty-three selected clones were then cultivated in liquid M9 glucose medium at 44°C to identify the most rapidly growing one. One prospective clone that grew faster than the others at the elevated temperature was designated strain 333 (Fig. 3). Sequencing showed the presence of five amino acid substitutions—S61T, E213V, I229T, N267D, and N271K—in MetAE. coli-333. To determine which amino acid residue is responsible for the improved thermostability seen, single amino acid substitutions corresponding to those found in MetAE. coli-333 were introduced into the wild-type enzyme by site-directed mutagenesis. We integrated all of the mutated metAE. coli genes into the E. coli JW3973(ΔmetA) chromosome to yield strains 333, T61, V213, T229, D267, and K271. To study the thermostability of these metA mutants, we cultivated them on solid M9 glucose plates at 44°C. In Fig. 4, the left panel shows that control strain WE, which harbors nonmutated metA on the chromosome, grew weakly at 44°C, in contrast to mutant strains 333 and WG. Among the single mutants, only two strains, T229 and D267, grew better than the control strain at the elevated temperature (Fig. 4). All of the strains grew normally at 37°C (Fig. 4, right panel), except WG, which grew more slowly than the others.
FIG. 3.
Selection of thermostable MetAE. coli-333 from a library of metAE. coli mutants. Cultures of 23 strains of E. coli JW3973 harboring plasmid pMetA with mutated metAE. coli genes and a nonmutated control were flask cultivated in M9 glucose medium at 44°C. The OD600 was measured every hour. Symbols: pMetA control, •; pMetA-333, ▾; pMetA-334, ○. The growth curves of other metAE. coli mutants were similar to that of the control strain.
Thermotolerant E. coli metA mutants grow faster at elevated temperatures.
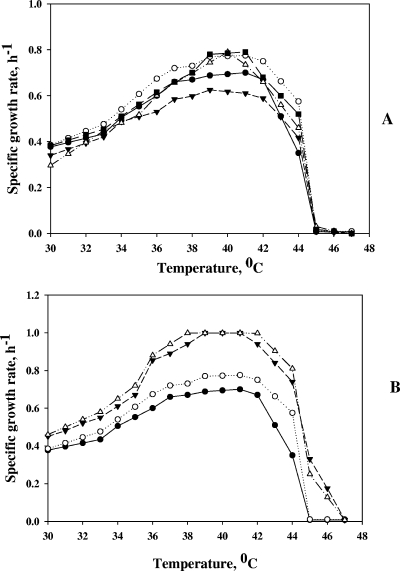
To study the growth of thermostable mutants at different temperatures, we cultivated them in a temperature gradient incubator. The results presented in Fig. 5A show that strain 333 grew 5 to 12% faster than nonmutated strain WE over a temperature range of 30 to 42°C. However, the difference in the specific growth rate between thermostable strain 333 and control strain WE increased to 30% at 43°C and to 64% at 44°C (Fig. 5A). metA single-mutant strains T229 and D267 grew 48 and 31% faster at 44°C and 18 and 10% faster at 43°C, respectively. Over a temperature range of 39 to 41°C, the difference between the specific growth rates of the single-site mutants and the control strain decreased to 4 to 12% and fell to zero over a temperature range of 30 to 38°C (Fig. 5A). No growth of any of the strains tested was detected at 45°C or higher temperatures.
FIG. 5.
Effects of different temperatures (A) and methionine supplementation (B) on the growth of E. coli metA mutants. Nonmutated and mutated metA genes were integrated into the chromosome of E. coli strain JW3973 instead of the kanamycin resistance gene. The strains were grown in M9 glucose medium in a temperature gradient incubator over a temperature range of 30 to 47°C for 12 h. Methionine was added to a final concentration of 50 μg ml−1. Symbols in panel A: WE, •; 333, ○; WG, ▾; D267, ▵; T229, ▪. Symbols in panel B: WE, •; 333, ○; WE plus methionine, ▾; 333 plus methionine, ▵.
The foreign metAGeo gene lowered host strain growth in liquid M9 medium at mild temperatures, 30 to 42°C, by approximately 10% (Fig. 5A). At 43°C, the specific growth rate of WG was the same as that of control strain WE, and at 44°C it grew 20% faster than the control strain (Fig. 5A).
We also cultivated the other single-site mutants at different temperatures. Two of them, V213 and K271, had lower specific growth rates than the wild type at 44°C (64 and 22% of that of the control strain) (data not shown) but grew 15 to 20% faster than the nonmutated strain at 36 to 41°C (data not shown). The single-site mutant T61 grew as well as the wild type at elevated temperatures, but its specific growth rate at 36 to 41°C was similar to that of mutants V213 and K271 (data not shown). Apparently, the metA multiple mutant 333 combined the abilities of the thermosensitive mutants to grow faster at mild temperatures and of the thermostable mutants to grow faster at higher temperatures.
In order to ascertain the influence of methionine on the growth of the thermostable E. coli 333 strain, we cultivated these bacteria with or without methionine at different temperatures with the nonmutated WE strain as a control (Fig. 5B). Despite the presence of the thermostable MetAE. coli-333 protein, this mutant could not attain the specific growth rate obtained in the presence of methionine. However, although the nonmutated strain grew 1.6 and 2 times slower in methionine-deficient medium at 43 and 44°C, respectively, the thermostable variant decreased this difference to 1.4-fold at both temperatures. This is apparently because another thermolabile protein is present in the methionine biosynthesis pathway. Previous investigations have shown that MetE, which catalyzes the final step in methionine biosynthesis, is also sensitive to elevated temperature (9).
Thermostable metA mutants are tolerant to acetic acid.
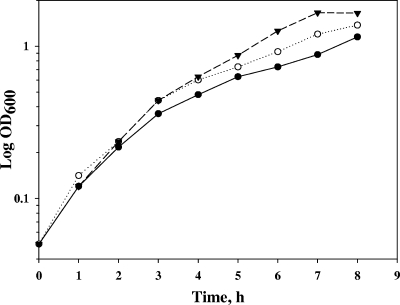
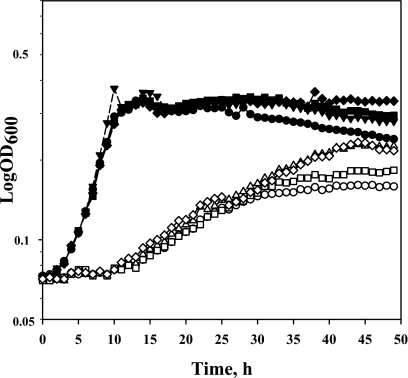
Since it is known that weak organic acids can destabilize MetAS. enterica (11), we supposed that thermostable metA mutants might also be resistant to acetic acid. Strains 333, D267, T229, and WE were cultivated in M9 glucose medium supplemented with 30 mM acetic acid at 30°C in a BioScreen C incubator (Labsystems). As shown in Fig. 6, the wild-type strain reached stationary phase 29 h after the beginning of cultivation. In contrast, thermostable E. coli strains 333 and T229 grew intensively for a further 11 h and achieved a 1.4 times greater cell density than the control strain. Another mutant, D267, grew slower than other thermostable strains, and its cell density was only 16% more than that of the control.
FIG. 6.
Influence of acetic acid on the growth of thermostable E. coli mutants. The strains were cultivated in 100 μl of M9 glucose medium at 30°C in a BioScreen C incubator for 49 h with 15-min intervals of shaking in the presence of 30 mM acetic acid (open symbols) or without it (solid symbols). Symbols: WE, circles; 333, triangles; D267, squares; T229, diamonds.
Thermostable MetA enzymes are less susceptible to aggregation in vivo under heat or acetic acid stress conditions.
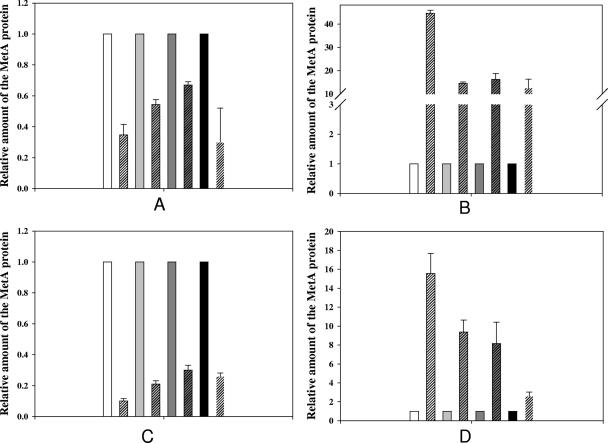
To determine the effect of elevated temperature or acetic acid treatment on thermostable MetAE. coli proteins in vivo, the mutant and control strains were heated from 30 to 45°C for 40 min or incubated with 30 mM acetic acid for 3 h at 30°C and the relative amount of MetAE. coli was evaluated by immunoblotting. Biran and coworkers (2) showed that E. coli cells contain no soluble MetAE. coli at temperatures above 44°C. Surprisingly, we found wild-type MetAE. coli protein in the soluble fraction after heat stress; its level was about 35% of that found in an unstressed culture (Fig. 7A). Perhaps part of the unfolded MetAE. coli protein partitioned into the soluble fraction. The relative amounts of the soluble thermostable MetAE. coli-333 and MetAE. coli-D267 proteins were 58 and 67% (Fig. 7A). In contrast, the level of soluble MetAE. coli-T229 protein at 45°C was only 29% of that in the unheated control (Fig. 7A). The relative content of insoluble nonmutated MetAE. coli protein increased 44 times after heating (Fig. 7B). The insoluble fractions of all of the mutants tested were only 13 to 19 times more abundant after high-temperature treatment than in unheated cultures (Fig. 7B). The data obtained in this experiment confirmed that the MetAE. coli-333 and MetAE. coli-D267 mutants are more resistant in forming aggregates after a heat challenge. For MetAE. coli-T229, we assume that resistance to aggregation is greater because of a more stable protein secondary structure.
FIG. 7.
Densitometric analysis of MetA proteins in heat (A, B)-stressed and acetic acid (C, D)-stressed cultures. Strains WE (white columns), 333 (gray columns), D267 (dark gray columns), and T229 (black columns) were grown in M9 glucose medium to exponential phase (OD600 = approximately 0.6) at 30°C and then shifted to 45°C for 40 min or incubated in the presence of 30 mM acetic acid for 3 h at 30°C. Soluble (A, C) and aggregated (B, D) fractions of MetA proteins were purified from 25-ml cultures as described in Materials and Methods. Three micrograms of total protein was subjected to 12% SDS-PAGE, followed by Western blotting with rabbit anti-MetA serum. MetA proteins were quantified by densitometry with LabWorks software and normalized to the total amount of protein. MetA proteins from the unstressed cultures were made equal to 1 (plain columns). An average of two independent experiments is presented.
Acetic acid challenge of exponentially growing cultures produced a relative level of soluble MetAE. coli 1.5 to 3 times higher in all of the mutants tested than in the wild-type strain (Fig. 7C). The insoluble MetAE. coli content was approximately 15 times greater in the wild-type strain, 8 to 9 times greater in the D267 and 333 strains, and 2.6 times greater in the T229 strain (Fig. 7D). These results confirm that the mutated MetAE. coli proteins are more resistant in forming aggregates than the wild-type protein when challenged with acetic acid.
Thermostable MetA enzymes have increased transition midpoints.
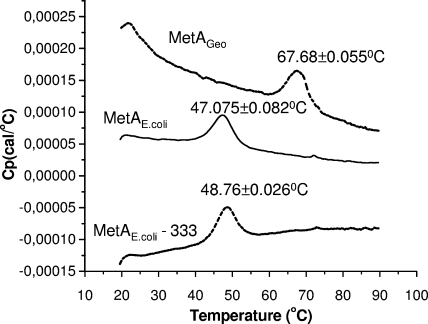
Differential scanning calorimetry was used to compare the transition midpoint (Tm) values of mutated and wild-type MetA proteins. The Tm is an indicator of thermostability, and proteins with higher Tm values are less susceptible to unfolding and denaturation. Nonmutated and mutated MetA proteins containing a C-terminal six-histidine tag were purified as described in Materials and Methods. As shown in Fig. 8, wild-type six-histidine-tagged MetAE. coli had a Tm of 47.075 ± 0082°C, while MetAE. coli-333 had a higher Tm (48.76 ± 0.026°C) (Fig. 8). The highest Tm (67.68 ± 0.055°C) was found for MetAGeo (Fig. 8). Although the single-site-mutated proteins MetAE. coli-D267 and MetAE. coli-T229 stimulated E. coli growth at elevated temperatures, they had Tm values very close to that of wild-type MetAE. coli (47.36 ± 0.014 and 46.99 ± 0.063°C, respectively). Maybe these mutant enzymes are not more stable than the wild type but more resistant in forming aggregates after heat challenge. This possibility will be investigated in the near future.
FIG. 8.
Thermograms of histidine-tagged MetA proteins obtained by differential scanning microcalorimetry. All of the proteins were scanned as described in Materials and Methods. The Tm indicated above the corresponding curve is the midpoint of the unfolding transition in differential scanning microcalorimetry; it was determined from three independent experiments. Cp, heat capacity at constant pressure.
Mutated MetA enzymes have enhanced activities at elevated temperatures and in the presence of acetic acid.
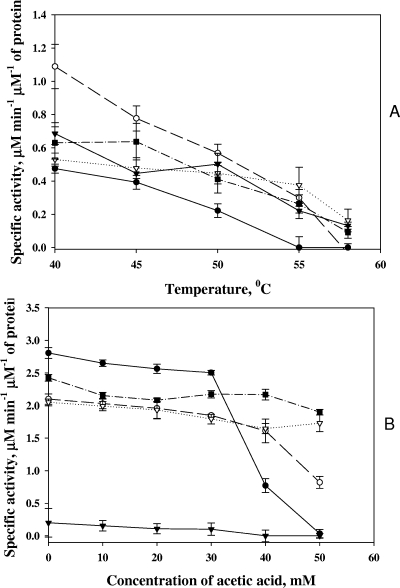
The activities of the six-histidine-tagged MetA enzymes purified as described in Materials and Methods were measured at temperatures between 40 and 58°C by monitoring the change in absorbance at 232 nm due to hydrolysis of the succinyl-CoA thioester bond. The data obtained for the thermostable MetA proteins are presented in the top panel of Fig. 9. MetAE. coli-333 was approximately 2.5 times more active than the nonmutated protein at 40 to 55°C but completely lost its activity at 58°C. In contrast to MetAE. coli-333, MetAGeo, MetAE. coli-D267, and MetAE. coli-T229 displayed lower activities at 40 to 45°C but at 50°C and higher temperatures, the profiles of all of the thermostable MetA proteins were similar. Surprisingly, the catalytic activity of MetAGeo gradually decreased at elevated temperatures, perhaps because of the thermal instability of succinyl-CoA.
FIG. 9.
Temperature dependence (A) and acetic acid tolerance (B) of histidine-tagged MetA proteins. The activities of the nonmutated MetAE. coli and MetAGeo proteins and the mutated MetAE. coli protein were measured by monitoring the decrease in absorbance at 232 nm caused by hydrolysis of the thioester bond of succinyl-CoA, the substrate for MetA, over a temperature range of 40 to 58°C (A) and at 25°C in the presence of acetic acid (B). An average of three independent measurements is presented for each point. Symbols: MetAE. coli, •; MetAE. coli-333, ○; MetAGeo, ▾; MetAE. coli-D267, ▿; MetAE. coli-T229, ▪.
We tested the catalytic activity of the mutated MetA proteins in the presence of acetic acid at 25°C. Acetic acid at a final concentration of 10 mM lowered the pH of the reaction buffer from 7.5 to 7.0, 20 mM lowered it to 6.6, 30 mM lowered it to 6.2, 40 mM lowered it to 5.8, and 50 mM lowered it to 5.4. Figure 9B shows that the activity of wild-type MetAE. coli decreased sharply when the acetic acid concentration was greater than 30 mM (pH 6.2) and was not detectable at 50 mM (pH 5.4). Our findings confirmed an earlier report that wild-type MetAE. coli activity significantly dropped over a pH range of 6.0 to 6.5 (3). Acetic acid also inhibited the activity of MetAE. coli-333, starting from 30 mM, but somewhat less severely than the wild-type enzyme. Moreover, MetAE. coli-333 remained active in 50 mM acetic acid (Fig. 9B). Two other enzymes, MetAE. coli-D267 and MetAE. coli-T229, had unchanged catalytic activities at all of the acetic acid concentrations tested (Fig. 9B). Interestingly, MetAGeo was almost inactive at 25°C (Fig. 9B).
Consequently, we compared the catalytic activities of the wild-type MetAE. coli and thermostable MetAE. coli mutant enzymes at 25 and 40°C (Fig. 9A and B). The wild-type enzyme activity was 80% lower at 40°C. In contrast, MetAE. coli-333 was 50% less active at 40°C, while MetAGeo was 3.5 times more active at the elevated temperature (Fig. 9A and B). These results of wild-type enzymes are consistent with previous findings; the activity of MetAE. coli decreased by 70% at 37 and 42°C (14).
DISCUSSION
The unusual instability of MetA, the first enzyme in the methionine biosynthesis pathway, is the main factor limiting E. coli growth at elevated temperatures (2, 5, 13, 14, 15). MetAE. coli activity is reduced at temperatures higher than 33°C due to the formation of aggregates (2). As addition of methionine or replacement with thermostable MetA relieved the growth inhibition at higher temperatures, directed evolution of MetAE. coli should widen the temperature range over which E. coli strains can grow. By directed evolution, we obtained the thermotolerant enzyme MetAE. coli-333 with five amino acid substitutions (S61T, E213V, I229T, N267D, and N271K), which accelerates the growth of the host strain not only at higher temperatures but also at the normal growth temperature, 37°C. The higher thermostability of MetAE. coli-333 was confirmed in vivo by differential scanning calorimetry data, and higher activity was demonstrated at elevated temperatures. MetAE. coli-333 was also classified as stable by the ProtParam software (http://ca.expasy.org/); it has an instability index of 39.79, in contrast to wild-type MetAE. coli, which has an instability index of 42.26. The instability index provides an estimate of protein stability. A protein whose instability index is lower than 40 is predicted to be stable, and a value above 40 predicts that the protein may be unstable (6).
Accelerated growth of the single-site metAE. coli mutants at 44°C allowed the identification of critical amino acid residues involved in the thermal instability of MetAE. coli. Replacement of asparagine 267 with aspartic acid was expected to increase thermostability because the MetA proteins known from thermophilic strains carry a positively charged amino acid at this position (see the multiple alignment in Fig. S1 in the supplemental material). In general, this finding confirms the earlier observation that proteins from thermophiles contain more charged amino acid residues than those from mesophiles (18). If isoleucine 229 were replaced with threonine is an “opposite” case, theoretically, it would not be expected to increase the thermostability of MetA because proteins from thermophilic strains usually contain lower levels of polar amino acids (18). Secondary structure prediction for MetAE. coli (http://www.igb.uci.edu/) showed that isoleucine 229 belongs to one of the beta strands. Several beta strands connected laterally by three or more hydrogen bonds generally form a twisted, pleated sheet (20). Association among beta sheets has been implicated in the formation of protein aggregates and fibrils observed in many human diseases, notably, the amyloidoses (10). We assume that the replacement of isoleucine 229 with threonine decreases beta strand length (http://www.igb.uci.edu/) and decreases the capacity of MetAE. coli to aggregate under stress conditions. These mutant enzymes may improve the growth of E. coli at higher temperatures not because they became more thermostable but because they became more resistant in forming aggregates after heat challenge.
Biran and coworkers (2) attempted to stabilize MetAE. coli. They showed that the amino-terminal part of the protein (the first 23 amino acids) was responsible for its instability and assumed that this sequence constituted a proteolytic site or a binding site for proteins that might convert MetAE. coli into a proteolytic substrate (2). On the basis of this investigation, we supposed that thermostable MetAE. coli might carry mutations in the N-terminal region. However, the thermostable MetAE. coli-333 mutant protein harbored no amino acid substitutions in that region.
Notably, thermostable E. coli MetA proteins were more resistant to acetic acid in vivo than the wild-type protein. In previous investigations, it has been shown that methionine relieves the inhibition of E. coli growth by acetic acid (7, 12). Extended exponential growth of the metAE. coli mutants in the presence of acetic acid might serve as additional evidence of their increased acid tolerance because mid-exponential-phase cultures were found to be more acid sensitive than stationary-phase cultures (1). Price-Carter and coworkers found that MetA from S. enterica was as unstable at elevated temperatures as in the presence of weak organic acids, including acetate, benzoate, and propionate (11). Earlier, Roe and coworkers (12) concluded that inhibition of E. coli growth by acetate treatment was due to accumulation of homocysteine, the substrate of MetE. We assume that instability in both of these enzymes may affect E. coli growth under heat and acetic acid stress conditions.
In this study, we showed that methionine stimulates E. coli growth not only at elevated temperatures (42 to 46°C) but also at the normal growth temperature (37°C). Thermotolerant MetAE. coli-333 significantly increased the growth rate of the host strain at 44°C but did not rescue it at 45°C. The same effect was found in strain WG, which harbors metA from the thermophilic bacterium G. kaustophilus. Supplementation with methionine produces additional E. coli growth at 45 and 46°C, indicating that there would be at least one more thermosensitive enzyme in the methionine biosynthesis pathway. We suppose that MetE, which catalyzes the final step in methionine biosynthesis, is a candidate because MetE has been shown to be sensitive to high temperature (9) and oxidative stress (8).
This paper describes the first experimental demonstration that E. coli growth is accelerated under normal and stress conditions by increasing the stability of a single cytosolic enzyme, MetA. This study paves the way to obtaining new E. coli strains that grow at a higher rate at the normal growth temperature, thus potentially improving microbial factory productivity. More directly, being able to grow E. coli cells at higher temperatures means reduced cooling, which is a considerable cost factor in bioprocesses.
Supplementary Material
Footnotes
Published ahead of print on 31 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arnold, K. W., and C. W. Kaspar. 1995. Starvation- and stationary phase-induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biran, D., E. Gur, L. Gollan, and E. Z. Ron. 2000. Control of methionine biosynthesis in Escherichia coli by proteolysis. Mol. Microbiol. 37:1436-1443. [DOI] [PubMed] [Google Scholar]
- 3.Born, T. L., and J. S. Blanchard. 1999. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry 38:14416-14423. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gur, E., D. Biran, E. Gazit, and E. Z. Ron. 2002. In vivo aggregation of a single enzyme limits growth of Escherichia coli at elevated temperature. Mol. Microbiol. 46:1391-1397. [DOI] [PubMed] [Google Scholar]
- 6.Guruprasad, K., B. V. B. Reddy, and M. W. Pandit. 1990. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 4:155-161. [DOI] [PubMed] [Google Scholar]
- 7.Han, K., J. Hong, and H. C. Lim. 1993. Relieving effects of glycine and methionine from acetic acid inhibition in Escherichia coli fermentation. Biotechnol. Bioeng. 41:316-324. [DOI] [PubMed] [Google Scholar]
- 8.Hondorp, E. R., and R. G. Matthews. 2004. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2:1738-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rüdiger, D. Röder, H. Langen, and B. Bukau. 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson, R., M. R. Sawaya, M. Balbirnie, A. O. Madsen, C. Riekel, R. Grothe, and D. Eisenberg. 2005. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price-Carter, M., T. G. Fazzio, E. I. Vallbona, and J. R. Roth. 2005. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J. Bacteriol. 187:3088-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. [DOI] [PubMed] [Google Scholar]
- 13.Ron, E. Z., and B. D. Davis. 1971. Growth rate of Escherichia coli at elevated temperatures: limitation by methionine. J. Bacteriol. 107:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ron, E. Z., and M. Shani. 1971. Growth rate of Escherichia coli at elevated temperatures: reversible inhibition of homoserine trans-succinylase. J. Bacteriol. 107:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ron, E. Z. 1975. Growth rate of Enterobacteriaceae at elevated temperatures: limitation by methionine. J. Bacteriol. 124:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ron, E. Z., S. Alajem, D. Biran, and N. Grossman. 1990. Adaptation of Escherichia coli to elevated temperatures: the metA gene product is a heat shock protein. Antonie van Leeuwenhoek 58:169-174. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Scandurra, R. V., V. Consalvi, R. Chiaraluce, L. Politi, and P. C. Engel. 2000. Protein stability in extremophilic archaea. Front. Biosci. 5:787-795. [DOI] [PubMed] [Google Scholar]
- 19.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397-413. [DOI] [PubMed] [Google Scholar]
- 20.Voet, D., and J. G. Voet. 2004. Biochemistry, 3rd ed. John Wiley & Sons, Inc., New York, NY.
- 21.White, D., R. J. Sharp, and F. G. Priest. 1993. A polyphasic taxonomic study of thermophilic bacilli from a wide geographical area. Antonie van Leeuwenhoek 64:357-386. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler, K., S. M. Noble, E. Mutumanje, B. Bishop, D. P. Huddler, and T. L. Born. 2007. Identification of catalytic cysteine, histidine, and lysine residues in Escherichia coli homoserine transsuccinylase. Biochemistry 46:2674-2683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.