Abstract
The yeast Saccharomyces cerevisiae has been successfully established as a commercially viable system for the production of recombinant proteins. Manipulation of chaperone gene expression has been utilized extensively to increase recombinant protein production from S. cerevisiae, focusing predominantly on the products of the protein disulfide isomerase gene PDI1 and the hsp70 gene KAR2. Here we show that the expression of the genes SIL1, LHS1, JEM1, and SCJ1, all of which are involved in regulating the ATPase cycle of Kar2p, is increased in a proprietary yeast strain, developed by several rounds of random mutagenesis and screening for increased production of recombinant human albumin (rHA). To establish whether this expression contributes to the enhanced-production phenotype, these genes were overexpressed both individually and in combination. The resultant strains showed significantly increased shake-flask production levels of rHA, granulocyte-macrophage colony-stimulating factor, and recombinant human transferrin.
The intrinsic commercial value of heterologous proteins has driven a wide range of studies of the optimization of yeast strains for use as production “cell factories.” A number of reviews exist covering the virtues of different systems for this purpose (12, 27, 31). While the yeasts Saccharomyces cerevisiae and Pichia pastoris do not compete with filamentous fungi such as Aspergillus or Trichoderma spp., or bacterial systems such as Escherichia coli or Bacillus spp., in terms of productivity, they provide a proven, safe alternative without the complexity or high costs of mammalian cell culture. Currently, out of 21 FDA-approved therapeutic proteins produced using yeasts, 19 are derived from S. cerevisiae (28).
Factors affecting heterologous protein production include the properties of the target protein, the host strain, vector system, promoter choice (mRNA availability), leader sequences, translation signals, processing and folding in the endoplasmic reticulum (ER), and secretion (24). If any one of these is suboptimal, it can create a bottleneck leading to poor production yields (21). Thus, although production from S. cerevisiae has been reported in the g/liter range, titers are often 100- to 1,000-fold lower (10, 32).
The use of different expression systems makes comparative assessment of strain improvement approaches difficult, although manipulation of certain groups of genes, and in particular the ER chaperones, has been shown to have widely beneficial effects on the secreted production of at least some heterologous proteins (21, 31). Data indicate that ER function is frequently a major bottleneck in production of secreted heterologous proteins, thus providing a target for optimization (37, 49). This will provide the focus of data discussed here.
Manipulation of the ER luminal environment has mainly focused on the products of the protein disulfide isomerase gene PDI1 and the hsp70 gene KAR2 (33). While overexpression of PDI1 has relatively consistent effects on protein production across a range of protein substrates, the effects of KAR2 overexpression are much more varied (32). This was also observed in our mutagenized, enhanced-production yeast strains, with PDI1 duplication and multicopy overexpression increasing secretion of human transferrin significantly (10, 17, 39), while KAR2 duplication had little or no effect (unpublished data).
Kar2p is the major hsp70 chaperone present in the ER lumen and participates in protein translocation and folding, ER-associated degradation (ERAD), and regulation of unfolded protein response (UPR) signaling (1, 9, 15, 18, 19, 25, 45, 48, 49). Essential to all of these activities is the ATP-dependent cycle of protein binding and release (8, 34). The ATPase cycle is promoted by two sets of cochaperones. The first, consisting of Sec63p, Jem1p, and Scj1p (hsp40-type chaperones present in the ER), promotes ATP hydrolysis, while Sil1p and Lhs1p promote nucleotide exchange. In a recent study, it was demonstrated that Kar2p and Lhs1p, which is also an ER-resident hsp70, share coordinated ATPase cycles (44).
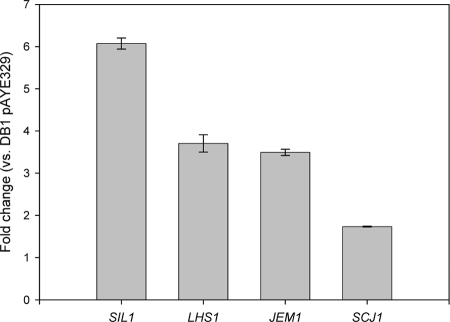
We hypothesized that stimulating the ATPase cycle of Kar2p via overexpression of the cochaperones listed above, both individually and in combination, might promote ER function for heterologous protein production. Further evidence that this might be the case was provided by an analysis of mRNA levels for these genes in our mutagenized, enhanced-production strains. With the exception of SEC63 (which was consequently excluded from subsequent analysis), transcript levels were significantly increased, with SIL1 showing a greater-than-sixfold increase relative to the progenitor strain.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains are summarized in Table 1. The yeast strains DB1 and DS569 have been described previously (39). DYB7 was derived from the strain D88 (17), which was in turn derived from DS569. DS569 and DYB7 contain unknown mutations resulting from chemical mutagenesis. Strains were routinely grown in buffered minimal medium with 2% (wt/vol) dextrose (BMMD) (41) or complete medium with 2% (wt/vol) dextrose (yeast extract-peptone-dextrose [YEPD]) at 200 rpm and 30°C. Log-phase cultures were grown to an optical density at 600 nm (OD600) of 2; stationary-phase cultures were grown for 4 to 5 days postinoculation, as specified. Analysis of heterologous protein production in stationary-phase cultures therefore represents an analysis of accumulated products, taking into account different growth phases up to the point of sampling.
TABLE 1.
Yeast strains used in this study
| Strain name | Genotype |
|---|---|
| DB1 | MATaleu2-3 leu2-112 |
| DB1 ura3 | MATaleu2-3 leu2-112 ura3 |
| DS569 | MATaleu2-3 leu2-112 |
| DYB7 | MATaleu2-3 leu2-112 ubc4 ura3 yap3::URA3 lys2 hsp150::LYS2 |
| DYB7 ura3 | MATaleu2-3 leu2-112 ubc4 ura3 yap3::ura3 lys2 hsp150::LYS2 |
Protein expression plasmids.
Plasmids are summarized in Table 2. All expression plasmids were of the disintegration format, being based on the native S. cerevisiae 2μm plasmid, whereby the E. coli origin of replication and ampicillin resistance marker are removed by site-specific recombination upon transformation into yeast (6). The recombinant human albumin (rHA) expression plasmid pAYE329 has been described previously and consists of the human serum albumin (HSA) cDNA under the control of the GPD1 promoter (40). The expression cassette for pDB2109 consists of PRB1 promoter, HSA/MFα-1 fusion secretion leader, granulocyte-macrophage colony-stimulating factor (GM-CSF) cDNA, and ADH1 terminator. The expression cassette for pDB3213 consists of PRB1 promoter, proprietary preleader sequence, human transferrin cDNA, and ADH1 terminator. In addition, pDB3213 contains a copy of the PDI1 gene cloned into the XcmI site in the inverted repeat region after REP2.
TABLE 2.
Plasmids used in this studya
| Plasmid | Description | Reference |
|---|---|---|
| pSAC35 | 2μm expression vector (2μm LEU2 ampR) | 42 |
| pAYE329 | 2μm rHA expression vector (2μm LEU2 ampR) | 45 |
| pDB2109 | 2μm GM-CSF expression vector (2μm LEU2 ampR) | |
| pDB3213 | 2μm rTf expression vector (2μm LEU2 PDI1 ampR) | |
| pBST+ | General purpose cloning vector (ampR) | 44 |
| YCplac33 | Yeast shuttle vector (CEN4 URA3) | 11 |
All plasmids except for YCplac33 were from Novozymes Biopharma UK.
Chaperone overexpression plasmids.
Chaperone overexpression constructs were initially made in the plasmid pTPA02. pTPA02 is based on pBST+ (41) but contains a polylinker containing the following restriction sites (5′ to 3′): MluI, AleI, BbvCI, FseI, NarI, PspOMI, RsrII, SbfI, XhoI, PacI, PmeI, and EcoRI. After constructs were completed in this plasmid, they were transferred to the centromeric plasmid YCplac33 (11) (also containing a polylinker to facilitate cloning). Primer sequences are shown in Table 3.
TABLE 3.
Oligonucleotide primers used for overexpression plasmid construction
| Primer name | Sequence (5′-3′)a |
|---|---|
| A01 | CTAGGTAACTTAATTAA(PacI)GGGTAAGCTGCCACAGCA |
| A02 | CTACGTACTCTAGA(XbaI)TGTTAATTCAGTAAATTTTC |
| A03 | CTAGACTCTAGA(XbaI)TCTCTGCTTTTGTGCGCG |
| A04 | CATGCTACGTTTAAAC(PmeI)GATGATCATATGATACAC |
| A05 | CTAGTCTCTAGA(XbaI)ATGGAAATGACTGATTTTGAAC |
| A06 | CTAGTCTAGA(XbaI)TCATGAAGTGATGAAGAAATC |
| A07 | CGATCACCGATGTG(AleI)GTTGTTTCCGGGTGTACAATATGG |
| A08 | CCTATAGCAACAAAAGCTGTTAAAAATAAAAGCCTTAAAACGTTTCGCATTGTATATGAGATAGTTGATTG |
| A09 | CGGTAGTACCTGCAGG(SbfI)AAGCAACAGGCGCGTTGGAC |
| A10 | GGCAACAACAATAAAGATAGTATCAAATGTATATATAATTTTGGAATCATTTTGTAATTAAAACTTAGATTAGATTGC |
| A11 | CACAATATTTCAAGCTATACCAAGCATACAATCAACTATCTCATATACAATGCGAAACGTTTTAAGGCT |
| A12 | GCATGCTGAGG(BbvCI)GTGCCACTATAATATTAATGTGC |
| A13 | CACGCTTACTGCTTTTTTCTTCCCAAGATCGAAAATTTACTGAATTAACAATGATTCCAAAATTATATATAC |
| A14 | GCATCTCGAG(XhoI)GACTTTGAGACCTGTGATC |
| A15 | GCATGGCCGGCC(FseI)ACCATATGGAGGATAAGTTGG |
| A16 | ACCTAGTCTAGA(XbaI)TTTGTTTTGTGTGTAAATTTAG |
| A17 | GCATGGGCCC(PspOMI)AGATTCCTGACTTCAACTCAAG |
| A18 | GATCTAGTCTAGA(XbaI)TGTTTTATATTTGTTGTAA |
| A19 | CTAGATCTCTAGA(XbaI)ATGGTCCGGATTCTTCC |
| A20 | GCATGGCGCC(NarI)CCACGGCAGGGCAGTTGGCAC |
| A21 | CTAGATCTCTAGA(XbaI)ATGATACTGATCTCGGG |
| A22 | CGATCGGTCCG(RsrII)AGGGAAATAAGGCAGATCAAAG |
Restriction endonuclease sites are underlined.
The ACT1 promoter and terminator were amplified by PCR from DB1 genomic DNA using the primers A01-A02 and A03-A04, respectively. PCR products were purified and restriction digested with PacI/XbaI (promoter) and XbaI/PmeI (terminator). The two fragments were gel extracted and ligated in a three-way ligation with PacI/PmeI-digested pTPA02 to create pTPA03.
The spliced form of HAC1 (HAC1i) was amplified by PCR from cDNA derived from DB1 using the primers A05 and A06. The PCR product was gel extracted, digested with XbaI, and repurified. This fragment was ligated with XbaI-digested pTPA03, resulting in insertion of HAC1i between the ACT1 promoter and terminator present in pTPA03, creating pTPC01. Restriction digests of putative plasmids were carried out to check for the correct orientation of the HAC1i open reading frame (ORF) relative to the ACT1 promoter/terminator.
The ADH1 and TEF1 promoters and LHS1 and SCJ1 ORFs (including native terminator regions) were amplified by PCR from DB1 genomic DNA using the primers A07-A08, A09-A10, A11-A12, and A13-A14, respectively. The promoters and ORF PCR products were then joined by overlap extension PCR (36). The promoter-ORF fusion fragments were digested with AleI/BbvCI for ADH1prom/LHS1 and SbfI/XhoI for TEF1prom/SCJ1 and subsequently ligated into pTPA02 digested with the same enzymes to create the plasmids pTPC03 and pTPC05.
The TDH1 and PGK1 promoters and SIL1 and JEM1 ORFs (including native terminator regions) were amplified by PCR from DB1 genomic DNA using the primers A15-A16, A17-A18, A19-A20, and A21-A22, respectively. All fragments were gel extracted and digested with the following enzymes: FseI/XbaI (TDH1prom), PspOMI/XbaI (PGK1prom), XbaI/NarI (SIL1 ORF), and XbaI/RsrII (JEM1 ORF). The promoter and ORFs were then ligated with pTPA02 and digested with the same enzymes (i.e., those present on promoter forward primer and ORF reverse primer) in a three-way ligation. The resultant plasmids were named pTPC02 (TDH1prom/SIL1) and pTPC04 (PGK1prom/JEM1).
The multiple chaperone overexpression plasmids pTPC06 to pTPC08 were created by adding promoter/ORF constructs from pTPC02, pTPC03, and pTPC05 to pTPC04 in a serial manner by digestion with the same restriction enzymes used to ligate fragments into pTPA02 (i.e., sites found in promoter forward primer and ORF reverse primer).
Finally, overexpression constructs were transferred to YCplac33 plasmids (containing a polylinker with an AleI site cloned between BamHI and EcoRI sites). Overexpression constructs were transferred to YCplac33 using AleI/BclI (BamHI-compatible site for BclI in YCplac33) and AleI/XhoI (SalI-compatible site for XhoI in YCplac33). The YCplac33 plasmids corresponding to each of pTPC01 to pTPC08 are named pTPC11 to pTPC18.
RNA extraction.
Yeast strains were grown to an OD600 of 2 in BMMD and harvested by centrifugation at 3,000 × g for 3 min. Supernatants were poured off, and pellets were resuspended in a small volume of growth medium (approximately 0.8 ml per 50-ml culture) and transferred to a microcentrifuge tube. Two-hundred-microliter aliquots were released drop by drop into caps from 20-ml universal tubes (Sterilin) containing liquid nitrogen and stored at −80°C. RNA extractions were carried out on cell pellets as described previously (14) using a Braun dismembrator and Trizol reagent (Invitrogen). RNA was quantified using A260 readings. RNA quality was assessed using an Agilent bioanalyzer, and samples with a 28S:18S rRNA ratio of greater than 1.8 were deemed satisfactory for quantitative reverse transcriptase-PCR (qRT-PCR).
DNase digestion/cDNA preparation.
Ten micrograms of total RNA was added to 10 μl of 10× DNase digestion buffer, 5 μl of RQ1 RNase-free DNase (Promega), and nuclease-free H2O to a 95-μl final volume. Samples were incubated at 37°C for 3 h. An additional 5 μl of RNase-free DNase was then added, and samples were incubated for a further 3 h. Digested RNA samples were frozen at −20°C and purified the following day using an RNeasy cleanup column (Qiagen). Columns were eluted with 40 μl of nuclease-free water. One microgram of DNase-digested total RNA was used in a 20-μl Superscript III (Invitrogen) reverse transcription reaction mixture as described in the manufacturer's instructions.
qRT-PCR.
TaqMan probes/primers were designed using the Applied Biosystems “assay-by-design” service with the exception of HAC1i, which was designed using Primer Express software (Applied Biosystems). The HAC1i probe binds across the exon-exon boundary formed upon splicing. Probe/primer sequences are shown in Table 4. All reagents were supplied by Applied Biosystems or Invitrogen (Ultrapure water). Reactions were set up with 25-μl volumes comprising 12.5 μl 2× Universal PCR master mix, 1.25 μl 20× TaqMan probe/primer mix, 5 μl cDNA template (original cDNA diluted 100×), and water to 25 μl. For HAC1i, instead of the 20× probe/primer mix, 2.5 μl of 2.5 μM probe and 2.25 μl of each 10 μM primer were added.
TABLE 4.
qRT-PCR probe/primer sequences
| Gene name | Forward primer (5′-3′) | Reverse primer (5′-3′) | Probe (5′-3′) | Probe binding coordinates (relative to start codon) |
|---|---|---|---|---|
| ACT1 | CCCAGAAGCTTTGTTCCATCCTT | ATGATGGAGTTGTAAGTAGTTTGGTCAA | CAGATTCCAAACCCAAAACA | 795-814 |
| LHS1 | ACACTACTCAGCCCGTTACAATAGA | GTAAACTTTGCACCACCTAGATGTG | ATTTGAAGGATATGGGTATAATC | 789-811 |
| SIL1 | GACATGTACGAAAATGACGATACAAATCT | TCGTTTGCCCACTCTTGCA | TTTGACGACCAATTCTC | 940-956 |
| SCJ1 | GGCGCAGGTGGATTCCA | CGCCAGGACCTCCATGAC | CATATTCGAACGGATGTTTC | 342-361 |
| JEM1 | CCTCTCCACGCACATCGA | TGCTTGTCGAGGATTGTTTCGTAAT | TCGTTAGCTGCTGCTATCA | 592-610 |
| HAC1i | ACAATTCAATTGATCTTGACAATTGG | TCAATTCAAATGAATCAAACCTGAC | CGTAATCCAGAAGCGCA | 652-668 |
Reactions were carried out on an Applied Biosystems 7500 system as described in the manufacturer's instructions. Data were analyzed using the relative standard curve method using ACT1 as the reference gene.
ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were performed using a human albumin ELISA quantitation kit (Bethyl Laboratories) as described in the manufacturer's instructions. Ninety-six-well plates were scanned using a SpectraMax plate reader (Molecular Devices), and data were interpreted using SoftMax Pro software (Molecular Devices).
SDS-polyacrylamide gel electrophoresis (PAGE)-densitometry.
For rHA gels, 20 μl of culture supernatant was run on a 12% (wt/vol) acrylamide nonreducing sodium dodecyl sulfate (SDS)-polyacrylamide gel (Invitrogen) in MOPS (morpholinepropanesulfonic acid) buffer at 200 V for 50 min next to standards of known concentrations. Quantification was performed using LabWorks software (UVP) by comparison to the rHA standard curve. Results were normalized to OD600 readings to account for differences in growth. For GM-CSF gels, 30 μl of culture supernatant was run on a 4 to 12% (wt/vol) acrylamide nonreducing SDS-polyacrylamide gel (Invitrogen) in morpholineethanesulfonic acid buffer at 200 V for 35 min. Relative quantification was performed using LabWorks software (UVP). Gels were stained using Gel-code blue stain (Pierce).
Rocket immunoelectrophoresis.
Culture supernatants were loaded at 5 μl per well in 1% (wt/vol) agarose gels containing goat polyclonal antitransferrin (human) antiserum (Calbiochem) at 40 μl per 50-ml gel (7). The rocket immunoelectrophoresis gels were run at 20 V/cm for 120 min using a Tris/Tricine-based system (22) and stained with PhastGel Blue R (Pharmacia Biotech). Standards were human plasma holotransferrin (Calbiochem) at 100, 50, 20, and 10 μg/ml (all added at 5 μl per well).
RESULTS
Mutagenized production strains show increased transcription of the cochaperone-encoding genes SIL1, LHS1, JEM1, and SCJ1.
Transcript levels for the genes SIL1, LHS1, JEM1, and SCJ1 were investigated using qRT-PCR (Fig. 1). All four genes showed increased expression in the strain DS569[pAYE329] compared to DB1[pAYE329]. This was not a result of increased protein production by DS569, as a comparable result was obtained by analysis of nonexpressing variants of the same strains (data not shown).
FIG. 1.
qRT-PCR analysis of chaperone gene expression in DS569[pAYE329]. All analysis was carried out on log-phase cells. Data are presented as changes from DB1[pAYE329] to DS569[pAYE329]. qRT-PCR data shown are normalized to ACT1 transcript levels. All values are averages of duplicate measurements on duplicate RNA samples, each of which contained RNA derived from three independent cultures. Error bars indicate standard deviations (n = 2).
Cochaperone overexpression results in reduced HAC1i transcript levels.
Centromeric plasmids based on YCplac33 were constructed for the overexpression of SIL1, LHS1, JEM1, and SCJ1 (see Materials and Methods). Promoters of known highly expressed genes were used to direct transcription. In addition, the spliced form of the unfolded protein response transcription factor HAC1 (HAC1i) was overexpressed individually. Overexpression of HAC1i has been shown previously to increase secreted production of some heterologous proteins in S. cerevisiae (47). Centromeric plasmids were then transformed into DB1 ura3[pAYE329]. The resultant strains follow the corresponding plasmid names (i.e., TPC13 is DB1 ura3[pAYE329 pTPC13]).
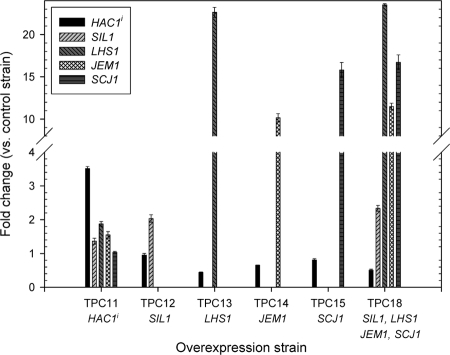
qRT-PCR was performed to confirm gene overexpression in strains overexpressing each gene individually and all four cochaperones together (Fig. 2). All genes were successfully overexpressed, though expression levels varied significantly. SIL1 showed the smallest increase in expression, with transcript levels increasing approximately twofold relative to the control strain, whereas LHS1 showed the largest increase, at approximately 23-fold. Overexpression of multiple genes from the same plasmid had no detrimental effect on expression levels relative to individual overexpression.
FIG. 2.
Confirmation of gene overexpression by qRT-PCR. All analysis was carried out on log-phase cells. Genes overexpressed in each of the TPC strains are given underneath strain names. Data shown are normalized to ACT1 transcript levels and presented as changes over the levels of the control strain, DB1 ura3[pAYE329, pTPA05]. All values are averages of duplicate measurements with exponential-phase cultures. Error bars indicate standard deviations (n = 2). The absence of a bar indicates that this transcript was not measured in this strain.
Overexpression of HAC1i resulted in induction of SIL1, LHS1, and JEM1 (TPC11, Fig. 2). LHS1 showed the most dramatic effect, with levels elevated approximately 1.9-fold in TPC11. Unexpectedly, overexpression of LHS1, JEM1, and all four cochaperones together reduced HAC1i transcript levels up to approximately twofold. This effect was strongest in the strain overexpressing LHS1 (TPC13).
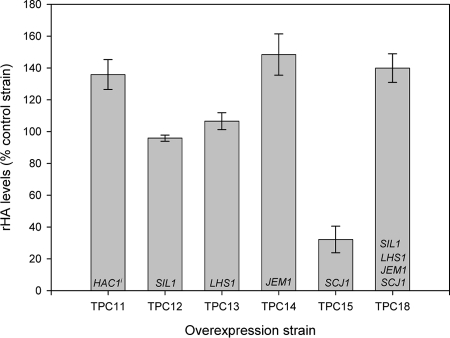
Cochaperone overexpression results in increased production of rHA at log phase.
To assess the effect of cochaperone overexpression on rHA production, culture supernatants were collected from TPC11 to TPC15 and TPC18 at mid-log phase (OD600 of 2). It should be noted that overexpression affected growth rate in a number of the strains used (data not shown). ELISAs were then performed on supernatants (Fig. 3). Overexpression of HAC1i, LHS1, and JEM1 individually and SIL1, LHS1, JEM1, and SCJ1 together had a positive effect on rHA production levels. Overexpression of SIL1 alone had a negligible effect on production, as might be expected due to low-level overexpression. SCJ1 overexpression had a detrimental effect on production with rHA levels reduced to less than 40% of those of the control strain.
FIG. 3.
Log-phase rHA secretion levels in chaperone overexpression strains. All supernatants analyzed were from cultures inoculated to an OD600 of 0.05 and grown to an OD600 of 2. rHA secretion was assessed by ELISA. All values shown are presented as percentages of rHA secretion compared to the control strain, DB1 ura3[pAYE329, pTPA05], and are averages of duplicate measurements. Error bars indicate standard deviations (n = 2). Gene names shown in bars indicate genes overexpressed by each strain.
Cochaperone overexpression results in increased production of rHA in stationary-phase cultures.
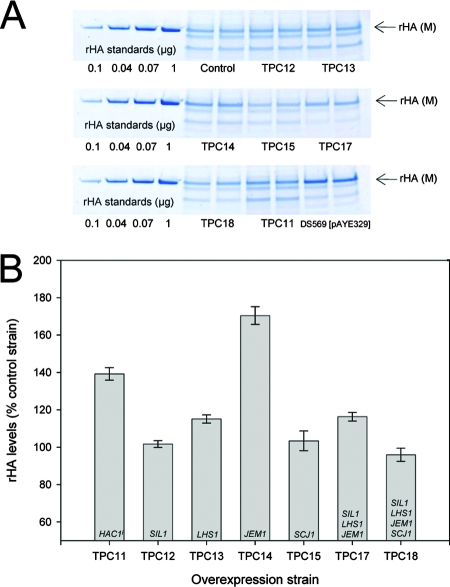
rHA production was also assessed in stationary-phase cultures. Culture supernatants were analyzed by SDS-PAGE (Fig. 4A), and rHA protein bands were quantified by densitometry and comparison to albumin standard curves (Fig. 4B). As final culture densities varied between strains (within the region of approximately 10%), rHA values obtained were normalized to OD600 readings.
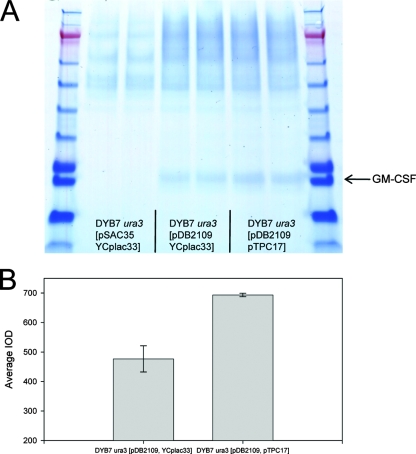
FIG. 4.
SDS-PAGE analysis of rHA production from stationary-phase cultures. All supernatants analyzed were from cultures grown for 5 days. The control strain is DB1 ura3[pAYE329, pTPA05]. (A) Twenty microliters of culture supernatant was run on 12% nonreducing SDS-polyacrylamide gels next to standards of known concentrations. Mature (M) rHA is indicated by arrows. (B) rHA secretion was assessed by densitometric analysis of SDS-polyacrylamide gels and comparison to an rHA standard curve. Due to differences in final cell densities between strains, values have been normalized to OD600 readings. Duplicates shown for each strain represent two individual shake-flask cultures from the same transformant. Error bars indicate standard deviations (n = 2). Gene names shown in bars indicate genes overexpressed by each strain.
Results for TPC11 to TPC14 were strikingly similar to those obtained from log-phase cultures. Due to the negative impact of SCJ1 overexpression on rHA production at log phase (TPC15, Fig. 3), a further strain overexpressing SIL1, LHS1, and JEM1 (TPC17) but not SCJ1 was included in the stationary-phase analysis.
In contrast to the log-phase data, SCJ1 had little impact on rHA production in stationary-phase cultures. Of the two multiple overexpression strains, only TPC17 showed an increase in rHA production. TPC18 showed no improvement in production in stationary-phase cultures.
Cochaperone overexpression results in increased production of rTf and GM-CSF from the further-enhanced-production strain DYB7.
Due to the promising results obtained with rHA production, cochaperone overexpression was investigated with two additional heterologous proteins, recombinant transferrin (rTf) and GM-CSF. As expression of these proteins was low relative to that of rHA in the strain DB1 (data not shown), expression was performed in the strain DYB7 ura3. DYB7 is part of the same series of mutagenized, enhanced-production strains as DS569. The known genotype (i.e., excluding mutations resulting from chemical mutagenesis) is shown in Materials and Methods.
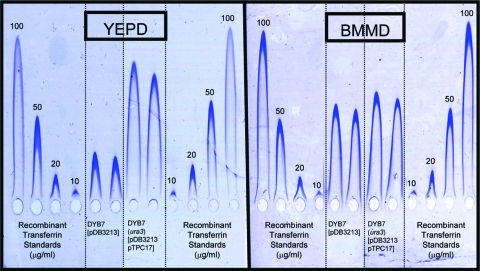
This part of the study was performed in parallel with the analysis of rHA production in stationary-phase culture. As a result, only pTPC17 (containing SIL1, LHS1, and JEM1) was selected, based on log-phase data. Levels of GM-CSF were elevated approximately 45% upon cochaperone overexpression (Fig. 5). rTf levels were increased even more markedly, although the extent of this effect was shown to be dependent on the growth medium (Fig. 6).
FIG. 5.
Increased expression of GM-CSF from DYB7. Two transformants for each strain were inoculated into 10 ml BMMD in 50-ml shake flasks in duplicate and incubated in an orbital shaker at 30°C and 200 rpm for 4 days. Culture supernatants were harvested, and the recombinant GM-CSF titers were compared by SDS-PAGE (A) and densitometric analysis (B). Data are presented as integrated optical density readings (IODs) produced by densitometry. Error bars indicate standard deviations (n = 2).
FIG. 6.
Increased expression of rTf from DYB7. Transformants of each strain were inoculated into 10 ml BMMD and 10 ml YEPD in 50-ml shake flasks and incubated in an orbital shaker at 30°C and 200 rpm for 4 days. Culture supernatants were harvested, and the rTf titers were compared by rocket immunoelectrophoresis.
DISCUSSION
The ER provides an environment suitable for the folding and initial maturation of secretory proteins. Two interrelated homeostatic systems, termed the UPR and ERAD, govern the flux of proteins through the ER. Both systems are induced by ER perturbation, either via chemical treatment (e.g., with dithiothreitol to inhibit disulfide bond formation or tunicamycin to prevent N-linked glycosylation) or by overexpression of certain heterologous proteins (4, 16, 46). The yeast ER protein-folding machinery consists of three distinct groups of proteins, hsp70 chaperones (e.g., Kar2p and Lhs1p), along with their cochaperones, the protein disulfide isomerase family (and accessory proteins such as Ero1p and Erv2p) and ERAD/ER quality control proteins (e.g., Der1p and Cne1p) (25).
Altering levels of components of the ER protein-folding machinery has been shown previously to affect UPR signaling. Strains overexpressing PDI1 showed reduced levels of HAC1 splicing, corresponding to reduced intracellular levels of scFv (49). This sets a precedent for the unexpected result shown in Fig. 2, indicating that overexpression of LHS1, JEM1, and all four cochaperones together reduced HAC1i transcript levels. This effect was most apparent with LHS1 (TPC13), where HAC1i transcript levels were reduced more than twofold.
Comparison of data from the analysis of HAC1i transcript levels and rHA production in overexpression strains (Fig. 2 and 3, respectively) showed that, with the exception of TPC11 (where HAC1i transcript levels are artificially elevated), strains showing increased rHA production also showed reduced HAC1i transcript levels. This suggests that ER protein-folding efficiency is improved in these strains.
Therefore, these results support an indirect link between cochaperones of Kar2p and HAC1 splicing. Maintaining Kar2p in its active, cycling state may both keep luminal load down and keep Ire1p in its inactive form. To complicate this picture, Lhs1p has been shown to have chaperone activity itself, aside from its interaction with Kar2p (29).
The accumulated secreted levels of rHA were analyzed at both log phase and at stationary phase (Fig. 3 and 4). Good correlation was seen between the two sets of results. Overexpression of HAC1i, LHS1, and JEM1 individually produced consistent results, increasing rHA production in both data sets. The strain TPC15, which overexpresses SCJ1, and the multiple overexpression strain TPC18 behaved less consistently. While SCJ1 overexpression appeared to be detrimental to rHA production in log-phase cultures, it had a negligible effect on rHA production in stationary-phase cultures. Conversely, TPC18 showed an approximately 40% increase in rHA production in log-phase cultures but a marginal decrease in stationary-phase cultures.
The reason for the differences in rHA production seen with SCJ1 and JEM1 overexpression is unknown (LHS1 and SIL1 data presented here are not comparable due to low overexpression of SIL1). Cells lacking JEM1 and SCJ1 are temperature sensitive and are defective in transport of constitutively misfolded carboxypeptidase Y (CPY*), suggesting overlapping function (26, 38). One aspect that may affect function is localization. Jem1p is an ER membrane protein (though the J domain is found in the ER lumen), whereas Scj1p is a soluble, ER luminal protein (26, 30).
Normalization to optical density readings was used during analysis of rHA production to indicate protein production on a “per-cell” basis. It is true, however, that transcription and translation are affected by growth rate (5). Thus, increased rHA production might be a secondary effect resulting from chaperone overexpression influencing growth rate. In the case of JEM1 overexpression (TPC14) the general pattern of growth was remarkably similar to that of the control strain, indicating that this is unlikely to be a major factor influencing increased protein production. Furthermore, transcription of GPD1 and PRB1 (promoters used in rHA and GM-CSF/rTf expression cassettes, respectively) was not shown to be significantly affected by growth rate (5).
Experimental data also demonstrated that GM-CSF and rTf production was significantly increased with cochaperone overexpression. Data were restricted to combined overexpression of SIL1, LHS1, and JEM1 in the strain DYB7. Many studies in this field have shown that the beneficial effects of chaperone overexpression are highly substrate dependent (13, 43). In this case all three proteins, with distinct characteristics (Table 5), showed increased production levels from shake-flask culture.
TABLE 5.
Characteristics of heterologous proteins
| Protein | Leader sequence | Proteolytic processing | Size (kDa) | No. of disulfide bonds | Known modification |
|---|---|---|---|---|---|
| rHA | HSA/mating factor α-prepro | Signal peptidase/Kex2p | 66.5 | 17 | |
| GM-CSF | HSA/mating factor α-prepro | Signal peptidase/Kex2p | 14.7 | 2 | N glycosylation |
| rTf | Proprietary (pre only) | Signal peptidase | 80 | 19 |
Analysis of data for rHA production in stationary-phase cultures (Fig. 4) allows dissection of the effects of the individual cochaperones. The data suggest that JEM1 is likely to be the main driver in increasing heterologous protein production. Due to the specific role of Sec63p as the hsp40 interacting with Kar2p during translocation, this indicates that the function of Kar2p, supporting protein production (as facilitated by JEM1), is unrelated to translocation (2, 3, 35).
As previously performed with PDI1 (10), it would be informative to incorporate JEM1 along with its native promoter into the 2μm expression plasmid to investigate whether this would increase protein production further. The plasmid used for rTf production already contains a PDI1 expression cassette and so demonstrates a combined effect of PDI1, SIL1, LHS1, and JEM1 overexpression. It is key to note that, like all studies of this nature, we are providing only a “snapshot” of the effects of chaperone overexpression on heterologous protein production. Ideally, the levels of gene expression for each overexpressed gene would be tunable to provide a more comprehensive assessment. Such an approach has been used successfully in filamentous fungi to identify optimal levels of BiP (fungal Kar2p) and Pdi1p for the production of the plant sweet protein thaumatin (20, 23).
rTf production from shake-flask culture varied significantly between different growth media (Fig. 6). Increased biomass formation with complete medium (YEPD) normally results in increased protein production relative to minimal medium (BMMD). In this case, the opposite was observed. This may be a result of reduced plasmid stability in nonselective YEPD. The effect of cochaperone overexpression could thus be twofold—increasing productive folding in the ER and reducing the deleterious effects of rTf production on cell physiology and/or growth. This may go some way to explaining the differential effects observed in the two media.
In conclusion, the large number of factors surrounding heterologous protein production, compounded by use of different substrate proteins, ensures that the optimization of the secretion pathway for heterologous protein production is necessarily complex. The term bottleneck is used frequently and is indicative of the problems associated with manipulating single components in a “balanced” system. In the future, large-scale experiments utilizing distinct classes of substrate proteins may be the way forward to gain a fuller understanding. In this way it might be possible to assess protein characteristics, secretory pathway modifications, and consequent production yields in an informative and constructive way.
Acknowledgments
This work was supported by Novozymes Biopharma UK Ltd. and the BBSRC in the form of a BBSRC CASE studentship awarded to T.P.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Blond-Elguindi, S., S. E. Cwirla, W. J. Dower, R. J. Lipshutz, S. R. Sprang, J. F. Sambrook, and M. J. H. Gething. 1993. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding-specificity of BiP. Cell 75:717-728. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky, J. L., J. Goeckeler, and R. Schekman. 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92:9643-9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodsky, J. L., and R. Schekman. 1993. A Sec63p-Bip complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5:729-735. [DOI] [PubMed] [Google Scholar]
- 5.Castrillo, J., L. Zeef, D. Hoyle, N. Zhang, A. Hayes, D. Gardner, M. Cornell, J. Petty, L. Hakes, L. Wardleworth, B. Rash, M. Brown, W. Dunn, D. Broadhurst, K. O'Donoghue, S. Hester, T. Dunkley, S. Hart, N. Swainston, P. Li, S. Gaskell, N. Paton, K. Lilley, D. Kell, and S. Oliver. 2007. Growth control of the eukaryote cell: a systems biology study in yeast. J. Biol. 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinery, S. A., and E. Hinchliffe. 1989. A novel class of vector for yeast transformation. Curr. Genet. 16:21-25. [DOI] [PubMed] [Google Scholar]
- 7.Cox, H., D. Mead, P. Sudbery, R. M. Eland, I. Mannazzu, and L. Evans. 2000. Constitutive expression of recombinant proteins in the methylotrophic yeast Hansenula polymorpha using the PMA1 promoter. Yeast 16:1191-1203. [DOI] [PubMed] [Google Scholar]
- 8.Dorner, A. J., L. C. Wasley, and R. J. Kaufman. 1990. Protein dissociation from Grp78 and secretion are blocked by depletion of cellular ATP levels. Proc. Natl. Acad. Sci. USA 87:7429-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fewell, S. W., K. J. Travers, J. S. Weissman, and J. L. Brodsky. 2001. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35:149-191. [DOI] [PubMed] [Google Scholar]
- 10.Finnis, C. A., G. Shuttleworth, and D. Sleep. 2005. High-level secretion of recombinant proteins from S. cerevisiae by co-expression of genes from 2-micron vectors, abstr. 2-9. Abstr. 22nd Int. Conf. Yeast Genet. Mol. Biol.
- 11.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking 6-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 12.Greene, J. J. 2004. Host cell compatibility in protein expression, p. 3-14. In P. Balbas and A. Lorence (ed.), Recombinant gene expression. Springer, New York, NY. [DOI] [PubMed]
- 13.Harmsen, M. M., M. I. Bruyne, H. A. Raue, and J. Maat. 1996. Over-expression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 14.Hauser, N. C., M. Vingron, M. Scheideler, B. Krems, K. Hellmuth, K. D. Entian, and J. D. Hoheisel. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14:1209-1221. [DOI] [PubMed] [Google Scholar]
- 15.Holkeri, H., E. Paunola, E. Jamsa, and M. Makarow. 1998. Dissection of the translocation and chaperoning functions of yeast BiP/Kar2p in vivo. J. Cell Sci. 111:749-757. [DOI] [PubMed] [Google Scholar]
- 16.Kauffman, K. J., E. M. Pridgen, F. J. Doyle, P. S. Dhurjati, and A. S. Robinson. 2002. Decreased protein expression and intermittent recoveries in BiP levels result from cellular stress during heterologous protein expression in Saccharomyces cerevisiae. Biotechnol. Prog. 18:942-950. [DOI] [PubMed] [Google Scholar]
- 17.Kerry-Williams, S. M., S. C. Gilbert, L. R. Evans, and D. J. Ballance. 1998. Disruption of the Saccharomyces cerevisiae YAP3 gene reduces the proteolytic degradation of secreted recombinant human albumin. Yeast 14:161-169. [DOI] [PubMed] [Google Scholar]
- 18.Kimata, Y., D. Oikawa, Y. Shimizu, Y. Ishiwata-Kimata, and K. Kohno. 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 167:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohno, K., K. Normington, J. Sambrook, M. J. Gething, and K. Mori. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13:877-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombraña, M., F. J. Moralejo, R. Pinto, and J. F. Martin. 2004. Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl. Environ. Microbiol. 70:5145-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattanovich, D., B. Gasser, H. Hohenblum, and M. Sauer. 2004. Stress in recombinant protein producing yeasts. J. Biotechnol. 113:121-135. [DOI] [PubMed] [Google Scholar]
- 22.Monthony, J. F., E. G. Wallace, and D. M. Allen. 1978. Non-barbital buffer for immunoelectrophoresis and zone electrophoresis in agarose gels. Clin. Chem. 24:1825-1827. [PubMed] [Google Scholar]
- 23.Moralejo, F. J., A. J. Watson, D. J. Jeenes, D. B. Archer, and J. F. Martin. 2001. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genomics 266:246-253. [DOI] [PubMed] [Google Scholar]
- 24.Niebauer, R. T., and A. S. Robinson. 2005. Saccharomyces cerevisiae protein expression: from protein production to protein engineering, p. 253-296. In F. Baneyx (ed.), Protein expression technologies. Horizon Press, Norwich, United Kingdom.
- 25.Nishikawa, S., J. L. Brodsky, and K. Nakatsukasa. 2005. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J. Biochem. 137:551-555. [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa, S., and T. Endo. 1997. The yeast Jem1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 272:12889-12892. [DOI] [PubMed] [Google Scholar]
- 27.Palomares, L. A., S. Estrada-Mondaca, and O. T. Ramirez. 2004. Production of recombinant proteins: challenges and solutions, p. 15-52. In P. Balbas and A. Lorence (ed.), Recombinant gene expression. Springer, New York, NY. [DOI] [PubMed]
- 28.Rader, R. A. 2007. Biopharmaceutical products in the US and European markets, 6th ed. BioPlan Associates Inc., Rockville, MD.
- 29.Saris, N., H. Holkeri, R. A. Craven, C. J. Stirling, and M. Makarow. 1997. The Hsp70 homologue Lhs1p is involved in a novel function of the yeast endoplasmic reticulum, refolding and stabilization of heat-denatured protein aggregates. J. Cell Biol. 137:813-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlenstedt, G., S. Harris, B. Risse, R. Lill, and P. A. Silver. 1995. A yeast DnaJ homolog, Scj1p, can function in the endoplasmic-reticulum with BiP/Kar2p via a conserved domain that specifies interactions with hsp70s. J. Cell Biol. 129:979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, F. R. 2004. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 65:363-372. [DOI] [PubMed] [Google Scholar]
- 32.Schroder, M. 2007. The cellular response to protein unfolding stress, p. 117-139. In G. D. Robson, P. van West, and G. M. Gadd (ed.), Exploitation of fungi. Cambridge University Press, Cambridge, United Kingdom.
- 33.Schroder, M. 2008. Engineering eukaryotic protein factories. Biotechnol. Lett. 30:187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder, M., and R. J. Kaufman. 2005. ER stress and the unfolded protein response. Mutat. Res. 569:29-63. [DOI] [PubMed] [Google Scholar]
- 35.Scidmore, M. A., H. H. Okamura, and M. D. Rose. 1993. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologs of DnaK and DnaJ in the endoplasmic reticulum. Mol. Biol. Cell 4:1145-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shusta, E. V., R. T. Raines, A. Pluckthun, and K. D. Wittrup. 1998. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16:773-777. [DOI] [PubMed] [Google Scholar]
- 38.Silberstein, S., G. Schlenstedt, P. A. Silver, and R. Gilmore. 1998. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J. Cell Biol. 143:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleep, D., G. P. Belfield, D. J. Ballance, J. Steven, S. Jones, L. R. Evans, P. D. Moir, and A. R. Goodey. 1991. Saccharomyces cerevisiae strains that overexpress heterologous proteins. Bio/Technology 9:183-187. [DOI] [PubMed] [Google Scholar]
- 40.Sleep, D., G. P. Belfield, and A. R. Goodey. 1990. The secretion of human-serum albumin from the yeast Saccharomyces cerevisiae using 5 different leader sequences. Bio/Technology 8:42-46. [DOI] [PubMed] [Google Scholar]
- 41.Sleep, D., C. Finnis, A. Turner, and L. Evans. 2001. Yeast 2μ plasmid copy number is elevated by a mutation in the nuclear gene UBC4. Yeast 18:403-421. [DOI] [PubMed] [Google Scholar]
- 42.Sleep, D., J. E. Ogden, N. A. Roberts, and A. R. Goodey. 1991. Cloning and characterization of the Saccharomyces cerevisiae glycerol-3-phosphate dehydrogenase (GUT2) promoter. Gene 101:89-96. [DOI] [PubMed] [Google Scholar]
- 43.Smith, J. D., B. C. Tang, and A. S. Robinson. 2004. Protein disulfide isomerase, but not binding protein, overexpression enhances secretion of a non-disulfide-bonded protein in yeast. Biotechnol. Bioeng. 85:340-350. [DOI] [PubMed] [Google Scholar]
- 44.Steel, G., D. Fullerton, J. Tyson, and C. Stirling. 2004. Coordinated activation of hsp70 chaperones. Science 303:98-101. [DOI] [PubMed] [Google Scholar]
- 45.Taxis, C., R. Hitt, S. H. Park, P. M. Deak, Z. Kostova, and D. H. Wolf. 2003. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278:35903-35913. [DOI] [PubMed] [Google Scholar]
- 46.Travers, K. J., C. K. Patil, and J. S. Weissman. 2001. Functional genomic approaches to understanding molecular chaperones and stress responses, Adv. Protein Chem. 56:345-390. [DOI] [PubMed] [Google Scholar]
- 47.Valkonen, M., M. Penttila, and M. Saloheimo. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel, J. P., L. M. Misra, and M. D. Rose. 1990. Loss of BiP/grp78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 110:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, P., D. Raden, F. J. Doyle, and A. S. Robinson. 2005. Analysis of unfolded protein response during single-chain antibody expression in Saccharomyces cerevisiae reveals different roles for BiP and PDI in folding. Metab. Eng. 7:269-279. [DOI] [PubMed] [Google Scholar]