Abstract
We describe a new enzymatic functionality for the surface layer (S-layer) of Lactobacillus acidophilus ATCC 4356, namely, an endopeptidase activity against the cell wall of Salmonella enterica serovar Newport, assayed via zymograms and identified by Western blotting. Based on amino acid sequence comparisons, the hydrolase activity was predicted to be located at the C terminus. Subsequent cloning and expression of the C-terminal domain in Bacillus subtilis resulted in the functional verification of the enzymatic activity.
Surface layers (S-layers) have been recognized ubiquitously in both Eubacteria and Archaea. Their respective structures and functionalities have been investigated intensively in the past (3, 4). Several species of the genus Lactobacillus possess an S-layer. The S-layer of Lactobacillus acidophilus ATCC 4356 is composed of a single S-protein (SA protein) of around 45 kDa, which has been extensively characterized by Peter H. Pouwels and coworkers (5, 6, 7, 8, 9, 23, 24, 25). The function of the S-layers of these organisms is unknown, but they may be important for bacterial adhesion to intestinal epithelial cells and extracellular matrix components (11, 12, 15, 16). Lactobacillus acidophilus is one of the main species of the genus Lactobacillus found in human and animal intestines. Several lactobacilli have been proposed to have probiotic characteristics. Probiotics are live microorganisms, usually contained in food, traditionally regarded as safe for human and animal use. When ingested in sufficient numbers, probiotics are believed to play an important role in the control of the host intestinal microbiota and in the modulation of host immune responses (26). An antagonist action of the Lactobacillus S-layer bearing S traits has been suggested (11, 15). Different groups have found probiotic activity in Lactobacillus acidophilus strains (13, 14, 15, 18), particularly in strain ATCC 4356, used in this study (12, 18, 21).
In an attempt to evaluate whether there was any cell wall polymer hydrolysis activity, purified S-layers extracted from L. acidophilus cells (statically grown overnight in MRS medium at 37°C) by using 6 M LiCl as described previously (6, 9) were subjected to a zymogram assay. For that purpose, 2% purified peptidoglycan (PG) was prepared from several species according to the method of Bousfield et al. (10) and was incorporated into a 12% polyacrylamide gel. Zymography was performed as described by Kakikawa et al. (17) and was used to detect the lytic activity. Gels were cast with only 0.01% sodium dodecyl sulfate (SDS). After the run, hydrolase activity was detected by a clear zone (27). We observed an intense clear band when PG from Salmonella enterica serovar Newport was used (Fig. 1C). The lytic band corresponds to the molecular weight of SA protein (Fig. 1A) and was confirmed by Western blotting (Fig. 1B) with an antibody against the S-layer. A polyclonal anti-S-layer antibody was produced by injecting the protein obtained from SDS-polyacrylamide gels into a mouse and was used at a 1:10,000 dilution. A biotin-conjugated anti-mouse antibody was detected, with a second biotinylated antibody conjugated to alkaline phosphatase, by chemiluminescence using CDP-Star (GE-Biosciences).
FIG. 1.
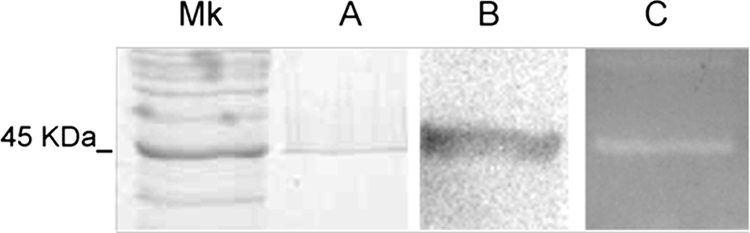
SDS, Western blot, and zymography profiles of the S-layer obtained from L. acidophilus. (A) SDS-polyacrylamide gel electrophoresis after isolation with LiCl and staining with Coomassie blue; (B) Western blot detection with the anti-S-layer antibody; (C) zymogram of the S-layer showing lytic activity over the cell wall from Salmonella serovar Newport. Mk, molecular mass standard.
Also, PGs from Escherichia coli and Micrococcus luteus gave positive results, whereas no activity was observed against PGs from Bacillus cereus, Lactobacillus casei, and L. acidophilus (data not shown). Although L. acidophilus ATCC 4356 has been extensively characterized by Peter H. Pouwels and coworkers (5, 6, 7, 8, 9, 23, 24, 25), this novel activity had not been visualized previously. Here we describe a true uncharacterized murein hydrolase activity associated with this S-layer, with lytic activity toward the cell walls of several Eubacteria. A role of murein hydrolases for an S-layer protein had previously been described only for Bacillus anthracis (1). This activity of the S-layer protein as a murein hydrolase is also expected for proteins sharing extensive homology with the SA protein in the carboxy-terminal portion, such as those described by Boot et al. (8).
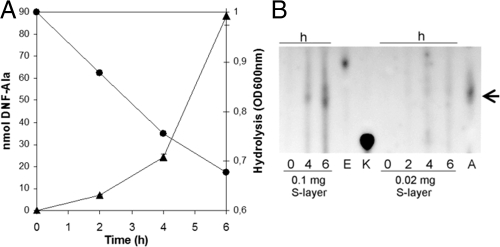
To compare this in vitro effect, in vivo assays with whole viable cells were also performed. Viable cells and cell walls from Salmonella serovar Newport were isolated, washed once with phosphate-buffered saline, and mixed with S-layer protein at the concentrations indicated. Lysis was monitored by the decrease in absorbance (optical density at 600 nm [OD600]) (Fig. 2A and B). With cell wall preparations, the best lytic performance was achieved at the lowest S-layer concentration (0.02 to 0.1 mg). Reassembling of S-proteins at high concentrations may mask the lytic activity, which would explain the decrease in the lytic activity observed (Fig. 2A). With viable cells at this low S-layer concentration, no significant decrease in the OD was observed (data not shown); this may be due to the presence of lipopolysaccharides in the Salmonella cell wall. However, at higher S-layer concentrations (0.5 to 1 mg), lysis was obtained but took longer.
FIG. 2.
Hydrolase activities against isolated cell walls and viable cells of Salmonella serovar Newport. Mixtures of cell walls (0.5 mg/ml) (A) or whole cells (OD660, 1) (B) and the S-layer at the indicated mass were incubated, and the OD600 was measured.
To determine the specificity of the lytic activity, we analyzed the newly exposed groups of the PG after hydrolysis with the S-layer protein. An increase in the number of sugar-reducing groups will be detected if the enzyme is a muramidase or a glucosaminidase, and an increase in the number of free amino groups will be detected if the enzyme is an amidase or an endopeptidase. Free amino groups released during hydrolysis of the cell walls were determined in the presence of 2,4-dinitrofluorobenzene (DNFB) and analyzed by thin-layer chromatography (TLC) as described previously (20). The lysis of the Salmonella serovar Newport cell walls was accompanied by an increase in the level of free amino groups; particularly, we were able to quantify the increase in the level of the dinitrophenylalanine (DNF-Ala) (Fig. 3), while there was no change in the number of reducing ends, determined, as described previously (20), with 8-amino-naphthalene-1,3,6-trisulfonic acid (ANTS; Molecular Probes) (data not shown). These results are consistent with the absence of activity toward the glycan strand of PG but allow us to postulate that this lytic activity works as an endopeptidase or even a PG amidase as defined by Vollmer et al. (28).
FIG. 3.
Correlation between hydrolysis and increased levels of DNFB-treated free amino groups. (A) Hydrolysis with 0.1 mg of S-layer protein was followed by determination of the OD600 (•) and the amount of DNF-Ala (▴) by TLC and densitometry. (B) TLC analysis of hydrolysis with two S-layer concentrations and different times (in hours) as indicated. DNF-amino acids (100 nmol) are Glu (E), Lys (K), and Ala (A). The arrow indicates the position of DNF-Ala.
In silico sequence analysis with the BLASTp tool (2) showed that the C-terminal motif has homology with lytic enzymes of several Lactobacillus strains. ClustalW alignments (http://www.ebi.ac.uk/Tools/clustalw2/index.html) are shown in Fig. 4.
FIG. 4.
Identities of the primary sequence of the SA protein to those of other lytic enzymes analyzed by ClustalW. The cloned C-terminal portion of the SA protein is underlined. Asterisks indicate residues identical in all sequences in the alignment; colons, conserved substitutions; periods, semiconserved substitutions. CAA61560, SA protein (Lactobacillus acidophilus ATCC 4356); YP_618401, putative amidase (Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842); YP_812313, N-acetylmuramoyl-l-alanine amidase (L. delbrueckii subsp. bulgaricus ATCC BAA-365).
Primers were designed to amplify the carboxy-terminal domain of the protein, and a 968-bp amplicon was obtained, cloned into the pGEM-T Easy vector (Promega), and sequenced. Primers 5′-CAGAAAATGCAGGTAAGACTGTTA-3′ (forward) and 5′-GCGGAATTCGAGCTCAGCGTTAGTGCTACGACT-3′ (reverse) were used. Complete homology with the previously reported sequence (EMBL accession number X89375) (4) was obtained. The 968-bp fragment (323 amino acids) in the pHCMC05 shuttle plasmid, kindly provided by the Bacillus Genetic Stock Center (19), was subcloned in order to express this fragment under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter, allowing us to produce the pC1C2 plasmid (recombinant plasmid of pHCMC05 containing the 968-bp insert). When introduced into E. coli JM109 (grown in LB medium at 37°C with aeration), the plasmid showed high instability and decreased growth, particularly when IPTG was present (data not shown). The lack of success in expressing the cloned hydrolase motif in E. coli was certainly related to the PG structure of this host bacterium, since we found that it was a substrate for this lytic activity. This may explain the instability that other authors (9, 25) encountered when they attempted to clone the entire slpA gene of ATCC 4356 into E. coli.
Therefore, we introduced the plasmid into Bacillus subtilis 168 competent cells. For expression, cells were induced by the addition of 0.5 mM IPTG. B. subtilis was grown in LB medium at 37°C with aeration in the presence of 1 M NaCl to avoid the presence of proteases (22) that might degrade the heterologous product. To prepare for the analysis of expression by zymograms and Western blotting, cells were disrupted by sonication after harvest at the indicated times. Western blot and zymogram analyses showed the predicted 33-kDa band when the plasmid was expressed in Bacillus subtilis (Fig. 5A). When IPTG was added to the growth medium, a lytic band corresponding to the molecular mass of this fragment was seen; expression of this fragment was confirmed by Western blotting with an antibody against the S-layer (Fig. 5B).
FIG. 5.
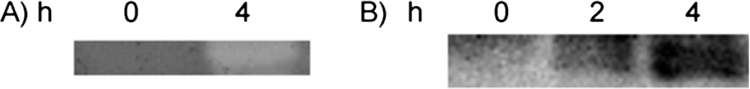
Heterologous expression. (A) Zymogram of the C terminus showing lytic activity over the Salmonella serovar Newport cell wall. (B) Western blot detection with anti-S-layer. Times (measured in hours) after the addition of 0.5 mM IPTG are shown.
Murein or PG hydrolases comprise a large family of enzymes with roles in daughter cell separation, PG turnover, autolysis, spore formation, and antibiotic-induced lysis within their own cells (27, 28, 29). However, these enzymes are also important for an adaptive response to environmental conditions that might result from an antagonistic relationship in the same ecological niche. The gastrointestinal tract will determine the coexistence of gram-negative and gram-positive species, dealing with high osmotic strength, pH gradients, anaerobic conditions, and nutrient variability, resulting in a competition for survival. The murein hydrolase activity provides S-layer-bearing strains of lactobacilli with an additional means to succeed and survive. Due to the lytic activity against whole cells of Salmonella serovar Newport that we observed, one might wonder if these S-layer characteristics may account for the probiotic properties of Lactobacillus acidophilus. In vivo assays would be required to address the question of whether the inhibition of adhesion of gram-negative bacteria and the competitive exclusion of pathogens that have been reported for S-layer-bearing strains (12, 13, 14) are associated with the antibacterial activity reported here. The precise characterization of murein hydrolase activity will be the aim of our future investigation.
Acknowledgments
We are grateful to D. R. Zeigler of the Bacillus Genetic Stock Center for the provision of strains and plasmids and to H. Goldman for the help in the preparation of the antiserum against the S-layer.
This work was supported by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and the University of Buenos Aires (UBA) in Argentina. During this work, M.P.-A., M.M.P., and M.C.A. benefited from CONICET fellowships. C.S.R. and S.M.R. are career investigators of CONICET.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Ahn, J. S., L. Chandramohan, L. E. Liou, and K. W. Bayles. 2006. Characterization of CidR-mediated regulation in Bacillus anthracis reveals a previously undetected role of S-layer proteins as murein hydrolases. Mol. Microbiol. 62:1158-1169. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., J. C. Wootton, E. M. Gertz, R. Agarwala, A. Morgulis, A. A. Schäffer, and Y. K. Yu. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Åvall-Jääskeläinen, S., and A. Palva. 2005. Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29:511-529. [DOI] [PubMed] [Google Scholar]
- 4.Beveridge, T. J., P. H. Pouwels, M. Sára, A. Kotiranta, K. Lounatmaa, K. Kari, E. Kerosuo, M. Haapasalo, E. M. Egelseer, I. Schocher, U. B. Sleytr, L. Morelli, M. L. Callegari, J. F. Nomellini, W. H. Bingle, J. Smit, E. Leibovitz, M. Lemaire, I. Miras, S. Salamitou, P. Béguin, H. Ohayon, P. Gounon, M. Matuschek, and S. F. Koval. 1997. Functions of S-layers. FEMS Microbiol. Rev. 20:99-149. [DOI] [PubMed] [Google Scholar]
- 5.Boot, H. J., C. P. Kolen, and P. H. Pouwels. 1995. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 177:7222-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot, H. J., C. P. Kolen, and P. H. Pouwels. 1996. Interchange of the active and silent S-layer protein genes of Lactobacillus acidophilus by inversion of the chromosomal slp segment. Mol. Microbiol. 21:799-809. [DOI] [PubMed] [Google Scholar]
- 7.Boot, H. J., C. P. Kolen, F. J. Andreadaki, R. J. Leer, and P. H. Pouwels. 1996. The Lactobacillus acidophilus S-layer protein gene expression site comprises two consensus promoter sequences, one of which directs transcription of stable mRNA. J. Bacteriol. 178:5388-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boot, H. J., C. P. Kolen, B. Pot, K. Kersters, and P. H. Pouwels. 1996. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology 142:2375-2384. [DOI] [PubMed] [Google Scholar]
- 9.Boot, H. J., C. P. Kolen, J. M. van Noort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J. Bacteriol. 175:6089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bousfield, I. J., R. M. Keddie, T. R. Dando, and S. Shaw. 1985. Simple rapid methods of cell wall analysis as an aid in the identification of aerobic coryneform bacteria, p. 221-236. In M. Goodfellow and D. E. Minnikin (ed.), Chemical methods in bacterial systematics. Academic Press, London, England.
- 11.Buck, B. L., E. Altermann, T. Svingerud, and T. R. Klaenhammer. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:8344-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X., J. Xu, J. Shuai, J. Chen, Z. Zhang, and W. Fang. 2007. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is [sic] responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115:307-312. [DOI] [PubMed] [Google Scholar]
- 13.Coconnier, M. H., V. Liévin, M. Lorrot, and A. L. Servin. 2000. Antagonistic activity of Lactobacillus acidophilus LB against intracellular Salmonella enterica serovar Typhimurium infecting human enterocyte-like Caco-2/TC-7 cells. Appl. Environ. Microbiol. 66:1152-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coconnier-Polter, M.-H., V. Lievin-Le Moal, and A. L. Servin. 2005. A Lactobacillus acidophilus strain of human gastrointestinal microbiota origin elicits killing of enterovirulent Salmonella enterica serovar Typhimurium by triggering lethal bacterial membrane damage. Appl. Environ. Microbiol. 71:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frece, J., B. Kos, I. K. Svetec, Z. Zgaga, V. Mrsa, and J. Susković. 2005. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 98:285-292. [DOI] [PubMed] [Google Scholar]
- 16.Golowczyc, M. A., P. Mobili, G. L. Garrote, A. G. Abraham, and G. L. De Antoni. 2007. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar Enteritidis. Int. J. Food Microbiol. 118:264-273. [DOI] [PubMed] [Google Scholar]
- 17.Kakikawa, M., K. J. Yokoi, H. Kimoto, M. Nakano, K. Kawasaki, A. Taketo, and K. Kodaira. 2002. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage fg1e. Gene 299:227-234. [DOI] [PubMed] [Google Scholar]
- 18.Kos, B., J. Susković, S. Vuković, M. Simpraga, J. Frece, and S. Matosić. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94:981-987. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen, H. D., Q. A. Nguyen, R. C. Ferreira, L. C. Ferreira, L. T. Tran, and W. Schumann. 2005. Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid 54:241-248. [DOI] [PubMed] [Google Scholar]
- 20.Prado Acosta, M., E. Valdman, F. Battaglini, S. G. F. Leite, and S. M. Ruzal. 2005. Biosorption of copper by Paenibacillus polymyxa cells and their exopolysaccharide. World J. Microbiol. Biotechnol. 21:1157-1163. [Google Scholar]
- 21.Resta-Lenert, S., and K. E. Barrett. 2003. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 52:988-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruzal, S. M., and C. Sánchez Rivas. 1998. In Bacillus subtilis DegU-P is a positive regulator of the osmotic response. Curr. Microbiol. 37:368-372. [DOI] [PubMed] [Google Scholar]
- 23.Smit, E., and P. H. Pouwels. 2002. One repeat of the cell wall binding domain is sufficient for anchoring the Lactobacillus acidophilus surface layer protein. J. Bacteriol. 184:4617-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smit, E., D. Jager, B. Martinez, F. J. Tielen, and P. H. Pouwels. 2002. Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein-protein interaction of two subdomains. J. Mol. Biol. 324:953-964. [DOI] [PubMed] [Google Scholar]
- 25.Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwels. 2001. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305:245-257. [DOI] [PubMed] [Google Scholar]
- 26.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valence, F., and S. Lortal. 1995. Zymogram and preliminary characterization of Lactobacillus helveticus autolysins. Appl. Environ. Microbiol. 61:3391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vollmer, W., B. Joris, P. Charlier, and S. Foster. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259-286. [DOI] [PubMed] [Google Scholar]
- 29.Yokoi, K. J., K. Kawasaki, A. Taketo, and K. Kodaira. 2004. Characterization of lytic enzyme activities of Lactobacillus gasseri with special reference to autolysis. Int. J. Food Microbiol. 96:273-279. [DOI] [PubMed] [Google Scholar]