Summary
gp130 is a shared receptor for at least nine cytokines, and can signal either as a homodimer, or as a heterodimer with Leukemia Inhibitory Factor Receptor (LIF-R). Here we biophysically and structurally characterize the full-length, transmembrane form of a quaternary cytokine receptor complex consisting of gp130, LIF-R, the cytokine Ciliary Neurotrophic Factor (CNTF), and its alpha receptor (CNTF-Rα). Thermodynamic analysis indicates that, unlike the cooperative assembly of the symmetric gp130/Interleukin-6/IL-6Rα hexameric complex, CNTF/CNTF-Rα heterodimerizes gp130 and LIF-R via non-cooperative energetics to form an asymmetric 1:1:1:1 complex. Single particle electron microscopic (EM) analysis of the full-length gp130/LIF-R/CNTF-Rα/CNTF quaternary complex elucidates an asymmetric structural arrangement, in which the receptor extracellular and transmembrane segments join as a continuous, rigid unit, poised to sensitively transduce ligand engagement to the membrane-proximal intracellular signaling regions. These studies also enumerate the organizing principles for assembly of the ‘tall’ class of gp130-family cytokine receptor complexes including LIF, IL-27, IL-12, and others.
Introduction
Cytokines and their receptors are essential to many aspects of cell growth and differentiation, across the immune, hematopoietic and nervous systems. Members of the short (Interleukin-2 type) and long chain (Interleukin-6 type) cytokine families exhibit a characteristic four-helix bundle fold and engage type I cytokine receptors, all of which contain a tandem pair of Fibronectin-III (FnIII) domains, known as “cytokine-binding homology regions” (CHR) (Boulanger and Garcia, 2004). The basic mechanistic paradigm for receptor activation is that the cytokine engages CHR domains, resulting in dimerization of the receptor ectodomains (de Vos et al., 1992). Cytokine receptor intracellular domains are constitutively bound to Janus kinases (JAKs) (Leonard and O'Shea, 1998) which, when brought into the proper orientation and proximity by receptor dimerization, trans-phosphorylate one another as well as the receptor (Heinrich et al., 2003). These phosphorylated receptor tyrosines then serve as docking sites for latent STAT (signal transducer and activator of transcription) molecules (Leonard and O'Shea, 1998), which, after being phosphorylated by JAKs, oligomerize and translocate to the nucleus to act as transcription factors for a range of cytokine-responsive genes.
gp130 is the founding member of the “IL-6/IL-12” family of tall receptors that includes Leukemia Inhibitory Factor Receptor (LIF-R), IL-12Rβ1, IL-12Rβ2, Granulocyte Colony-Stimulating Factor Receptor (GCSF-R), and Oncostatin-M Receptor (OSM-R), and serves as a shared signal transducing subunit for IL-6, IL-11, Leukemia Inhibitory Factor (LIF), Oncostatin-M (OSM), Cilliary Neutrophic Factor (CNTF), Cardiotrophin-1 (CT-1), Cardiotrophin-like cytokine (CLC), and IL-27 (Benigni et al., 1996; Hibi et al., 1990; Pflanz et al., 2004; Senaldi et al., 1999). The biological roles of gp130 cytokines are highly diverse, ranging from immune regulation (IL-6), to neural growth (CNTF), maintenance of stem cell pluripotency (LIF), and cardiovascular functions (CT-1) (Nicola and Hilton, 1998).
Some “gp130-cytokines” homodimerize gp130 (IL-6 and IL-11), while a second class of gp130-cytokines (CNTF, LIF, OSM, CLC, CT-1) heterodimerize gp130 with the shared receptor LIF-R. (Heinrich et al., 2003). These “tall” cytokine receptors contain an additional three membrane-proximal FnIII domains in comparison to other type I receptors, and these domains are necessary for signaling (Hammacher et al., 2000; Kurth et al., 2000). The tall receptors require both the conserved two-domain CHR and a top-mounted Ig-domain for activation, in contrast to the short receptors (e.g. hGH, EPO, IL-2) which only require the CHR. In keeping with the increased structural complexity of their receptors, the cytokine ligands for the tall receptors have three receptor binding epitopes, compared to two for most other type I cytokines (Bravo and Heath, 2000). A crystal structure of the top three domains of gp130 in complex with IL-6 and IL-6Rα revealed that the site 1 and site 2 of the cytokines engaged IL-6Rα and gp130 in a manner analogous to the hGH/EPO paradigm, but that the site 3 engaged the Ig-domain to bridge two heterotrimeric complexes into a dyad-symmetric hexamer (Boulanger et al., 2003b). This structure raised the question of how a heterodimeric gp130/LIF-R signaling complex can be formed, since it would seem to require breaking the dyad symmetry necessary for receptor dimerization. Recently, the structure of a binary complex of the top five domains of LIF-R in complex with LIF revealed an interaction analogous to the site 3 (Huyton et al., 2007), as seen in the IL-6 hexamer (Boulanger et al., 2003b), and together with a prior structure of LIF bound to gp130 (Boulanger et al., 2003a), the ternary signaling complex was modeled as an asymmetric structure (Huyton et al., 2007). However, direct structural evidence for an asymmetric gp130/LIF-R signaling complex is lacking.
The structural mechanisms for how cytokine recognition is coupled to JAK and STAT activation also remain enigmatic. Currently no structural information exists for full-length cytokine receptors containing transmembrane (TM) or intracellular domains (ICDs), nor any structural information on cytokine receptor ICDs alone. Precise receptor subunit conformations and complex architectures are apparently necessary for activation of intracellular signaling. For example, gp130 activation requires two different anti-gp130 monoclonal antibodies delivered in tandem, presumably to both dimerize the receptor and induce the appropriate signaling conformation(s) (Autissier et al., 1998; Muller-Newen et al., 2000). In the Growth Hormone (hGH) and Erythropoetin (EPO) receptor systems, the previous assumption that simple receptor homodimerization was sufficient for activation has now been superceded by a body of data showing that these receptors exist as preformed, quiescent dimers in the membrane, and the role of ligand is to precisely re-orient the dimer (Brown et al., 2005; Constantinescu et al., 2001b; Livnah et al., 1999). EPO receptor complexes induced by agonist peptides have also shown that the receptor-receptor orientation angles varied in accord with ligand potency (Livnah et al., 1998; Livnah et al., 1996). In the hGH, EPO and gp130 systems, insertion of alanine spacer residues into the membrane proximal ICD regions revealed an apparent pitch-dependence of intracellular phosphorylation consistent with a rigid α-helix (Brown et al., 2005; Constantinescu et al., 2001a; Greiser et al., 2002), highlighting an intimate structural communication between the extracellular cytokine complex and the ICDs bound to JAKs.
With regards to tall receptor activation, a single particle EM reconstruction of the hexameric gp130/IL-6Rα/IL-6 complex containing the entire six-domain gp130 extracellular domain (ECD) revealed the gp130 D4-D6 FnIII ‘legs’ exhibited significant conformational variation (Skiniotis et al., 2005). A majority of the particles exhibited legs that bent towards one another at the level the receptor would enter the membrane, presumably reflecting the ‘active’ state of the receptor, in which the TM and ICDs would be in close proximity. The structural variability of the gp130 legs has also been suggested by another EM study of the gp130/IL-11 hexameric complex (Matadeen et al., 2007). These studies were both conducted using soluble ectodomain complexes, so direct structural data on the full-length receptors remains an important goal for visualizing the bona fide receptor signaling complex architecture.
In the present study we address the issues of both gp130/LIF-R assembly and the resulting full-length signaling complex architecture. We find that, in contrast to the homodimeric gp130 signaling complexes, the gp130/LIF-R/CNTF-Rα/CNTF complex is asymmetric (1:1:1:1), and gp130 and LIF-R heterodimerize in a non-cooperative fashion. The structure of this asymmetric assembly is resolved using a combination of X-ray crystallography and EM, resulting in visualization of the overall architecture of the full-length quaternary receptor complex containing TM and ICD regions. We find that the full-length receptor complex appears to lack segmental flexibility between the ECD, TM and ICD regions, and speculate that this rigidity may serve to sensitively transmit ligand-induced conformational changes across the membrane to the ICDs, which appear unstructured in the absence of associated JAK molecules.
Results
Assembly of the quaternary gp130/LIF-R/CNTF-Rα/CNTF complex
We first sought to delineate the assembly pathway and stoichiometry of a gp130/LIF-R heterodimeric ectodomain complex. Prior biochemical and structural studies showed that IL-6, IL-6Rα and gp130 cooperatively assemble into a hexameric signaling complex (Boulanger et al., 2003b). In this sequence, IL-6 first engages IL-6Rα through site 1 to form a binary complex, followed by low affinity recruitment of the gp130 CHR through a composite site 2 to form a non-signaling ternary complex. The final step, a high affinity dimerization of two gp130/IL-6Rα/IL-6 heterotrimers via site 3 is dependent on prior engagement of site 1 by the IL-6Rα, and site 2 by the gp130 CHR. In this fashion, the site 3 interaction, which has undetectably low affinity for gp130 IgD alone, is stabilized by the avidity afforded by the “two-point attachment” between opposing, antiparallel gp130 trimeric complexes.
In order to carry out an analogous study on assembly of the CNTF receptor complex, which is known to require gp130/LIF-R heterodimerization, we expressed a range of soluble ligand-binding fragments of CNTF, CNTF-Rα, gp130 and LIF-R (Figure S1). For both gp130 and CNTF-Rα we expressed both CHR-only (D2D3) versions and D1D2D3 versions to assess the role of the Ig-domains in complex assembly. LIF-R differs from gp130 in that it contains an additional two-domain CHR on “top” (D1D2) while the IgD, thought to engage site 3 across the tall family, resides at the D3 position (see Figure 1A). Therefore we expressed a five-domain version of LIF-R containing both CHRs (D1D2 and D4D5) and the IgD (D3). Using these proteins, we carried out a series of binary, ternary, and quaternary titrations to form the quaternary complex. For the purposes of this discussion, we do not elaborate on the specifics of the thermodynamics of the various interactions, but instead focus on the stoichiometries and binding affinities of different combinations of cytokine and receptors.
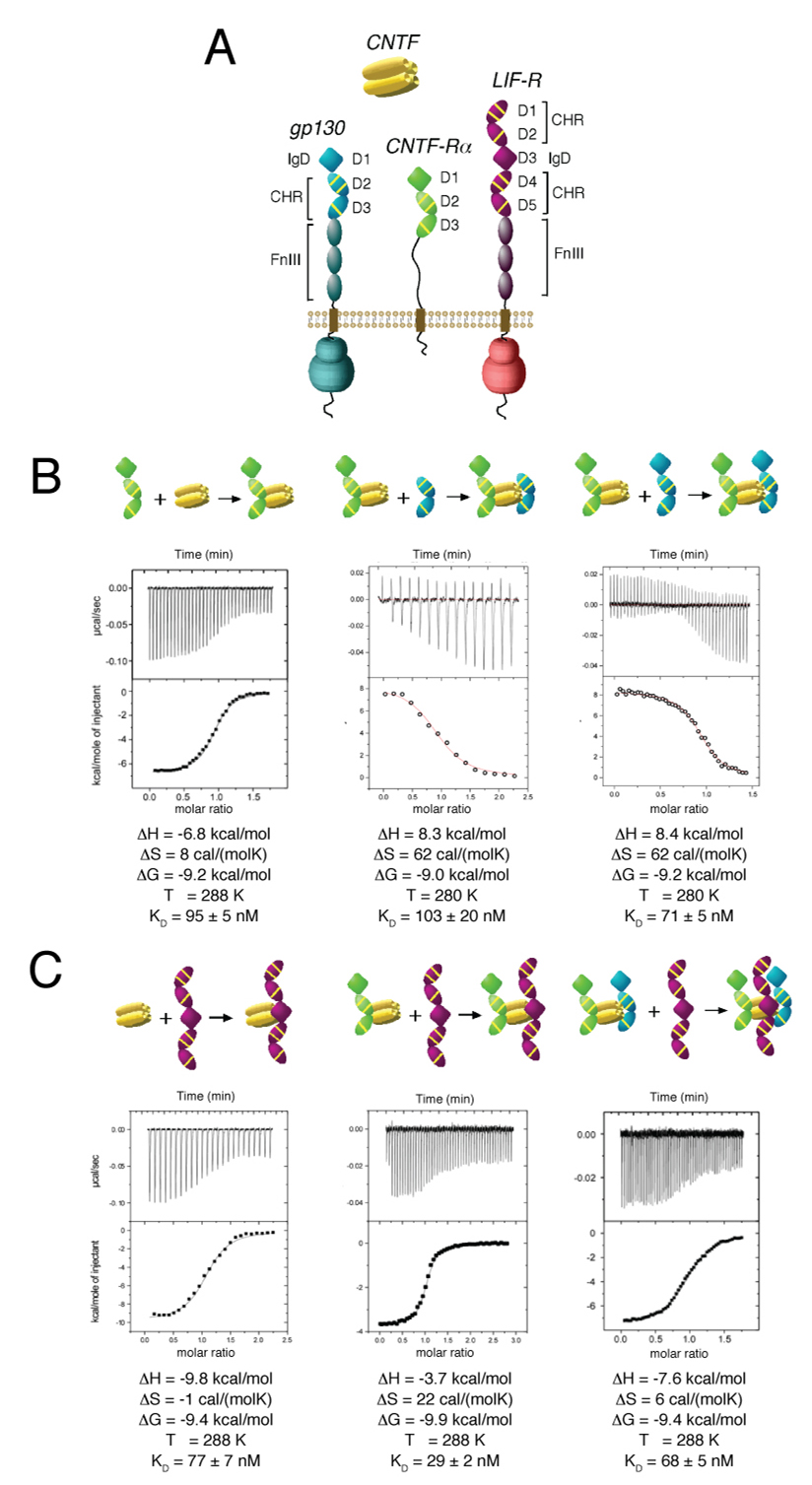
Figure 1. Thermodynamic analysis of the assembly of the gp130/LIF-R/CNTF-Rα/CNTF complex.
(A) Schematic representation of gp130, LIF-R, CNTF-Rα, and CNTF. IgD denotes “Ig-like domain”, while CHR denotes “cytokine-binding homology region” and FnIII denotes “Fibronectin-type III domain”. Each CHR contains three yellow lines to represent conserved disulfides and a WSXWS motif. (B) Isothermal titration calorimetry illustrating the assembly of CNTF and CNTF-Rα, followed by assembly of the CNTF-Rα/CNTF binary complex with gp130 minus or plus the D1 IgD. (C) Titration of LIF-R with CNTF, the CNTF-Rα/CNTF binary complex, and the gp130/CNTF-Rα/CNTF ternary complex. Inset tables list the thermodynamic parameters for the representative binding isotherms.
Similar to the gp130/IL-6Rα/IL-6 complex, we find that the two and three-domain (minus and plus IgD, respectively) versions of CNTF-Rα bind to the CNTF site 1 with roughly equal affinities, and that CNTF requires pre-complexation with CNTF-Rα to then engage gp130 CHR via site 2 (Figure 1B, and data not shown). However, in contrast to the IL-6 system, we find that the CNTF/CNTF-Rα complex binds to a two-domain gp130 fragment (CHR only) with similar affinity and stoichiometry as the three-domain gp130 fragment (IgD plus CHR) (Figure 1B). In the IL-6 system, inclusion of the gp130 IgD results in a two orders-of-magnitude affinity increase and conversion of the heterotrimeric complex to a hexamer (Boulanger et al., 2003b). Size exclusion chromatography also indicates that both gp130 constructs form a 1:1:1 ternary complex with CNTF and CNTF-Rα (Figure S1). Thus, the IgD of gp130, which engages the site 3 of IL-6 in the symmetric hexamer, does not appear to participate in a site 3 interaction with CNTF. Strikingly, titration of either CNTF, CNTF/CNTF-Rα or CNTF/CNTF-Rα/gp130 with LIF-R all showed similar binding affinities and 1:1, 2:1, and 3:1 stoichiometries, respectively, albeit with small deviations (Figure 1C and Figure S1). Thus, pre-complexation of CNTF sites 1 and 2 with CNTF-Rα and gp130, respectively, does not appear to be a prerequisite for LIF-R binding, and CNTF is competent to bind LIF-R either alone, as a binary (CNTF/CNTF-Rα), or a ternary (gp130/CNTF/CNTF-Rα) complex. Collectively, we find that CNTF can form a quaternary complex (1:1:1:1) through several redundant pathways and that recruitment of LIF-R occurs via a non-cooperative high-affinity site 3 interaction with CNTF independent of gp130 and CNTF-Rα. We surmise that this non-cooperativity is a result of the high affinity of the site 3 CNTF/LIF-R contact as an independent binary interaction that is unperturbed by the other receptors, as has been shown for LIF/LIF-R (Boulanger et al., 2003a; Huyton et al., 2007). The implications of this are that CNTF, as well as other cytokines that likely heterodimerize LIF-R/gp130 into an asymmetric signaling complex (e.g. OSM, LIF, CLC, CT-1), do not require the two-point attachment (i.e. “doubling”) to stabilize a weak site 3/gp130 interaction that is necessary for IL-6 and IL-11 mediated homodimerization of gp130. Instead, the high affinity site 3 in the gp130/LIF-R cytokines negates the need for avidity enhancement through homodimerization.
Structure of unliganded LIF-R
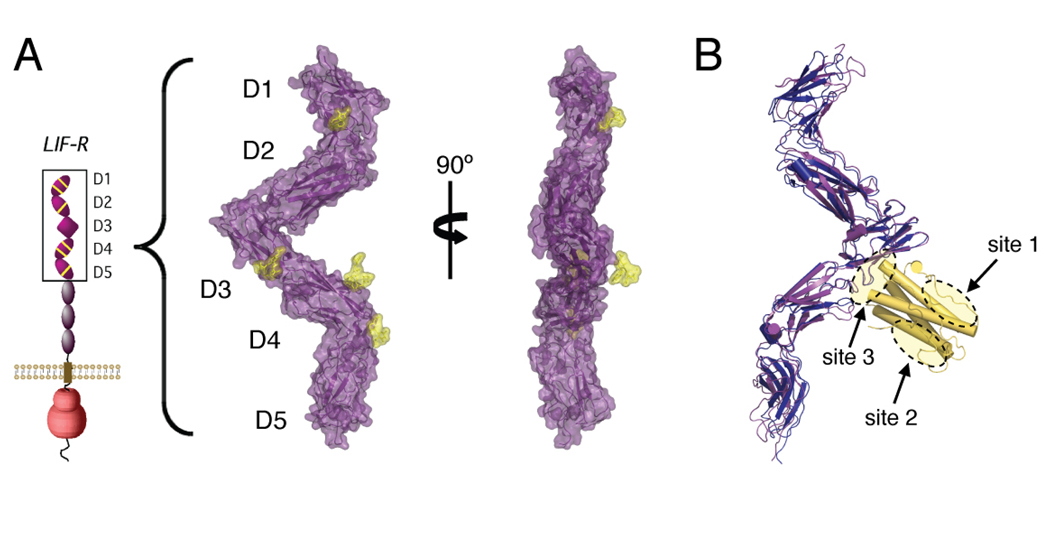
To investigate the structure of the cytokine-binding region of LIF-R, we crystallized a partially glycosylated (see Experimental Procedures) version of domains D1 to D5 of LIF-R, expressed in insect cells, and solved the structure at a resolution of 3.1Å (Table S1). Unliganded human LIF-R adopts an extended ‘flying V’ structure ~150Å long (Figure 2A), similar to that seen in the 4.0Å structure of the mouse LIF/LIF-R complex (Huyton et al., 2007). Overall, the D1D2 and D4D5 CHR modules adopt the canonical bent elbow shape commonly seen in other cytokine receptors, while the D3 Ig domain is positioned in the middle at the base of the central ‘V’. Overlay of our LIF-R structure with the mouse LIF-R using the D3 IgD as an anchor point shows very slight differences in overall geometry (Figure 2B), and superimposition of the cytokine-binding D3 Ig domains results in an rmsd for α carbon atoms of 0.72 Å. An interdomain disulfide bond between Cys194 and Cys251 fixes the D2 at nearly a right angle to the D3 Ig domain. This disulfide bond fixes the top CHR to extend up and away from the binding site of a potentially side-on cytokine α-receptor subunit, possibly preventing D1-D2 of LIF-R from sterically blocking complex formation with the cytokine/α-receptor. A comparison of the D3 Ig domain ligand-binding interfaces of unliganded human LIF-R and mouse LIF-R bound to human LIF (Huyton et al., 2007) shows sidechain positions that are remarkably similar (Figure S3), indicating that only minor structural rearrangement occurs in the LIF-R binding interface in response to ligand binding.
Figure 2. Structure of unliganded LIF-R and comparison with the LIF-R/LIF complex.
(A) Surface representations of unliganded human LIF-R D1-D5. Sugar residues modeled into the structure are colored yellow. (B) Overlay of human LIF-R onto mouse LIF-R/LIF structure. The D3 Ig domain of human LIF-R (purple) was aligned with the D3 Ig domain of mouse LIF-R (blue), with the cytokine LIF shown in gold. Receptor interaction sites 1, 2, and 3 on LIF-R are circled.
Purification and 2D projection analysis of the gp130/LIF-R/CNTF-Rα/CNTF complex
In order to investigate the architecture of the extracellular domains of the gp130/LIF-R/CNTF-Rα/CNTF complex as well as the overall architecture of a full-length cytokine/receptor complex containing TM and intracellular regions, we co-expressed full-length gp130 and LIF-R in insect cells in the presence of a single-chain fusion consisting of domains 1 to 3 of CNTF-Rα linked to CNTF (‘hyper-CNTF’) (Marz et al., 2002). The quaternary receptor complex was purified as an intact membrane protein in n-dodecyl-β-D-maltoside (DDM) using a monoclonal antibody anti-peptide affinity column followed by size exclusion chromatography to isolate monodisperse receptor complexes for electron microscopic (EM) imaging (Figure 3A). In order to directly visualize the full-length gp130/LIF-R/CNTF-Rα/CNTF assembly, we recorded EM images of the complex in negative stain. The EM images revealed mono-disperse particles with variable sizes and shapes (Figure S4). The differences in shape of the particles could represent different orientations of the complexes on the carbon film, but could also reflect variability in the configurations adopted by the individual subunits. As all three receptors are composed of contiguous modules of Ig-like and FnIII domains, structural variability was expected at the inter-domain boundaries as seen, for example, in a VEGF receptor complex (Ruch et al., 2007). To analyze the level of structural heterogeneity due to the flexibility of the receptor chains, we interactively selected 23,648 particles from 83 images and classified them into 400 different conformational classes. The majority of the class averages revealed an elongated and overall asymmetric particle, suggesting that most of the complexes adsorbed to the carbon support film in a single preferred orientation. To improve the alignment, we performed a second multi-reference alignment step using only the 12,711 particles in the classes showing well-defined averages and specifying 90 output averages (Figure S5). As a result of the better quality of the reduced data set and the increased number of particles per class we obtained 2D averages with better definition of the stain-excluding areas representing protein domains.
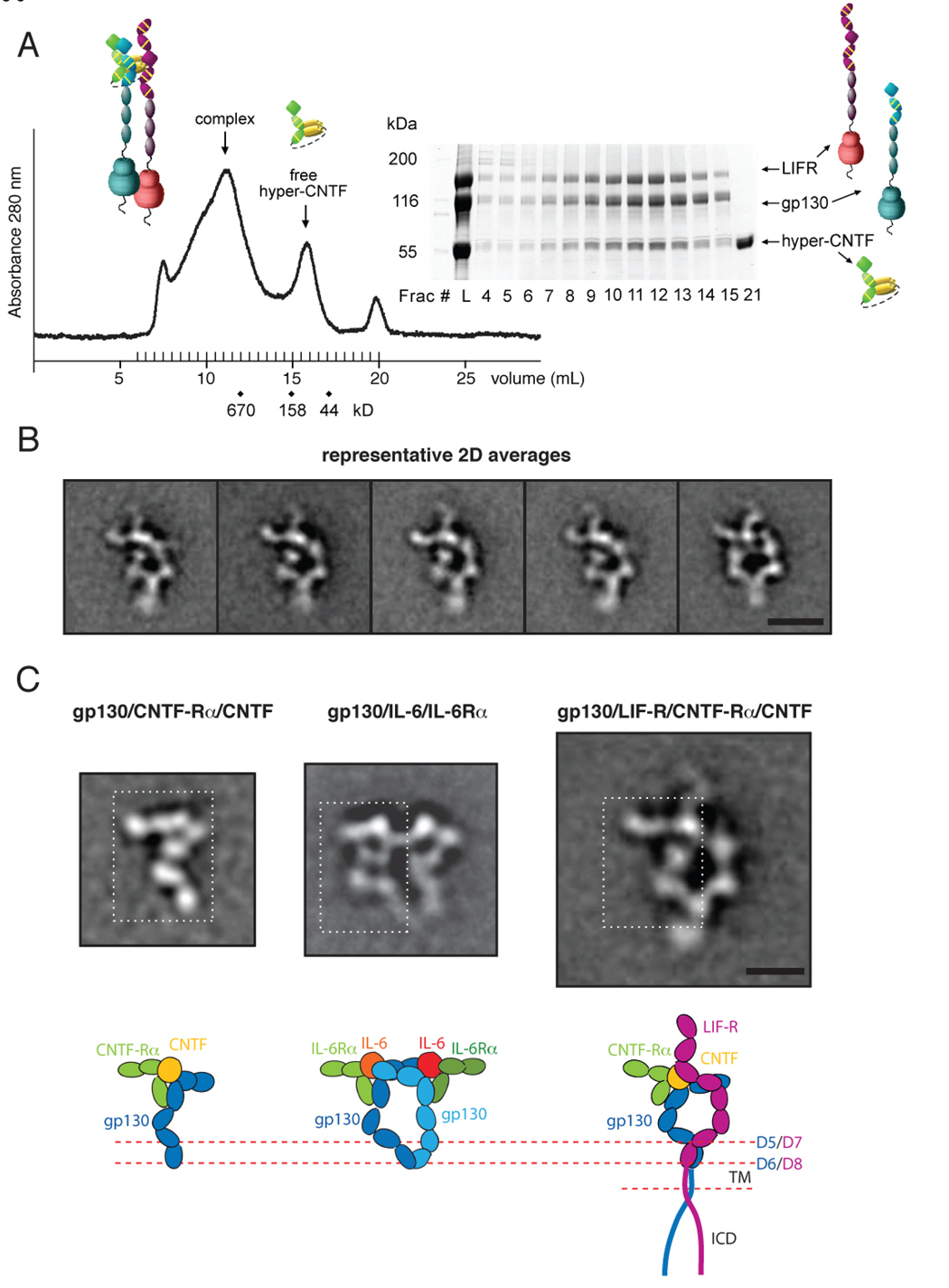
Figure 3. Purification and 2D EM averages of the full-length gp130/LIF-R/CNTF-Rα/CNTF complex.
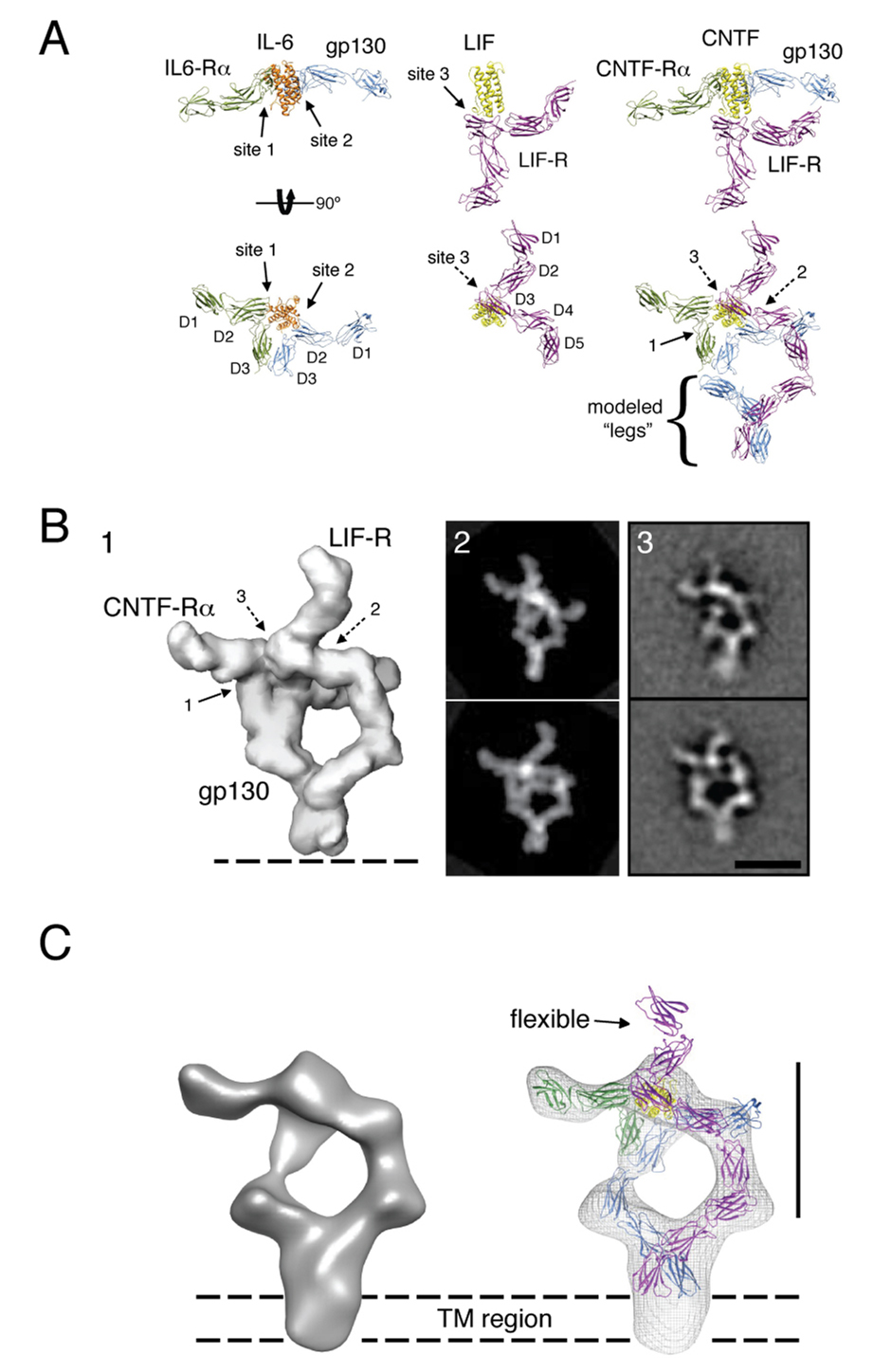
(A) gp130/LIF-R/hyper-CNTF complex was purified by size exclusion chromatography on a Superose 6 column equilibrated in 0.02% DDM, and fractions analyzed by SDS-PAGE. Fraction 12 corresponding to the UV absorbance peak at ~11.5 mL were subsequently used for EM imaging. (B) Representative 2D averages of negatively stained gp130/LIF-R/hyper-CNTF particles. The scale bar equals 20 nm. (C) Comparison of 2D averages and domain organization between the extracellular assembly of gp130/ hyper-CNTF sub-complex (left), the hexameric gp130/IL-6/IL-6-Rα complex (middle) and the tetrameric full-length gp130/LIF-R/hyper-CNTF complex (right). The white dotted rectangles indicate the regions of similarity between the gp130/ hyper-CNTF sub-complex with either side of the symmetric gp130 homodimeric complex and the left side of the gp130/LIF-R heterodimeric complex. This similarity suggests that the gp130/CNTF-Rα/CNTF sub-complex constitutes the left side of the gp130/LIF-R complex, with the density projecting to the left corresponding to the D1-D2 domains of CNTF-Rα. The legs of the gp130/LIF-R heterodimer appear to be joining at a higher level than the legs in the homodimeric gp130 complex. Below the fuzzy TM region of the heterodimer, no distinct protein density is observed. The scale bar equals 10 nm.
The class averages of the gp130/LIF-R/CNTF-Rα/CNTF complex reveal the same view of an asymmetric assembly with a central stain accumulation, suggesting a separation of the receptor chains at this location (Figure 3B and Figure S5). The ‘lower part’ of the complex is formed by two quasi-symmetric rod-like protein densities, which join together at the bottom, ending in a region that often appears fuzzy or smeared, which presumably represents the structural heterogeneity of the ICDs as they emanate from the TM boundary. The ‘top part’ of the assembly appears asymmetric and more convoluted. Two protein densities appear to be joining at the top-left of the complex, from where two elongated protrusions emanate, one towards the left and the other upwards (Figure 3B). All class averages with well-defined features showed the complex in the same conformation. While some of the less well-defined averages may potentially result from complexes in different conformations, none of these conformations were sufficiently populated to produce an average with clear structural features, suggesting that there is only a single ‘stable’ conformation of the complex.
To interpret the projection averages and to assign individual densities to the different protein subunits in the gp130/LIF-R/CNTF-Rα/CNTF assembly, we prepared (Fig. S6) and calculated 2D averages of a ternary sub-complex of gp130 with CNTF-Rα/CNTF (Figure 3C, left, and Figure S6) missing LIF-R, for comparison to the full quaternary complex containing LIF-R. The homology of CNTF with IL-6, CNTF-Rα with IL-6Rα, and gp130 with LIF-R allows us to compare the 2D average of the gp130/CNTF-Rα/CNTF sub-complex not only with those of the gp130/LIF-R/CNTF-Rα/CNTF assembly but also with those of the extracellular gp130/IL-6Rα/IL-6 complex (Skiniotis et al., 2005). The structure of the gp130/CNTF-Rα/CNTF sub-complex should resemble half the structure of the symmetric hexamer of the gp130/IL-6Rα/IL-6 complex (Figure 3C, middle) and also part of the asymmetric gp130/LIF-R/CNTF-Rα/CNTF complex (Figure 3C, right). Indeed, the gp130/CNTF-Rα/CNTF sub-complex appears similar to both sides of the symmetric gp130/IL-6Rα/IL-6 complex (Figure 3C; white rectangles) and also to the left side of the gp130/LIF-R/CNTF-Rα/CNTF assembly, except for the density projecting from the top (see below). Consequently, the right side of the average of the gp130/LIF-R/CNTF-Rα/CNTF assembly represents LIF-R. LIF-R appears to interact with CNTF at the level of the D3 (IgD), corresponding to the point of the ‘flying V’ seen in the LIF-R crystal structure (Figure 2), and consistent with the crystal structure of the LIF/LIF-R complex (Huyton et al., 2007). The unengaged density projecting from the top of the LIF/LIF-R interaction site corresponds to LIF-R domains D1 and D2 that constitute the N-terminal CHR, which is not present in gp130.
At the bottom half of the assembly, the FnIII legs of both gp130 (D4-D6) and LIF-R (D6-D8) bend towards each other until they come together, and further continue downwards as a single density. The legs of the full-length gp130/LIF-R heterodimer unambiguously join at the level of gp130 D5/LIF-R D7 in all averages, in contrast to the gp130 homodimer that was found to interact mostly at the level of D6 (Figure 3C, middle and (Skiniotis et al., 2005)). Importantly, we found a significant amount of heterogeneity in the positioning of the FnIII ‘leg’ domains of gp130 in the gp130/CNTF-Rα/CNTF averages (Fig S6), indicating that heterodimerization with LIF-R stabilizes the conformations of the membrane proximal domains of gp130. Below the densities representing the D6 domain of gp130 and the D8 domain of LIF-R, at the level of the TM and ICDs, the protein regions appear fuzzy, with no discernable or definable protein density observed. Given the close association of the membrane-proximal FnIII domains of gp130 and LIF-R, their neighboring TMs may be associated; however, this remains uncertain at the resolution of these projection averages. The lack of clear density C-terminal to the TM regions suggests that the intracellular part of gp130 and LIF-R either exist as largely unstructured polypeptides or are too flexible to produce a signal in the averages. However, the uniformity of the positions of the ‘legs + TM + ICD’ unit seen in the negative stain strongly suggests conformational rigidity and a lack of segmental flexibility in the hinges connecting these regions.
NMR structural characterization of the gp130 intracellular region
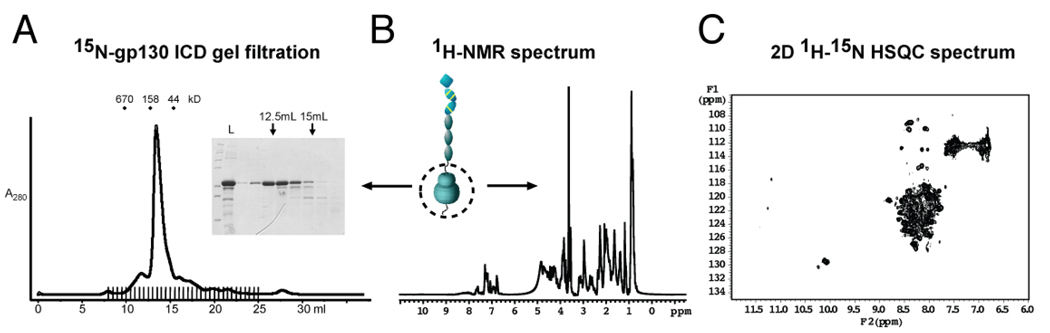
To investigate whether the intracellular regions of the gp130/LIF-R heterodimer are unstructured or too flexible to be visible in the EM averages obtained, we expressed and purified the intracellular domain (ICD) of gp130 from E. coli and collected 1H and 1H-15N HSQC (Heteronuclear Single Quantum Coherence) NMR spectra to analyze its structure. In the 1D spectrum (Figure 4B), very little chemical dispersion of the aliphatic and amide protons was observed, and no up-shifted methyls are present which would indicate the presence of a stable hydrophobic core. The 2D-HSQC spectrum shows little dispersion in both the proton and nitrogen dimensions (Figure 4C), indicating the amide groups of the peptide backbone are disordered. Additionally, the 34kD purified gp130 ICD migrated on a size exclusion column at an apparent size of ~100kD (Figure 4A) and is extremely sensitive to trypsin and carboxypeptidase digestion (data not shown), indicating that the ICD may exist in a loosely packed, molten globule-like state. Based on these results, it appears the gp130 ICD is unstructured in the absence of bound signaling proteins, consistent with the lack of distinct protein densities in the EM averages. It is possible that the gp130 ICD may assume more structure when paired with the LIF-R ICD, but we think this is unlikely given that the gp130 ICD alone is present in the IL-6 and IL-11 gp130 homodimeric signaling complexes. This suggests that the isolated fragment studied here can be extrapolated to the heterodimeric situation. Similar to gp130, the ICD of LIF-R is predicted to be random coil in ~70–80% of its sequence.
Figure 4. NMR analysis of the intracellular domain of gp130 reveals a flexible state.
(A) 15N-labelled gp130 intracellular domain (amino acids 642-918, 34 kDa) exhibited monodisperse behavior and eluted at an approximate size of 100 kDa from a size exclusion column. Fractions were analyzed by SDS-PAGE (inset). (B) 1H-NMR spectrum and (C) 1H-15N HSQC spectrum of the gp130 intracellular domain.
Molecular modeling and 3D reconstruction of the gp130/LIF-R/CNTF-Rα/CNTF complex
To obtain a comprehensive understanding of the architecture of the entire full-length gp130/LIF-R/CNTF-Rα/CNTF complex, we used crystal structures of corresponding actual or homologous proteins and complexes, together with our calorimetry data, EM 2D class averages and 3D reconstruction to model the ligand/receptor assembly. For this strategy, we first constructed an atomic model of the gp130/LIF-R/CNTF-Rα/CNTF ectodomain complex using components of several crystal structures (Figure 5A), informed by the site 1, 2 and 3 assembly data from the ITC experiments. Next, we created a ‘density volume’ of this model for which projections were calculated and compared to our actual EM data. Finally, we calculated a 3D reconstruction of the full-length gp130/LIF-R/CNTF-Rα/CNTF complex, which was used to validate and refine our model.
Figure 5. Architecture of the gp130/LIF-R/CNTF-Rα/CNTF complex.
(A) The 3D structure of the ectodomain ‘headpiece’ of the gp130/LIF-R/CNTF-Rα/CNTF complex was modeled based on the crystal structures of gp130/IL-6Rα/IL-6 and the LIF-R/LIF complexes. The gp130 and LIF-R ‘legs’ (D4-D6 and D6-D8, respectively) were modeled according to the densities observed in the 2D EM averages as shown in Figure 3C (gp130: blue, IL-6Rα: green, IL-6: orange, LIF-R: magenta, LIF: yellow, CNTF-Rα: green, CNTF̣: yellow). (B) The model of the full extracellular gp130/LIFR/CNTF-Rα/CNTF complex was filtered to 30Å resolution (panel 1), and was subsequently used to generate projections at regular angular intervals. The projections (panel 2) most similar to the experimental 2D averages were identified by cross-correlation against two of the most representative class averages (panel 3). (C) 3D reconstruction of the gp130/LIF-R/CNTF-Rα/CNTF complex and fit crystal structures. The 3D map shows clearly density corresponding to the transmembrane region of gp130/LIFR below the level of the C-terminal extracellular domains. The scale bars in B and C are 20 nm.
To model the gp130(D1D2D3)/CNTF-Rα (D1D2D3)/CNTF ternary complex module we used the crystallographic coordinates of the gp130/IL-6Rα/IL-6 ternary complex (Boulanger et al., 2003b) (Figure 5A). The LIF-R model was derived from the crystal structure here and the structure of LIF-R/LIF (Huyton et al., 2007). Based on our calorimetry experiments (Figure 1), and the recently determined crystal structure of LIF-R in complex with LIF (Huyton et al., 2007), the IgD (D3) domain of LIF-R interacts directly with site 3 of CNTF, as shown in Figure 5A. In contrast, the IgD (D1) domain of gp130 does not engage in complex formation, as also indicated by the 2D averages of the complex (Figure 3B). Therefore, by aligning the LIF coordinates obtained from the LIF-R/LIF structure to the IL-6 coordinates in the gp130/IL-6Rα/IL-6 structure we obtained an approximation of the gp130/LIF-R/CNTF-Rα/CNTF membrane-distal region (i.e. ‘the headpiece’). To better match the resulting model with the densities shown in our 2D projections we slightly adjusted the position of LIF-R D4 and D5, which are likely somewhat dynamic based on the flexible hinge between the D3 Ig domain and the D4D5 CHR domain. To model the three membrane-proximal FnIII domains in gp130 and LIF-R, we used coordinates extracted from the structure of the human fibronectin 7–10 fragment (domains 7–9) (Leahy et al., 1996). Using the 2D averages as a guide, the FnIII domains were modeled to approximate the structure for the membrane proximal D6-D8 of LIF-R and D4-6 of gp130. In contrast to our model for the full extracellular assembly of gp130/IL-6Rα/IL-6 (Figure 6A), the FnIII ‘legs’ of LIF-R and gp130 were modeled to engage at the level of the two last C-terminal FnIII domains (D5-D6 for gp130 and D7-D8 for LIF-R), based on the negative stain EM data presented in Figure 3.
Figure 6. The organizing principles of the gp130 family of cytokine/receptor complexes.
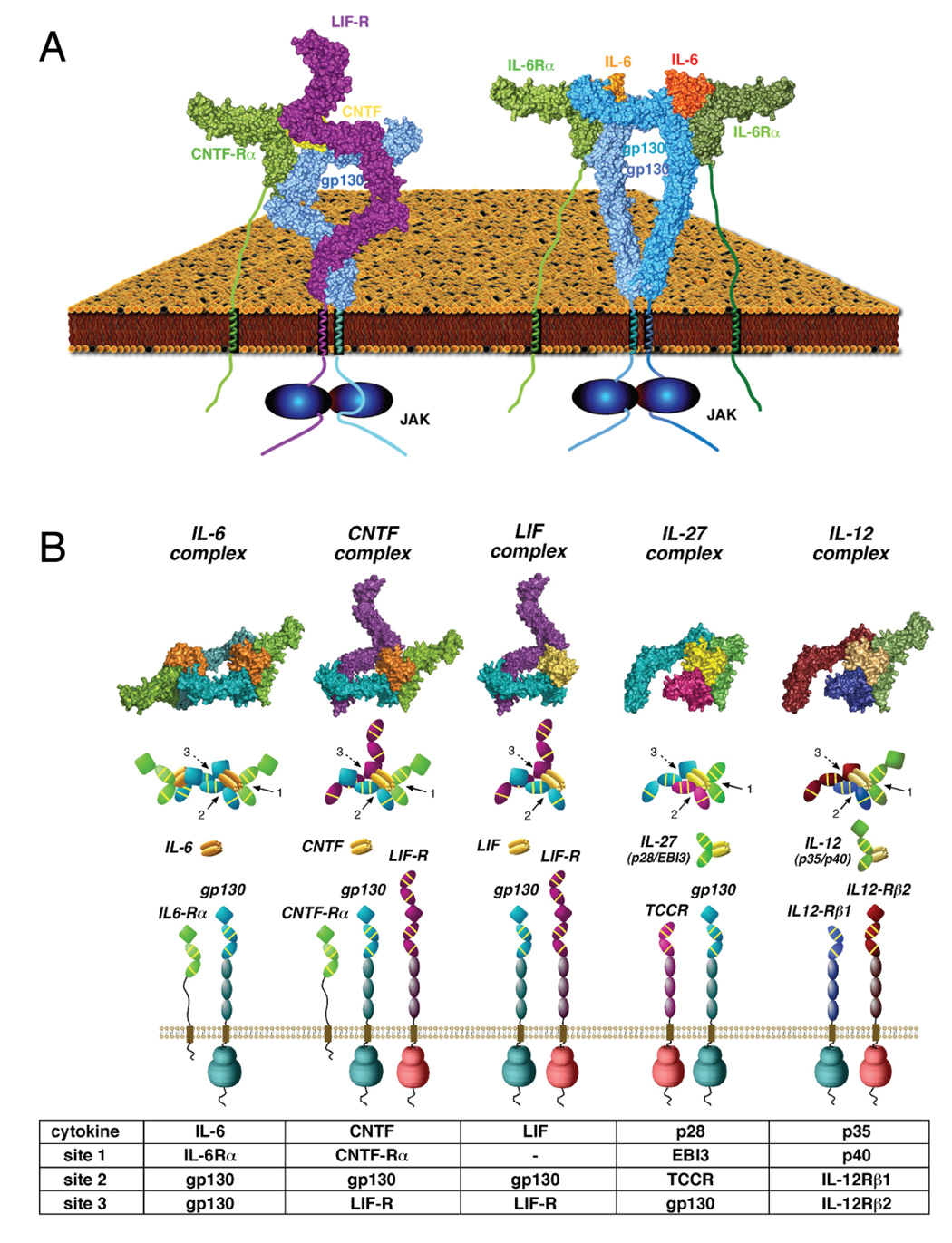
(A) Structural comparison of homodimeric and heterodimeric gp130 receptor complexes. The structure to the left is the 1:1:1:1 gp130/LIF-R/CNTF-Rα/CNTF complex, and the image to the right is the 2:2:2 gp130/IL-6Rα/IL-6 complex (Skiniotis et al., 2005). In both cases, the bending of the ‘legs’ allows gp130 and LIF-R to form intimate receptor-receptor contact prior to entering the cell membrane, enforcing a close apposition of the receptor dimer. (B) Models were constructed for the cytokine-binding regions of representative classes of the gp130 ‘tall’ receptor family. gp130 homodimerization induced by IL-6 and IL-11 is represented by the structure of the gp130/IL-6Rα/IL-6 hexamer (panel 1). CNTF-induced heterodimerization of gp130 and LIF-R requires CNTF-Rα (which interacts with site 1), while LIF can heterodimerize gp130 and LIF-R in the absence of an alpha receptor. For IL-27, gp130 heterodimerizes with TCCR/WSX-1 (lacking IgD), with TCCR/WSX-1 binding to site 2, and gp130 interacting with site 3 on the p28 subunit. For the IL-12 cytokine receptor complex (panel 5), IL-12Rβ1 (lacking IgD) interacts with site 2 on the p35 subunit of the IL-12 heterodimer, while IL-12Rβ2 (contains IgD) interacts with site 3. All models were generated in COOT and PYMOL using known structures for gp130/IL-6Rα/IL-6 (PDB ID 1P9M), unliganded IL-6Rα (PDB ID 1N26), LIF-R/LIF (PDB ID 2Q7N), and unliganded LIF-R (PDB ID 3E0G).
To test our chain assignment and modeling, the crystallographic model was converted into a density volume filtered to a resolution of 30 Å (Figure 5B, panel 1), which was then used to generate projections at an angular interval of 5°. The projections most similar to the experimental 2D averages were identified by cross-correlation against two of the most representative class averages that showed slightly different arrangements of the complex (Figure 5B, panel 3). The projections from our model that presented the highest cross-correlation coefficients (Figure 5B, panel 2) were indeed almost identical to the class averages (Figure 5B, compare panels 2 and 3). This result suggests that the small variations in the 2D averages are due to slightly different views of the complex and confirms that our model is a good approximation of the 3D architecture of the gp130/LIF-R/CNTF-Rα/CNTF complex.
To further validate our model, we calculated a 3D reconstruction of the full-length gp130/LIF-R/CNTF-Rα/CNTF complex (Figure 5C and Figure S7) using the random conical tilt approach (Radermacher et al., 1987). The final 3D reconstruction was based on combining a total of 3,654 projections from 60°/0° image tilt pairs. Despite the relatively low nominal resolution of the reconstruction (~35 Å), the 3D map confirms the asymmetric nature of the complex, with the two parallel densities corresponding to the receptor chains joining at the top and bottom of the EM density, and the density of CNTF-Rα projecting perpendicular to the assembly at only one side of the EM map (Figure 5C and Figure S7). Our initial model fit well into the 3D reconstruction, but small-scale adjustments, mainly of the flexible leg domains, further improved the fit. The 3D reconstruction shows only an indication for the two N-terminal domains of LIF-R (D1-D2) that protrude from the top of the headpiece, presumably due to their high flexibility which is also evidenced by their weak density in the 2D projections (Figure 3B). Below the level of the last FnIII domains of gp130 (D6) and LIF-R (D8), the 3D reconstruction shows additional density that represents the transmembrane regions of the receptors, consistent with the 2D class averages.
Discussion
gp130 is the founding member of the family of “tall” cytokine receptors and has the unique ability to signal in response to a diverse group of cytokines. In order to achieve such diversity in signaling inputs while maintaining specificity, gp130 has evolved both a degenerate binding interface (Boulanger et al., 2003a) as well as the ability to homodimerize or heterodimerize with several other members of the tall receptor family, including LIF-R, OSM-R and TCCR/WSX-1. Up to this point, only the architecture of the cytokine binding region of symmetric homodimeric complexes of gp130 with IL-6 or IL-11 have been elucidated (Boulanger et al., 2003b; Chow et al., 2001; Matadeen et al., 2007). The question has remained how gp130 is able to heterodimerize with other receptors when the symmetry, and consequent energetic advantage provided by two site 3 binding epitopes is not present. While structural evidence exists that gp130 engages LIF-R cytokines via the site 2 and LIF-R engages the same cytokines via site 3 (Boulanger et al., 2003a; Huyton et al., 2007), the architecture of a bona fide heterodimeric complex has remained an important unresolved question in the field.
To answer this question, we first examined the binding energetics of the gp130/LIF-R/CNTF-Rα/CNTF complex using calorimetry. We found that only the D2D3 CHR of gp130 is required to engage CNTF-Rα/CNTF complex, indicating gp130 engages site 2 on the cytokine. Secondly, we found that LIF-R could engage CNTF alone, the CNTF-Rα/CNTF dimer, or the trimeric gp130/CNTF-Rα/CNTF complex with similar energetics, indicating that LIF-R engages site 3 on CNTF as a structurally independent binding epitope. We conclude from these studies that in the gp130/LIF-R/CNTF-Rα/CNTF complex, the gp130/CNTF-Rα/CNTF “site 1–2” ternary complex closely resembles the gp130/IL-6Rα/IL-6 ternary complex, but that LIF-R forms a single point of attachment to the ternary complex via the site 3 of CNTF, which is of high enough affinity to stabilize the heterocomplex. The crystal structure of LIF-R combined with the EM 2D averages of the gp130/CNTF-Rα/CNTF sub-complex and the gp130/LIF-R/CNTF-Rα/CNTF complex support the conclusion that this complex is a 1:1:1:1 tetramer. Alignment of the gp130/IL-6Rα/IL-6 trimer with the LIF-R/LIF structure using the cytokine as the anchor point provides a model that corresponds well with both the EM 2D averages and 3D reconstruction of the complex (Figure 5). While there appears to be some flexibility in the extracellular domains of the complex (Figure S5), nearly all averages display the “X” shape adopted by the cytokine binding regions of gp130, LIF-R, and CNTF-Rα.
Organizing principles for assembly of gp130 family receptor complexes
Given the conservation of the locations of the receptor binding epitopes on the cytokines (i.e. sites 1, 2 and 3), and the corresponding functional domains of the tall receptors (i.e. IgD, CHR and membrane proximal FnIII ‘legs’), resolution of the architecture of the gp130/LIF-R/CNTF-Rα/CNTF complex allows us to now model the architectures of all heterodimeric complexes in the ‘gp130 family’ (Figure 6B). The modeling is driven by the presence, or absence, of the IgD on one of the two signaling receptors that engages site 3 of the cytokine, thereby imposing a geometric restraint for assembly of the remainder of the complex. For example, while all of the cytokines contain site 3’s, in most of the heterodimeric receptor complexes, only one of the two signaling receptors contain an IgD to bind to the site 3 (e.g. gp130, IL-12Rβ2) – thus the presence of the Ig-domain ‘marks’ that tall receptor as engaging in a site 3 cytokine contact. With the IgD/site 3 interaction identified (e.g. CNTF/LIF-R, LIF/LIF-R, p28/gp130, p35/IL-12Rβ1), the cytokine site 1 is left to then engage a non-signaling alpha receptor (e.g. CNTF/CNTF-Rα, p28/EBI3, p35/p40, LIF has no alpha receptor) in the stereotypical growth hormone-type side-on site 1 fashion, leaving site 2 to engage the CHR of the second signaling receptor (lacking the IgD) (e.g. CNTF/gp130, LIF/gp130, p28/TCCR, p35/IL-12Rβ1). Sites 2 and/or 3 can be utilized by gp130 to generate both homodimeric complexes (represented by IL-6, IL-11, viral IL-6) as well as two types of heterodimeric signaling complexes (represented by LIF, CNTF and IL-27). In the case of IL-27 (p28/EBI3) and IL-12 (p35/p40), the gp130-like signaling subunits WSX-1/TCCR (Chen et al., 2000; Sprecher et al., 1998) and IL-12Rβ1 (Chua et al., 1994) lack N-terminal Ig domains to interact with site 3, and instead engage site 2 of their respective cytokines (p28 and p35). This arrangement leaves gp130 and IL-12Rβ2 to engage site 3 via their Ig domains. The gp130 paradigm can also be extended to other homodimeric members of the tall receptor family such as GCSF/GCSF-R and Leptin/OB-R (Tamada et al., 2006; Tartaglia et al., 1995).
Imaging of an activated cyokine-receptor complex
The 2D averages and 3D reconstruction of the gp130/LIF-R complex presented here provide us with a first snapshot of a full-length cytokine receptor ectodomain complex in an ‘on’ state, albeit in the absence of associated JAK molecules, and allows us to address several questions relating to cytokine receptor activation. One question has been whether the juxtamembrane, transmembrane and intracellular domains are closely associated in the full-length receptor complex. From the earlier single particle EM analysis of the IL-6 ectodomain complex, it was clear the gp130 membrane-proximal D4-D6 ‘legs’ were in close proximity in many class averages, although there were particles where the legs angled apart (Skiniotis et al., 2005). What is notable about the current full-length gp130/LIF-R class averages is that the receptor legs are uniformly joined, with connection of the legs occurring near D5 of gp130 and D7 of the LIF-R ECDs. These results are consistent with functional studies of the gp130/LIF-R signaling complex suggesting an interaction between the D5 of gp130 and D7 of LIF-R facilitates signaling (Timmermann et al., 2002). Interestingly, the EM averages show very little structural heterogeneity in the juxtamembrane and TM regions, which indicates they are closely associated at all times after ligand engagement. These structural observations are in accord with cellular studies of the gp130 and EPO juxtamembrane segments, where it was suggested they are rigid units, not flexible linkers that would absorb, and consequently dampen, the ability to relay structural perturbations induced by ligand binding to the ECDs, (Constantinescu et al., 2001a; Greiser et al., 2002; Seubert et al., 2003). We speculate that the rigid conformation of the membrane proximal regions of the gp130/LIF-R complex serves to sensitively transmit structural information through the membrane to activate receptor-bound JAK kinases. It is not known at this point whether CNTF binding induces gp130/LIF-R receptor association, or rather alters the orientation of a pre-formed receptor dimer to activate signaling, as seen in the hGH system (Brown et al., 2005). What is clear is that the conformation of the membrane-proximal regions triggered by ligand binding is a structurally stable state, and likely represents their disposition in the active signaling complex.
Another question is whether the intracellular domains of cytokine receptors fold into defined tertiary structures, as so far there is no structural information bearing on this issue. Despite contributing one-fifth of the total mass of the complex (~500 amino acids), we observed that the intracellular domains of gp130 and LIF-R provide no discernable density in the EM averages of the complex. This finding suggested to us that the intracellular domains lacked the structure necessary to provide clear density in the EM averages. To address this possibility, we performed 1H and 1H-15N HSQC NMR analysis on the purified intracellular domain (ICD) of gp130. These results indicate that the gp130 ICD lacks definable tertiary structure. Interestingly, the gp130 ICD is stable and monodisperse on a size exclusion column, suggesting that it likely exists in a weakly ordered, molten globule-like state. Although we did not analyze the ICD of LIF-R by NMR, secondary structure prediction algorithms predict that like gp130, the LIF-R ICD is also unstructured (data not shown). One caveat to these interpretations is that cytokine receptor ICD’s are constitutively bound to JAKs, and we analyzed an apo- form. It is possible that the ICDs will assume a substantially more ordered structure when bound to JAKs. However, the JAK binding regions, known as ‘box 1’ and ‘box 2’ in the membrane-proximal regions of the receptor ICD’s have been mapped to two short stretches of amino acids (~20 amino acids) (Murakami et al., 1991). Thus, while box 1 and box 2 may bind to JAK and be rigidified, it is likely the remaining ICD polypeptide chain is highly flexible. We hypothesize that the intracellular domains of all cytokine receptors that engage JAK kinases may exist in a weakly ordered state to facilitate ligand-induced conformational changes required for JAK reorientation and signaling. gp130, for example, contains five tyrosine phosphorylation motifs dispersed over the C-terminal 200 amino acids of the intracellular domain, so flexibility would seem to be important to allow the JAK kinases, STAT transcription factors, and other signaling and regulatory proteins unhindered access to these sites.
Experimental Procedures
Soluble protein expression and purification
Human LIF-R (D1-D5), gp130 (D1-D3 or D2-D3), CNTF-Rα (D1-D3), and CNTF as well as the CNTF-RαD1-D3/CNTF fusion (hyper-CNTF) were expressed in HiFive insect cells. The intracellular domain of human gp130 was expressed in E.coli strain BL21(DE3) and induced in M9 minimal media containing 15NH4Cl. All constructs were purified by Ni-affinity chromatography followed by size exclusion chromatography.
Purification of full-length receptor complexes
Full-length human gp130 and LIF-R were cloned into the vector pACSG2 containing the GP67 signal sequence and a C-terminal EE epitope tag (EYMPME). HiFive cells were infected with gp130 virus alone or co-infected with gp130 and LIF-R virus and incubated with supernatant from cells expressing hyper-CNTF to allow for formation of the receptor complex in situ. After cell lysis and centrifugation, membrane pellets were solubilized in buffer containing 1% n-dodecyl-β-D-maltoside (DDM), and the supernatant was incubated with EE antibody-coupled CNBR-sepharose. The gp130/hyper-CNTF complex or gp130/LIF-R/hyper-CNTF complex were eluted from EE-sepharose with buffer containing 200 µg/mL EE peptide, and further purified in 0.02% DDM by gel filtration.
Isothermal titration calorimetry
Titrations were performed on a VP-ITC calorimeter (MicroCal, Northhampton, MA) at the given temperatures. Data were processed with the MicroCal Origin software. After the run was complete, the contents of the cell were run on a Superdex 200 gel filtration column and analyzed by SDS-PAGE to confirm that all proteins were present in the complexes (Figure S1).
NMR
15N-labeled gp130 (amino acids 642-918) was concentrated to 0.4 mM and D2O was added to 10%. 1D and 2D-HSQC NMR spectra were acquired using an 800 MHz Varian Inova spectrometer at 25°C.
Crystallization and data collection of LIF-R
A mutant version of LIF-R (D1-D5), with five of thirteen potential N-linked glycosylation sites mutated (Asn64/85/143/191/243 to Gln), was over-expressed in Hi-Five insect cells, and purified via Ni-affinity and gel filtration chromatography. Crystals were grown in 100 mM sodium citrate pH 5.0, 100 mM lithium sulfate, and 6–8% PEG 3350. Data was collected at beamline 11-1 of Stanford Synchotron Radiation Laboratory (SSRL), and diffraction data was processed using MOSFLM and integrated using SCALA and TRUNCATE. Data processing statistics are listed in Table S1.
Structure determination and refinement of LIF-R
The structure of LIF-R (D1-D5) was solved by molecular replacement using as search model individual domains of mouse LIF-R from the LIF-R/LIF structure (PDB ID 2Q7N) (Huyton et al., 2007). The resolution of the final refined structure was 3.1Å with R and Rfree factors 25.5% and 30.9% respectively. The final model contains Asp52 (residue 1 in the model) through Ser534 of LIF-R, with eleven sugar moieties modeled at four N-linked glycosylation sites (Asn131, Asn303, Asn407, and Asn426). Refinement statistics are listed in Table S1. The structure has been deposited in the PDB under ID 3E0G.
Electron microscopy
Receptor complexes were prepared for electron microscopy using the conventional negative staining protocol (Ohi et al., 2004), and imaged at room temperature with a Tecnai T12 electron microscope operated at 120 kV. Images of specimens were recorded using low-dose procedures at a magnification of 52,000x and a defocus value of about −1.5 µm.
Image processing
Multi-reference alignment and classification for the projection analysis of the gp130/LIF-R/CNTF-Rα/CNTF complex (Figure S5) and the gp130/hyper-CNTF sub-complex (Figure S6) was carried out using the SPIDER image processing suite (Frank et al., 1996).
For the 3D reconstruction we used the random conical tilt technique (Radermacher et al., 1987) to calculate a first backprojection map, which, after angular refinement in SPIDER, served as a reference for a combination of 3,654 projections (60° and 0° images of particles from 3 similar classes, marked with a yellow dot in Figure S8B) to calculate the final 3D reconstruction with the program FREALIGN (Stewart and Grigorieff, 2004).
Molecular modeling of the gp130/LIF-R/CNTF-Rα/CNTF complex
The models of the gp130/LIF-R/CNTF-Rα/CNTF complex were generated using the program O (Jones et al., 1991). The model was further refined by mainly adjusting the FNIII legs to obtain a better fit of the coordinates into the EM density map (Figure S7).
Supplementary Material
The Supplemental Data include one table, eight figures, and detailed experimental procedures and can be found with this article online at http://www.molecule.org/.
Acknowledgments
We thank Sean Juo for assistance with structure determination, and Luca Varani and Corey Liu at the Stanford Magnetic Resonance Laboratory for help with NMR data collection and analysis. We thank Stefan Rose-John for helpful discussion and gift of the hyper-CNTF cDNA. The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from NIH-GM62580 (TW). GS and PJL are Damon Runyon Fellows, supported by the Damon Runyon Cancer Research Foundation (DRG-1824-04 to GS and DRG-1928-06 to PJL). This work was funded by NIH grant (AI51321) and support from the Keck Foundation to KCG. KCG and TW are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Autissier P, De Vos J, Liautard J, Tupitsyn N, Jacquet C, Chavdia N, Klein B, Brochier J, Gaillard JP. Dimerization and activation of the common transducing chain (gp130) of the cytokines of the IL-6 family by mAb. Int. Immunol. 1998;10:1881–1889. doi: 10.1093/intimm/10.12.1881. [DOI] [PubMed] [Google Scholar]
- Benigni F, Fantuzzi G, Sacco S, Sironi M, Pozzi P, Dinarello CA, Sipe JD, Poli V, Cappelletti M, Paonessa G, et al. Six different cytokines that share GP130 as a receptor subunit, induce serum amyloid A and potentiate the induction of interleukin-6 and the activation of the hypothalamus-pituitary-adrenal axis by interleukin-1. Blood. 1996;87:1851–1854. [PubMed] [Google Scholar]
- Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol. Cell. 2003a;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003b;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- Boulanger MJ, Garcia KC. Shared cytokine signaling receptors: structural insights from the gp130 system. Adv. Protein Chem. 2004;68:107–146. doi: 10.1016/S0065-3233(04)68004-1. [DOI] [PubMed] [Google Scholar]
- Bravo J, Heath JK. Receptor recognition by gp130 cytokines. EMBO J. 2000;19:2399–2411. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat. Struct. Mol. Biol. 2005;12:814–821. doi: 10.1038/nsmb977. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ghilardi N, Wang H, Baker T, Xie MH, Gurney A, Grewal IS, de Sauvage FJ. Development of Th1-type immune responses requires the type I cytokine receptor TCCR. Nature. 2000;407:916–920. doi: 10.1038/35038103. [DOI] [PubMed] [Google Scholar]
- Chow D, He X, Snow AL, Rose-John S, Garcia KC. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–2155. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- Chua AO, Chizzonite R, Desai BB, Truitt TP, Nunes P, Minetti LJ, Warrier RR, Presky DH, Levine JF, Gately MK. Expression cloning of a human IL-12 receptor component. A new member of the cytokine receptor superfamily with strong homology to gp130. J. Immunol. 1994;153:128–136. [PubMed] [Google Scholar]
- Constantinescu SN, Huang LJ, Nam H, Lodish HF. The erythropoietin receptor cytosolic juxtamembrane domain contains an essential, precisely oriented, hydrophobic motif. Mol. Cell. 2001a;7:377–385. doi: 10.1016/s1097-2765(01)00185-x. [DOI] [PubMed] [Google Scholar]
- Constantinescu SN, Keren T, Socolovsky M, Nam H, Henis YI, Lodish HF. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc. Natl. Acad. Sci. U S A. 2001b;98:4379–4384. doi: 10.1073/pnas.081069198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Greiser JS, Stross C, Heinrich PC, Behrmann I, Hermanns HM. Orientational constraints of the gp130 intracellular juxtamembrane domain for signaling. J. Biol. Chem. 2002;277:26959–26965. doi: 10.1074/jbc.M204113200. [DOI] [PubMed] [Google Scholar]
- Hammacher A, Wijdenes J, Hilton DJ, Nicola NA, Simpson RJ, Layton JE. Ligand-specific utilization of the extracellular membrane-proximal region of the gp130-related signalling receptors. Biochem. J. 2000;345(Pt 1):25–32. [PMC free article] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M, Murakami M, Saito M, Hirano T, Taga T, Kishimoto T. Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell. 1990;63:1149–1157. doi: 10.1016/0092-8674(90)90411-7. [DOI] [PubMed] [Google Scholar]
- Huyton T, Zhang JG, Luo CS, Lou MZ, Hilton DJ, Nicola NA, Garrett TP. An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc. Nat. Acad. Sci. U S A. 2007;104:12737–12742. doi: 10.1073/pnas.0705577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kurth I, Horsten U, Pflanz S, Timmermann A, Kuster A, Dahmen H, Tacken I, Heinrich PC, Muller-Newen G. Importance of the membrane-proximal extracellular domains for activation of the signal transducer glycoprotein 130. J. Immunol. 2000;164:273–282. doi: 10.4049/jimmunol.164.1.273. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, O'Shea JJ. Jaks and STATs: biological implications. Ann. Rev. Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Livnah O, Johnson DL, Stura EA, Farrell FX, Barbone FP, You Y, Liu KD, Goldsmith MA, He W, Krause CD, et al. An antagonist peptide-EPO receptor complex suggests that receptor dimerization is not sufficient for activation. Nat. Struct. Biol. 1998;5:993–1004. doi: 10.1038/2965. [DOI] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Johnson DL, Middleton SA, Mulcahy LS, Wrighton NC, Dower WJ, Jolliffe LK, Wilson IA. Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [see comment] [DOI] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- Marz P, Ozbek S, Fischer M, Voltz N, Otten U, Rose-John S. Differential response of neuronal cells to a fusion protein of ciliary neurotrophic factor/soluble CNTF-receptor and leukemia inhibitory factor. Eur. J. Biochem. 2002;269:3023–3031. doi: 10.1046/j.1432-1033.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- Matadeen R, Hon WC, Heath JK, Jones EY, Fuller S. The dynamics of signal triggering in a gp130-receptor complex. Structure. 2007;15:441–448. doi: 10.1016/j.str.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Newen G, Kuster A, Wijdenes J, Schaper F, Heinrich PC. Studies on the interleukin-6-type cytokine signal transducer gp130 reveal a novel mechanism of receptor activation by monoclonal antibodies. J. Biol. Chem. 2000;275:4579–4586. doi: 10.1074/jbc.275.7.4579. [DOI] [PubMed] [Google Scholar]
- Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc. Nat. Acad. Sci. U S A. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola NA, Hilton DJ. General classes and functions of four-helix bundle cytokines. Adv. Protein Chem. 1998;52:1–65. doi: 10.1016/s0065-3233(08)60432-5. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflanz S, Hibbert L, Mattson J, Rosales R, Vaisberg E, Bazan JF, Phillips JH, McClanahan TK, de Waal Malefyt R, Kastelein RA. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J. Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. Journal of Microscopy. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat. Struct. Mol. Biol. 2007;14:249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- Senaldi G, Varnum BC, Sarmiento U, Starnes C, Lile J, Scully S, Guo J, Elliott G, McNinch J, Shaklee CL, et al. Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc. Nat. Acad. Sci. U S A. 1999;96:11458–11463. doi: 10.1073/pnas.96.20.11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seubert N, Royer Y, Staerk J, Kubatzky KF, Moucadel V, Krishnakumar S, Smith SO, Constantinescu SN. Active and inactive orientations of the transmembrane and cytosolic domains of the erythropoietin receptor dimer. Mol. Cell. 2003;12:1239–1250. doi: 10.1016/s1097-2765(03)00389-7. [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Boulanger MJ, Garcia KC, Walz T. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat. Struct. Mol. Biol. 2005;12:545–551. doi: 10.1038/nsmb941. [DOI] [PubMed] [Google Scholar]
- Sprecher CA, Grant FJ, Baumgartner JW, Presnell SR, Schrader SK, Yamagiwa T, Whitmore TE, O'Hara PJ, Foster DF. Cloning and characterization of a novel class I cytokine receptor. Biochem. Biophys. Res. Comm. 1998;246:82–90. doi: 10.1006/bbrc.1998.8576. [DOI] [PubMed] [Google Scholar]
- Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, Kuroki R. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. Proc. Nat. Acad. Sci. U S A. 2006;103:3135–3140. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- Timmermann A, Kuster A, Kurth I, Heinrich PC, Muller-Newen G. A functional role of the membrane-proximal extracellular domains of the signal transducer gp130 in heterodimerization with the leukemia inhibitory factor receptor. Eur. J. Biochem. 2002;269:2716–2726. doi: 10.1046/j.1432-1033.2002.02941.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplemental Data include one table, eight figures, and detailed experimental procedures and can be found with this article online at http://www.molecule.org/.