Abstract
Background and purpose:
Maintenance of poly(ADP-ribose) (PAR) polymers at homoeostatic levels by PAR glycohydrolase (PARG) is central in cell functioning and survival. Yet the pharmacological relevance of PARG inhibitors is still debated. Gallotannin, a complex mixture of hydrolysable tannins from oak gall, inhibits PARG but which of its constituents is responsible for the inhibition and whether the pharmacodynamic properties are due to its antioxidant properties, has not yet been established.
Experimental approach:
A structure–activity relationship study was conducted on different natural and synthetic tannins/galloyl derivatives as potential PARG inhibitors, using a novel in vitro enzymic assay. Cytotoxicity was assayed in cultured HeLa cells.
Key results:
Mono-galloyl glucose compounds were potent inhibitors of PARG, with activities similar to that of ADP-(hydroxymethyl) pyrrolidinediol, the most potent PARG inhibitor yet identified. When tested on HeLa cells exposed to the PAR polymerase (PARP)-1-activating compound 1-methyl-3-nitro-1-nitrosoguanidine (MNNG), 3-galloyl glucose weakly inhibited PAR degradation. Conversely, the more lipophilic, 3-galloyl-1,2-O-isopropylidene glucose, despite being inactive on the pure enzyme, efficiently prolonged the half-life of the polymers in intact HeLa cells. Also, PARG inhibitors, but not radical scavengers, reduced, in part, cell death caused by MNNG.
Conclusions and implications:
Taken together, our findings identify mono-galloyl glucose derivatives as potent PARG inhibitors, and emphasize the active function of this enzyme in cell death.
Keywords: PARG, PARP, tannins, poly(ADP-ribose), cell death
Introduction
Among the post-translational modification of proteins, poly(ADP-ribosyl)ation has recently emerged as a key event in the maintenance of cellular homoeostasis, as well as pathogenesis of various disorders. Poly(ADP-ribosyl)ation consists of the synthesis of polymers (up to 200 U) of poly(ADP-ribose) (PAR) from nicotinimade adenine dinucleotide (NAD+), catalysed by a family of enzymes called PAR polymerases (PARPs) (Gagne et al., 2006). PARP-1 is the oldest and best characterized member of the PARP family, and is responsible for almost 80% of cellular PAR formation, which typically takes place in the nucleus (Shieh et al., 1998). Other non-nuclear PARPs have also been identified (Ame et al., 2004), with tankyrase having a key function in mitotic spindle assembly (Chang et al., 2005). Because of the high negative charge of the polymers, functions of numerous proteins are profoundly affected by PAR both by direct polymerization at specific glutamate residues and electrostatic interaction through non-covalent modification (D'Amours et al., 1999; Pleschke et al., 2000; Ahel et al., 2008).
In the light of the impact of PAR on protein functions, intracellular contents of the polymers are kept under tight control. This control is manifested by very rapid degradation (half-life of about 1 min) by PAR glycohydrolase (PARG), an enzyme exhibiting both exo- and endoglycosidic activity, and therefore releasing monomers of ADP-ribose or oligomers of PAR (Davidovich et al., 2001; Gagne et al., 2006). In contrast to the numerous PARPs involved in poly(ADP-ribosyl)ation, PARG is thought to be the main enzyme capable of degrading PAR. Indeed, although the polymer can be rapidly hydrolysed by phosphodiesterases in vitro, their function in vivo is still debated. In addition, an ADP-ribose hydrolase-like protein named ARH3 has been demonstrated to behave as a PARG (Oka et al., 2006). PARG is ubiquitously expressed in mammalian cells, and nuclear localization/exportation signals in its amino-acid sequence allow fast nucleo-cytoplasmic shuttling and efficient PAR catabolism (Bonicalzi et al., 2003). At present, various PARG isoforms have been identified, although only a single gene is present in human and rodent genome; PARG isoforms may arise from complex, alternative initiation of transcription, splicing and/or translation (Cortes et al., 2004; Meyer et al., 2007). Interestingly, the different isoforms have different kinetic properties, cellular localization and, possibly, functions (Cortes et al., 2004; Gagne et al., 2006; Meyer et al., 2007). Regardless of the specific function(s) of PARG isoforms, complete suppression of the parg gene causes early embryonic lethality (Koh et al., 2004). Conversely, mice selectively lacking the 110-kDa isoform of PARG (PARG110−/−) are viable, develop normally, despite being more susceptible to genotoxic and endotoxic shock, as well as to ischaemic brain injury (Cortes et al., 2004; Cozzi et al., 2006). PARG110−/− mice, however, are protected from intestinal ischaemia and inflammatory bowel disease (Cuzzocrea et al., 2005, 2007). Additional studies report a function for PARG in cell cycle regulation (Ohashi et al., 2003), mitotic spindle assembly (Chang et al., 2004), development (Hanai et al., 2004; Koh et al., 2004), differentiation (Di Meglio et al., 2003) and cell death (Affar et al., 2001; Ying et al., 2001; Hanai et al., 2004; Blenn et al., 2006).
On this basis, a great deal of effort has been directed at developing potent chemicals able to selectively inhibit PARG activity. It is worth mentioning, indeed, that gene deletion studies are not always informative about the real significance of a protein to pharmacotherapy. Conversely, they may be misleading because gene deletion provides chronic rather than acute, inhibition of this protein's activity. At present, several studies have reported the inhibition of PARG by different compounds such as hydrolysable tannins, ethacridine, the ADP-ribose analogue ADP-(hydroxymethyl)pyrrolidinediol (ADP-HPD), N-bis-(3-phenyl-propyl)9-oxo-fluorene-2,7-diamide (GPI 16552) (see Zhang and Li, 2002), and the modified PAR containing the etheno moiety (Shirato et al., 2007). Although these compounds showed pharmacodynamic properties consistent with PARG inhibition (Ying et al., 2001; Lu et al., 2003; Rapizzi et al., 2004; Cuzzocrea et al., 2005), their use as tools able to inhibit PARG activity has been recently revisited. In particular, gallotannin (also known as tannic acid) exhibits nonspecific effects and appears unable to efficiently cross the plasma membrane, and GPI 16552 has proved to be of low potency (Falsig et al., 2004).
Hydrolysable tannins are found in nature as multiple esters of gallic acid with glucose. The molecular weight of these compounds can vary from 332 (mono-galloyl glucose) to over 10 000 Da, with more than 500 different glucogalloyl derivatives being identified (Haslam, 1996). Although these compounds are of growing interest to consumers, food manufacturers and the pharmaceutical industry (Haslam, 1996; Maatta-Riihinen et al., 2004; Romani et al., 2005; Arapitsas et al., 2007), the lack of pure, structurally defined and commercially available standards renders the study of their biological activities difficult. This is probably the main reason why various researchers use tannins available in the market (such as gallotannin) or home-made extracts (Khan et al., 2000; Labieniec and Gabryelak, 2003; Ono et al., 2004).
By means of an array of glucogalloyl derivatives obtained from the market or synthesized/isolated in our laboratories, in this study we aimed to gather structure–activity information on tannins and their pharmacophore for PARG inhibition, as well as furthering our understanding of the function of PARG in cell death.
Methods
NMR analysis
1H NMR spectra at 200 and 400 MHz, and 13C NMR spectra at 50 and 100 MHz, were obtained on a Varian Gemini 2000 spectrometer and a Varian Mercury Plus 400 spectrometer, respectively. For the NMR experiments, CDCl3 and CD3OD were used as solvents. Chemical shifts are reported in parts per million (p.p.m.) relative to the solvent non-deuterated residue. The following abbreviations are used: singlet (s), doublet (d), triplet (t), multiplet (m). High-resolution mass spectrometry (HRMS) electrospray ionization (ESI) was measured on LTQ Orbitrap-Ultimate 3000-(Finnigan LTQ), operated at the Interdepartmental Centre of Mass Spectrometry (CISM, Florence, Italy).
HPLC analysis
HPLC/DAD/MS analyses were carried out as reported earlier (Arapitsas et al., 2007). Briefly, a HP 1100L liquid chromatograph equipped with a DAD detector (Agilent Technologies, CA, USA); linked to a HP 1100 MSD mass spectrometer with an API/ESI interface (Agilent Technologies, Santa Clara, CA, USA) was used. The mass spectrometer operating conditions were: gas temperature, 350 °C; nitrogen flow rate, 10.5 L min−1; nebulizer pressure, 40 p.s.i.; quadrupole temperature, 40 °C and capillary voltage, 3500 V. A Luna C18(2) 250 mm × 4.60 mm, 5 μm (Phenomenex, Torrance, CA, USA) column was used, operating at 27 °C. HPLC/DAD/MS/MS analyses of tannic acid were carried out using a HP 1100L liquid chromatograph equipped with a DAD detector (Agilent Technologies) linked to a API 3000 linear triple quadruple mass spectrometer (Applied Biosystems, MDS Sciex, Toronto, ON, Canada) equipped with a pneumatically assisted ESI interface. Optimization for the parameters of the ESI/MS and ESI/MS/MS analyses of the gallotannin was performed by preparing a solution of tannic acid (10 μg mL−1) in acetonitrile/water/formic acid (50:49.9:0.1 v/v/v). The solution was infused into the ES ionization source using a syringe pump at a flow rate 20 μL min−1. The optimum conditions found for the analysis were: negative ion scan mode, nebulizer gas 6 p.s.i., curtain gas 8 p.s.i., collision gas 2.5 × 10−5 torr, ion spray voltage (IS) −4500 V, source temperature 200 °C, focusing potential −400 V, entrance potential −10 V, declustering potential −180 V, collision energy 60 V and collision cell exit potential −15 V. Nitrogen gas was used for curtain, collision and nebulizer. The enhanced MS scan mode in the mass range of 150–2100 U was performed. The operating system was Analyst 1.4. The column used was a 3.0 μm Discovery HS PEG, 10 cm × 2.1 mm i.d. (Supelco, Bellefonte, PA, USA), operating at 27 °C and with a flow rate of 0.2 mL min−1. The mobile phase was a multistep linear solvent gradient system consisting of (A) 0.1% formic acid aqueous solution and (B) acetonitrile. The elution profile was: 3 min 95% A, then the solvent B was increased to 35% in 5 min, maintained for 2 min, further increased to 40% in 3 min and to 95% in 5 min. After 5 min, buffer B was decreased to 5% in 5 min.
Organic syntheses
Solvents were dried by usual laboratory techniques. Unless otherwise noted, all air and moisture sensitive reactions were performed under a positive nitrogen atmosphere.
3-(3,4,5-Tribenzyloxy)galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose
A mixture of 1,2,5,6-di-O-isopropylidene-α-D-glucose (250 mg, 1.0 mmol), 3,4,5-tribenzyloxy gallic acid (500 mg, 1.14 mmol) and 4-dimethylaminopyridine (250 mg, 1.20 mmol) in dry CH2Cl2 (20 mL) was refluxed for 1 h under nitrogen, then dicyclohexylcarbodiimide (160 mg, 1.20mmol) was added and the mixture reacted with stirring at room temperature for 16 h. Later, the mixture was kept at −18 °C for 30 min and filtered through a pad of celite. The organic phase was washed with HCl 3% (1 × 20 mL), and brine (2 × 20 mL) dried over Na2SO4, concentrated and purified by flash chromatography (eluent petroleum ether (PE)/ethyl acetate (EtOAc)=6:1) to obtain 3-(3,4,5-tribenzyloxy)galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose (510 mg, 80% yield) as a white powder. 1H NMR (CDCl3, 200 MHz) δ (p.p.m.): 1.29 (s, 4H, CCH3), 1.34 (s, 4H, CCH3), 1.44 (s, 4H, CCH3), 1.58 (s, 4H, CCH3), 4.04–4.24 (m, 2H, H5+6), 4.30 (dd, J=7.6 and 2.8 Hz, 1H, H4), 4.59 (d, J=3.6 Hz, 1H, H2), 5.16 (s, 4H, 2 × CH2Bn), 5.18 (s, 2H, CH2Bn), 5.48 (d, J=2.8 Hz, 1H, H3), 5.94 (d, J=0.4 Hz, 1H, H1), 7.22–7.57 (m, 17H, CHPh). 13C NMR (CDCl3, 50 MHz) δ (p.p.m.): 22.2 (CH4), 26.2 (CH4), 26.7 (CH4), 26.8 (CH4), 67.0 (C6), 71.1 (CH2Bn), 72.4 (C3), 75.0 (C5), 76.4 (CH4), 76.5 (CH4), 79.7 (C2), 83.1 (C4), 104.9 (C1), 109.1 (2 × CPh), 112.1 (2 × CPh), 124.1 (2 × CH of Bn), 127.1 (2 × CH of Bn), 127.8 (2 × CH of Bn), 128.0 (2 × CH of Bn), 128.2 (2 × CH of Bn), 128.3 (2 × CH of Bn), 136.2 (2 × C of Bn), 137.0 (2 × C of Bn), 142.5 (CPh), 152.2 (2 × CPh), 164.3 (CO).
3-Galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose
A solution of 3-(3,4,5-tribenzyloxy)galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose (900 mg, 0.76 mmol) in 10 mL of EtOAc/methanol (MeOH) (3:1) was hydrogenated over Pd/C 10% (0.1 equiv) at room temperature with a hydrogen balloon. After 2 h, the mixture was filtered over a pad of celite and evaporated in vacuum: The grey–green glassy solid obtained was triturated with three portions of diethylether (Et2O) and three portions of n-hexane and purified by flash chromatography (eluent EtOAc/PE/CH2Cl2=4:2:1) to obtain 3-galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose (471 mg, 87% yield) as a colourless glassy solid. 1H NMR (CD3OD, 200 MHz) δ (p.p.m.): 1.27 (s, 4H, CCH3), 1.32 (s, 4H, CCH3), 1.39 (s, 4H, CCH3), 1.51 (s, 4H, CCH3), 3.58–3.84 (m, 2H, H6), 3.84–3.99 (m, 1H, H5), 4.28 (dd, J=8.2 and 2.8 Hz, 1H, H4), 4.62 (d, J=3.8 Hz, 1H, H2), 5.32 (d, J=2.8 Hz, 1H, H3), 5.95 (d, J=3.8 Hz, 1H, H1), 7.05 (s, 2H, HPh). 13C NMR (CD3OD, 50 MHz) δ (p.p.m.): 25.4 (CH4), 26.0 (CH4), 26.4 (CH4), 27.0 (CH4), 68.0 (C6), 74.0 (C3), 77.4 (C5), 81.2 (C2), 84.6 (C4), 106.5 (C1), 110.0 (2 × CPh), 113.2 (CPh), 139.9 (CPh), 146.4 (2 × CPh), 166.6 (CO). HRMS (ESI): calculated for C19H24O10 (M +Na)+, 435.1266; found, 435.1260.
3-Galloyl-1,2-O-isopropylidene-α-D-glucose (4)
A solution of 3-galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose (100 mg, 0.24 mmol) was stirred for 2 h at room temperature in 10 mL of CHCl3. The solution was concentrated and the crude residue was purified by trituration with three portions of Et2O and three portions of n-hexane to obtain 3-galloyl-1,2-O-isopropylidene-α-D-glucose (4) (80 mg, 90% yield) as a glassy solid. 1H NMR (CD3OD, 400 MHz) δ (p.p.m.): 1.31 (s, 4H, CCH3), 1.51 (s, 4H, CCH3), 3.63 (dd, A part of an AB system, J=11.6 and 5.6 Hz, 1H, H6), 3.80 (dd, B part of an AB system, J=11.6 and 2.8 Hz, 1H, H6), 3.86–3.96 (m, 1H, H5), 4.28 (dd, J=9.6 and 2.8 Hz, 1H, H4), 4.62 (d, J=3.6 Hz, 1H, H2), 5.37 (d, J=2.8 Hz, 1H, H3), 5.94 (d, J=3.6 Hz, 1H, H1), 7.07 (s, 2H, HPh). 13C NMR (CD3OD, 100 MHz) δ (p.p.m.): 26.4 (CH4), 26.9 (CH4), 65.2 (C6), 70.0 (C3), 77.5 (C5), 80.1 (C2), 84.5 (C4), 106.6 (C1), 110.2 (2 × CPh), 113.2 (CPh), 121.0 (CPh), 140.1 (CPh), 146.4 (CPh), 167.2 (CO). HRMS (ESI): calculated for C16H20O10 (M +Na)+, 395.0953; found, 395.0949.
α- and β-3-galloyl-D-glucose (5)
A solution of 3-galloyl-1,2,5,6-di-O-isopropylidene-α-D-glucose (250 mg, 0.61 mmol) was stirred for 2 h at room temperature in CF3COOH (2 mL). The solution was concentrated and the crude residue was purified by trituration with three portions of Et2O and three portions of n-hexane to obtain 3-galloyl-D-glucose as a two-third mixture of α- and β-anomer (98 mg, 50% yield). 1H NMR (CD3OD, 400 MHz) δ (p.p.m.): 3.34–3.43 (m, 2H, H6), 3.54–3.64 (m, 3H, Hβ5+α6), 3.67–3.77 (m, 2H, Hβ4+α5), 3.80 (dd, J=12.0 and 2.4 Hz, 1H, Hβ2), 3.87 (dd, J=12.0 and 2.4 Hz, 1H, Hα2), 3.93 (m, 1H, Hα4), 4.61 (d, J=7.6 Hz, 1H, Hα1), 5.10 (at, J=9.6 Hz, 1H, Hα3), 5.17 (d, J=4.0 Hz, 1H, Hβ1), 5.39 (at, J=9.6 Hz, 1H, Hβ3), 7.13 (s, 2H, HαPh), 7.13 (s, 2H, HβPh). 13C NMR (CD3OD, 100 MHz) δ (p.p.m.): 62.4 (Cs), 62.6 (Cs), 66.9 (Cs), 70.0 (Cs), 70.1 (Cs), 72.4 (Cs), 73.0 (Cs), 74.8 (Cs), 77.3 (Cs), 78.0 (Cs), 79.4 (Cs), 94.1 (Cβ1), 98.3 (Cα1), 110.4 (6 × CPh), 121.9 (CPh), 122.0 (CPh), 146.4 (2 × CPh), 168.3 (CO), 168.6 (CO). HRMS (ESI): calculated for C13H16O10 (M +Na)+, 356.0640; found, 356.0634.
Extraction/isolation of hydrolysable tannins
The extraction of raspberries was achieved as reported earlier (Arapitsas et al., 2007). Briefly, 40 g of fresh raspberries in 300 mL of ethanol/water 70:30 was extracted for 18 h at room temperature. Later, the extract was filtered, concentrated, dissolved in ethanol/water 70:30 (20 mL) and directly analysed by HPLC/DAD/MS. The final volume of the extract was lyophilized and the powder, dissolved in 1 mL of ethanol/water 70:30, was loaded in a chromatographic column Sephadex LH-20 (30 cm × 1 cm). Stepwise elution was then performed using the following MeOH/acetone/H2O mixture as mobile phase: 50 mL (90:0:10), 20 mL (70:20:10), 20 mL (30:50:20), 50 mL (0:80:20) and 50 mL (0:80:20). From this last fraction, 10 mg of sanguin H-6 was obtained. Hydrolysable tannins are absorbed to the stationary phase when methanol, ethanol and alcohols in general are used as mobile phase and they are released and eluted with aqueous acetone. For the isolation of 15 mg 1,2,3,4,6-pentagalloyl-β-D-glucose, and 5 mg trigalloyl-β-D-glucose, from 100 mg of tannic acid, the same column was used, but the mobile phase was isocratic acetone/water. The characterization of all compounds was made by HPLC/DAD/MS/MS and NMR analysis. To calculate the purity of each isolated and synthetic compound, 1 mg was separately dissolved in 1 mL of ethanol/water 70:30 and directly analysed by HPLC/DAD/MS.
PARG activity assay
3H-PAR was synthesized in vitro according to Banasik et al. (1992). Briefly, 0.05 U of purified bovine PARP-1 was incubated at 37 °C in a final volume of 100 μL 50 mM Tris-HCl buffer (pH 7.4), containing 5 mM MgCl2, 2 mM DL-dithiothreitol, 10 μg sonicated DNA, 0.5 mg mL−1 bovine albumin, 10 μM NAD and 1 μL 3H-NAD (35.5 nmol). After 60 min, PARP-1 activity was blocked with 30 μM 6(5H)-phenanthridinone (PHE) and different units of PARG were added to the mixture in the presence or absence of potential or known PARG inhibitors. After 240 min incubation at 37 °C, the reaction was stopped by adding 1 mL of trichloroacetic acid (TCA) 10% (w/v). Then, the mixture was centrifuged (15 min at 12 000 g at 4 °C), and the pellet containing 3H-PAR was washed twice with water and resuspended in 0.1 mM NaOH. Radioactivity was measured by scintillation counting (Tri-Carb 1900 TR; Packard, Meriden, CT, USA).
Cells and culture conditions
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 2 mM glutamine, 10% foetal bovine serum and antibiotics. Cultures were brought to 50–70% confluence and exposed to 1-methyl-3-nitro-1-nitrosoguanidine (MNNG) with or without pretreatment with different potential PARG inhibitors. Cell viability was evaluated by measuring lactate dehydrogenase release in the incubating media or reduction of methylthiazolyl tetrazolium as described earlier (Fossati et al., 2007).
Measurement of NAD, western blotting and immunocytochemistry
Measurement of intracellular NAD contents, western blotting, staining with Hoechst 33258 and immunocytochemistry for PAR were performed as described earlier (Cipriani et al., 2005). HeLa cells were grown on coverslips and challenged with different compounds. Cells were then washed twice with phosphate-buffered saline (PBS) and fixed with cold PBS/4% paraformaldehyde for 10 min. Cells were incubated for 1 h in PBS/0.1% Triton X-100/bovine serum albumin (5 mg mL−1) at room temperature, rinsed in PBS and incubated for 1 h with a polyclonal antiapoptosis-inducing factor (AIF) antibody (Santa Cruz Biotechnology, Tamecula, CA, USA) dissolved 1:200 in PBS/BSA (5 mg mL−1). After washing with PBS, slides were incubated for 1 h with a Cy-3-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories, Westgrove, PA, USA) dissolved 1:200 in PBS/5 mg/mL BSA. Fluorescence was examined using a rhodamine filter (Ex 520nm, Em >580nm). Specificity of immune detection was established by the omission of the primary antibody.
Cell counting
In Hoechst 33258-stained cultures, 30 different microscopic fields ( × 20) for each treatment were acquired by a charge coupled device camera. Cell nuclei were counted in each image and their number expressed as the mean±s.e.mean.
Comet assay
DNA strand breaks were evaluated by the comet assay, as described earlier (Giovannelli et al., 2003). Briefly, centrifuged cells were suspended in agarose and layered on microscopic slides to be run through the comet assay. Microscopic analysis was carried out using a Labophot-2 microscope (Nikon, Tokyo, Japan) provided with epifluorescence. Each experimental point was run in duplicate and the images of 50 randomly chosen nuclei per slide were captured and analysed using a custom-made imaging software coupled with a charge coupled device camera (model C5985; Hamamatsu, Japan). The program calculated the total fluorescence distribution along the longer axis of the nucleus and the fluorescence distribution of the head and tail of the comet, respectively, for each image. Data were expressed as a percentage of total fluorescence migrated in the tail for each nucleus (% DNA in tail), a parameter linearly related to the number of DNA breaks. The mean % DNA in tail of 50 nuclei per slide was calculated, and the duplicate values were further averaged. The value of % DNA in tail obtained estimated the basal number of DNA strand breaks.
Statistical analysis
Evaluation of significant differences among groups was performed using the Student's t-test or ANOVA followed by Tukey's w-test. Statistical difference was considered for P-values <0.05.
Materials
MeOH, PE, EtOAc, dichloromethane (CH2Cl2), n-hexane and Et2O were from Carlo Erba Reagenti SpA (Milano, Italy). Methyl gallate, CH3CN, PHE, tritiated NAD (0.93 TBq mmol−1 25.0 Ci mmol−1) was from Perkin Elmer (Milan, Italy). Dicyclohexylcarbodiimide, N,N,-dimethylformamide HPLC grade, Hoechst 33258, bovine albumin, TCA, MNNG, MgCl2, DL-dithiothreitol, 4-dimethylaminopyridine, CF3COOH, Pd/C 10%, Na2SO4, CDCl3, CD3OD, H3PO3, Pb(AcO)4, tannic acid (1), gallic acid (2), methyl gallate (3), 1,2,5,6-di-O-isopropylidene-α-D-glucose and gallotannin were from Sigma-Aldrich (St Louis, MO, USA). Acetonitrile (HPLC grade) and formic acid were purchased from Aldrich Company Inc. (Milwaukee, WI, USA). 2,3-Digalloyl-α,β-D-glucose (8), 2,3-digalloyl-α,β-O-methyl-D-glucose (9), 2-galloyl-α,β-O-methyl-D-glucose (7), 3-galloyl-α,β-O-methyl-D-glucose (6), 2,3-hexahydroxydiphenoyl-α,β-D-glucose (10), 2,3-di(3-galloyl,4,5-dihydroxy-benzoyl)-α,β-D-glucose (11) and 3,4,5-tribenzyloxy gallic acid were prepared as described by Arapitsas et al. (2007). Calf thymus DNA (10 mg mL−1) was from Invitrogen (San Diego, CA, USA). PARG bovine, PARP-1 and anti-PAR LP96-10 were from Alexis (Vinci Biochem, Vinci, Italy). ADP-HPD dehydrate ammonium salt was from Calbiochem (San Diego, CA, USA). Insta-gel plus was from Packard Instrument Co., Inc. (Downes Grove, IL, USA).
Results
Development of an in vitro PARG activity assay
Classic methods for screening of compounds as inhibitor of PARG activity are measurement of 32P-PAR degradation by thin layer chromatography or quantitation of nuclear PAR levels by immunocytochemistry in cultured cells exposed to PARP-1-activating compounds (Tavassoli et al., 1985; Tsai et al., 1992; Falsig et al., 2004; Keil et al., 2004; Shirato et al., 2007). Recently, a non-radiometric assay for high-throughput screening of PARG inhibitors has been reported (Putt and Hergenrother, 2004). In this study, we developed a very rapid and simple method to evaluate inhibition of PARG activity by measuring the hydrolytic activity of purified bovine PARG on 3H-PAR. As shown in Figure 1a, incubation of pure PARP-1 with 3H-NAD in the presence of sonicated DNA (containing histones) and bovine albumin led to the incorporation of radioactivity in TCA-precipitated proteins. Radioactivity incorporation was time dependent, tended to reach a plateau at 60-min incubation and was completely prevented by the presence of the PARP-1 inhibitor 6(5H)-phenanthridinone (Banasik et al., 1992) (Figure 1a). It is noted that the addition of PHE after 60 min blocked the residual incorporation of radioactivity, with the latter remaining stable for at least 3 h after PARP-1 inhibition (Figure 1a). These results indicate that radioactivity was incorporated into proteins by PARP-1, and that these polymers were stable under our experimental settings. Conversely, addition of pure PARG to the mixture after 60 min reduced the radioactivity measured in the TCA-precipitated pellet. When PARG was added together with PHE, an almost complete reduction of precipitable radioactivity was recorded (Figure 1b), suggesting extensive degradation of PAR, by the exo/endoglycosidic activity of PARG. PAR degradation by PARG was concentration dependent, reaching a maximiun at 0.05 U (Figure 1c).
Figure 1.
Synthesis and degradation of poly(ADP-ribose) (PAR) by pure poly(ADP-ribose) polymerase (PARP-1) and poly(ADP-ribose) glycohydrolase (PARG). (a) Time-dependent incorporation of radioactivity from 3H-NAD into trichloroacetic acid (TCA)-precipitated proteins in the absence or presence of 30 μM phenanthridinone (PHE; a PARP-1 inhibitor) at time 0 or 60 min. (b) Effect of pure PARG (0.05 U) added after 60 min on incorporation of radioactivity from 3H-NAD into TCA-precipitated proteins in the absence or presence of 30 μM PHE. (c) PARG-dependent reduction of radioactivity incorporation is concentration dependent. Each point/column represents the mean±s.e.mean of three (a, b) and two (c) experiments conducted in duplicate.
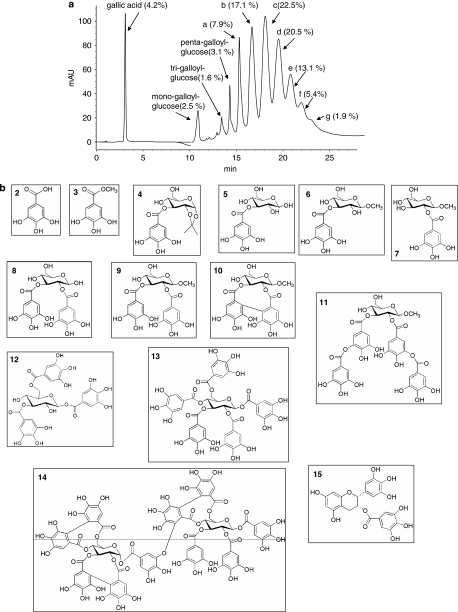
Inhibition of PARG activity by gallolyl-glucose derivatives
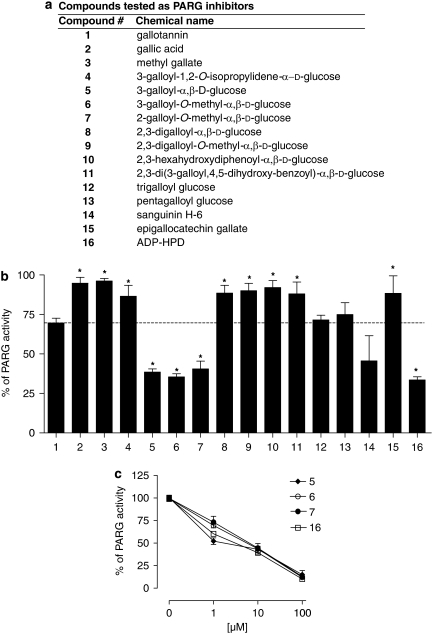
Gallotannin is classically used as a PARG inhibitor despite being a mixture of different compounds. Indeed, as shown in Figure 2a, chromatographic analysis coupled to mass spectrometry of solubilized gallotannin revealed the presence of gallic acid and various galloyl glucose derivatives containing up to 12 galloyl residues (see legend of Figure 2a). Although the presence of methyl-gallate, di- and tetra-galloyl glucose in the gallotannin mixture has been reported (Romani et al., 2006; Arapitsas et al., 2007), under our experimental conditions their concentrations were below detection limits. We therefore next evaluated the effect of the galloyl-glucose derivatives on PAR degradation by PARG in the above-mentioned enzymatic assay. We tested galloyl-glucose derivatives with increasing complexity (Figures 2b and 3a) to understand which constituent of gallotannin is mostly responsible for PARG inhibition, as well as to gather information on their structure–activity relationship. To this end, we tested compounds available in the market as well as molecules synthesized ex novo or from precursors extracted from gallotannin (Figures 2 and 3a, and see Methods). As shown in Figure 3b, gallotannin (1, see also Figure 3a) led to a 25% inhibition of PARG activity when tested at 10 μM, in good agreement with its reported IC50 of about 30 μM (Tsai et al., 1992). Gallic acid (2), a component of gallotannin, had no effects on the enzyme activity or its derivative methyl-gallate (3) (Figures 2 and 3b), synthesized to increase the lipophilicity of gallic acid. Similarly, 3-galloyl-1,2-O-isopropylidene glucose (4), intermediate of the synthesis of 3-galloyl-glucose, did not inhibit PARG at 10 and 100 μM (Figure 3b and not shown). An inhibition of about 65% of PAR degradation was obtained with 3-galloyl-α,β-D-glucose (5), 3-galloyl-O-methyl-α,β-D-glucose (6) and 2-galloyl-O-methyl-α,β-D-glucose (7) when tested at 10 μM (Figure 3b). These compounds showed IC50 values on PARG activity of 0.95±0.02, 7.1±0.05 and 7.2±0.03 μM, respectively. It is noted that their inhibitory potency was in the same range of that of ADP-HPD (16), that under our experimental settings had an IC50 of 3.2±0.08 μM (Figure 3c).
Figure 2.
Chemical constituents of gallotannin and other galloyl-containing molecules tested as poly(ADP-ribose) glycohydrolase (PARG) inhibitors. (a) Representative HPLC/DAD chromatographic profile of gallotannin, reported at 280 nm. The presence and % of the different galloyl derivatives are shown. a–g represent the esa-, epta-, octa-, ena-, deca-, endeca-, dodeca- and galloyl glucose derivatives, respectively. (b) Structures of galloyl derivatives tested as PARG inhibitors.
Figure 3.
Mono-galloyl glucose derivatives are potent poly(ADP-ribose) glycohydrolase (PARG) inhibitors. (a) List with chemical names and numbers of the compounds tested as PARG inhibitors. (b) Histogram showing the relative inhibitory potency of the compounds tested at 10 μM on PARG activity. (c) Inhibitory activity on PARG of compounds (5), (6), (7) and (16). In (b, c), each column/point represents the mean±s.e.mean of three experiments conducted in duplicate. *P<0.05 vs no. 1 (ANOVA+Tukey's post hoc test).
By increasing the number of gallic residues from one to two such as in compounds (8) and (9) (Figure 2), the inhibitory activity on PARG decreased (Figure 3b). Neither the presence of a C–C bond between the two gallic acid residues (hexahydroxydiphenoyl group) as in compound (10) nor the presence of depsidic moieties as in compound (11) led to significant PARG inhibition (Figure 3b). Conversely, an increase in gallic residues bound to glucose as in tri- and pentagalloyl glucose (12 and 13, respectively) augmented the inhibitory potency on PARG when compared with digalloyl substitutes (Figure 3b). We also tested the complex hydrolysable tannin sanguinin H-6 (14), which, at 10 μM, caused about 50% inhibition (Figure 3b). The gallic acid-containing, antioxidant compound epigallocatechin gallate (15) had no effects on PARG activity when tested at 10 μM, whereas at the same concentrations, the potent PARG inhibitor ADP-HPD (16) reduced the PAR-hydrolysing activity of PARG by ∼70% (Figure 3b).
Effects of mono-galloyl glucose derivatives on PAR content in intact cells
Among the galloyl glucose derivatives, the most potent were 3-galloyl-glucose, 3-galloyl-O-methyl-glucose and 2-galloyl-glucose (5, 6 and 7, respectively). We next evaluated whether these chemicals could inhibit PARG activity in intact cells. To this end, we used HeLa cells, a cell line characterized by massive synthesis of PAR on exposure to alkylating agents, and rapid polymer degradation by PARG (Rossi et al., 2002; Cipriani et al., 2005). Cells were exposed to the methylating agent MNNG and PAR contents were evaluated by western blotting or immunocytochemistry. As shown in Figure 4a, PAR content significantly increased 5 min after exposure to MNNG, and rapidly returned to basal level after 15 and 30 min. Incubation with 3-galloyl-glucose (5) 30 min before and during the challenge with MNNG did not provoke any apparent change in the reduction of PAR contents at 10 and 15 min (Figure 4a). In keeping with previous reports (Alvarez-Gonzalez and Althaus, 1989), these findings indicate that PAR degradation by PARG is efficiently activated on polymer synthesis. They also suggest that 3-galloyl-glucose (5), despite being a potent inhibitor in vitro (Figures 2b and c), exerts weak inhibition of PAR degradation in cultured cells. Given the high hydrophilicity of 3-galloyl-glucose, we reasoned that, akin to gallotannin (Falsig et al., 2004), its low inhibitory potency in intact cells could be ascribed to a low permeability through the plasma membrane. To address this issue, we took advantage of the availability of 3-galloyl-1,2-O-isopropylidene glucose (4), a protected precursor in the synthesis of 3-galloyl-glucose containing a isopropyldene group (Figure 2). We reasoned that such a compound should have higher lipophilicity and hence a facilitated passage through the plasma membrane. Also, once inside the cell, it could be readily transformed into 3-galloyl-glucose (5) by nonspecific hydrolases. In good agreement with this hypothesis, western blotting showed that 3-galloyl-1,2-O-isopropylidene glucose (4), despite being inactive per se (Figure 3b), increased the basal contents of PAR when added to the culture medium at concentrations of 10 or 100 μM. Notably, 3-galloyl-1,2-O-isopropylidene glucose (4) also counteracted the reduction of PAR content at 15 and 30 min after MNNG exposure (Figure 4b).
Figure 4.
Effects of mono-galloyl glucose on poly(ADP-ribose) glycohydrolase (PARG) activity in intact cells. (a) HeLa cells were exposed to the poly(ADP-ribose) polymerase (PARP-1)-activating compound 1-methyl-3-nitro-1-nitrosoguanidine (MNNG) (100 μM) in the absence or presence of 10 μM 3-galloyl-glucose (5) and poly(ADP-ribose) (PAR) levels were evaluated by western blotting at different times. β-Actin is shown as a loading control. (b) HeLa cells were exposed to the PARP-1-activating compound MNNG (100 μM) in the absence or presence of 3-galloyl-1,2-O-isopropylidene glucose (4) (10 or 100 μM) and PAR levels were evaluated by western blotting at different time points. β-Actin is shown as a loading control. Compound (4) was pre-incubated for 30 min. (c) Immunocytochemical evaluation of intracellular PAR contents in cells exposed to MNNG in the absence or presence of 3-galloyl-1,2-O-isopropylidene glucose (4, 100 μM). (d) Nicotinimade adenine dinucleotide (NAD) levels in cells exposed 30 min to 30 μM 6(5H)-phenanthridinone (PHE) or 30 and 60 min to 100 μM 3-galloyl-1,2-O-isopropylidene glucose (4). (e) NAD contents in cells exposed to 100 μM MNNG in the presence or absence of PHE (30 μM) or 3-galloyl-1,2-O-isopropylidene glucose (4, 100 μM). A blot/experiment representative of two (a–c) or three (d) is shown. Bar=20 μm. Columns are the mean±s.e.mean of two experiments (e). *P<0.05, **P<0.01 vs control (C) (ANOVA+Tukey's post hoc test).
These findings suggest that 3-galloyl-1,2-O-isopropylidene glucose (4) is a cell-permeable precursor of the potent PARG inhibitor 3-galloyl-glucose. Immunocytochemistry confirmed these data by showing that the nuclear content of PAR was higher in cells exposed 15 and 30 min to MNNG plus 3-galloyl-1,2-O-isopropylidene glucose (4), relative to those challenged with MNNG only (Figure 4c). In keeping with the basal levels of PAR in resting HeLa cells (indicative of ongoing poly(ADP-ribosyl)ation, first lane of Figure 4b), exposure to PHE significantly increased intracellular NAD content (Figure 4d). The latter also increased on exposure to 3-galloyl-1,2-O-isopropylidene glucose (4) (Figure 4d), indicating that the increased content of PAR found in cells challenged with the compound (first three lanes of Figure 4b) is not due to PARP-1 activation. Conversely, we ascribe the increased contents of NAD in cells exposed to 3-galloyl-1,2-O-isopropylidene glucose (4) to the ability of the latter to inhibit PARG and the ensuing inhibition of PARP-1 activity because of excessive auto-(ADP-ribosyl)ation (Davidovich et al., 2001; Rapizzi et al., 2004). In contrast to the results with PHE, however, 3-galloyl-1,2-O-isopropylidene glucose (4) had no effect on intracellular NAD depletion prompted by MNNG (Figure 4e), suggesting that it does not affect hyper-poly(ADP-ribosyl)ation on DNA damage.
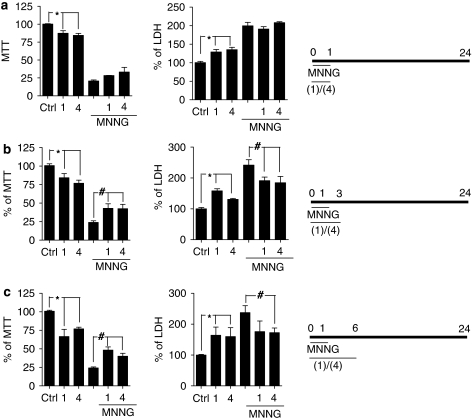
Effect of gallotannin, 3-galloyl-1,2-O-isopropylidene glucose or radical scavengers on cell death induced by MNNG
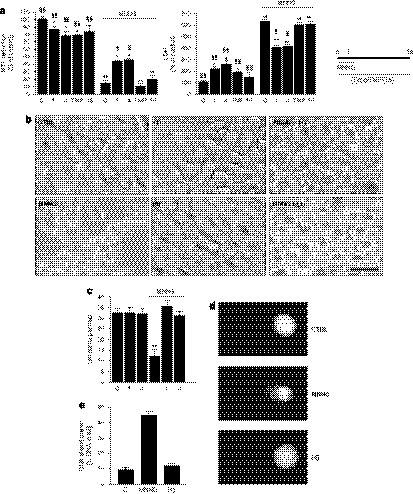
It has been reported that PARG inhibition/suppression exerts protection against cell death in models of PARP-1 hyperactivation (Ying et al., 2001; Lu et al., 2003; Cuzzocrea et al., 2005, 2007; Patel et al., 2005; Blenn et al., 2006). However, the study by Falsig et al. (2004) demonstrated that the cytoprotective effects of the PARG inhibitor gallotannin (1) were related to its antioxidant properties and not to the inhibition of PARG. To gather further information on PARG inhibition and cell death, HeLa cells were exposed to gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) in the presence or absence of MNNG and cell death was evaluated 24 h later by means of methylthiazolyl tetrazolium reduction or lactate dehydrogenase release assays. As reported earlier (Cipriani et al., 2005), MNNG-dependent cell death was completely prevented by two inhibitors of PARP-1 such as PJ34 and PHE (not shown), indicating that the enzyme's hyperactivation is causative in cell death. The results in Figure 5 show that, under our conditions, both PARG inhibitors caused cytotoxicity when present in the incubating media for 1, 3 and 6 h. Yet cell death evaluated 23 h after 1 h exposure to MNNG was in part reduced by gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) when present in the incubating media for 3 or 6 h (Figures 5b and c) but not 12 or 24 h (not shown). However, a 12 h exposure to the PARG inhibitors afforded protection from MNNG when cell death was evaluated at this time point (Figure 6a). To understand whether partial prevention of cell death showed by the PARG inhibitors were due to radical scavenging, we tested the effects of the well-known radical scavengers 4-hydroxy-TEMPO and epigallocatechin gallate (15) on PARP-1-dependent cell death triggered by MNNG. Notably, the presence of the two antioxidants in the culture medium had no effects on MNNG-dependent cell death evaluated 11 h after 1 h exposure to the alkylating agent (Figure 6a). These findings taken together suggest that PARG inhibition rather than radical scavenging underlies cytoprotection by gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) during PARP-1 hyperactivation. Analysis of cell morphology revealed that cells exposed for 1 h to MNNG acquired a round appearance and detached from the plate 11 h later. In contrast, cells exposed to gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) during MNNG and for the following 11 h were still adherent to the plate. However, they appeared unhealthy being swollen and with cytoplasmic vacuoles. Cells challenged with gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) for 12 h in the absence of MNNG did not show morphological abnormalities (Figure 6b). Cell counting confirmed these findings by showing that the number of cells was reduced almost to 50% after exposure to MNNG (100 μM; 12 h) but was unchanged in cultures exposed to the two PARG inhibitors alone or in combination with MNNG (Figure 6c). By means of the comet assay, we also checked whether 3-galloyl-1,2-O-isopropylidene glucose (4) induced DNA damage. As shown in Figures 6d and e, we found that MNNG (100 μM) but not the galloyl glucose derivative triggered significant DNA breaks. To further investigate cytoprotection from PARP-1 overactivation afforded by gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4), we next analysed their effects on two typical events occurring during PARP-1-dependent cell death, namely, release of AIF from mitochondria and chromatin condensation. In keeping with prior work (Cipriani et al., 2005), AIF immunoreactivity lost the typical punctate distribution present in resting cells and appeared diffuse in the cytosol and nucleus (indicating mitochondrial release of AIF) 3 h after exposing HeLa cells to a 1 h challenge to MNNG. AIF immunoreactivity was not significantly modified in cells exposed for 4 h to gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4). However, mitochondrial AIF release was prevented in part in cells exposed to MNNG (1 h) plus gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) (Figure 7a). Cell nuclei appeared shrunken often showing pyknosis and chromatin condensation 8 h after exposure to MNNG (1 h). The presence of gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) did not affect nuclear morphology. Yet nuclear alterations of MNNG-challenged cells were partially prevented by gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) present in the medium for the 8-h period. Specifically, the two compounds prevented nuclear condensation triggered by MNNG but nuclei were still shrunken with irregular surfaces (Figure 7b).
Figure 5.
Effect of gallotannin or 3-galloyl-1,2-O-isopropylidene glucose on cell death triggered by 24 h exposure to 1-methyl-3-nitro-1-nitrosoguanidine (MNNG). Cells were exposed to 100 μM gallotannin (1) or 100 μM 3-galloyl-1,2-O-isopropylidene glucose (4) in the presence or absence of 100 μM MNNG for 1 h and cell death was evaluated 24 h later by means of methylthiazolyl tetrazolium (MTT) reduction or lactate dehydrogenase (LDH) release assays. Both PARG inhibitors were present in the incubating media for 1 (a), 3 (b), 6 h (c) (see time lines on right of a–c). Bars are the mean±s.e.mean of three experiments conducted in duplicate. *P<0.05 vs control (ctrl), #P<0.05 vs MNNG (ANOVA+Tukey's post hoc test).
Figure 6.
Effect of gallotannin, 3-galloyl-1,2-O-isopropylidene glucose, 4-hydroxy-TEMPO (TMP) or epigallocatechin gallate on cell death triggered by 12 h exposure to 1-methyl-3-nitro-1-nitrosoguanidine (MNNG). (a) HeLa cells were exposed to 100 μM gallotannin (1), 100 μM 3-galloyl-1,2-O-isopropylidene glucose (4), 1 mM TMP or 100 μM epigallocatechin gallate (15) for 12 h and cell death was evaluated by methylthiazolyl tetrazolium (MTT) reduction and lactate dehydrogenase (LDH) release. Columns represent the mean±s.e.mean of three experiments conducted in duplicate. *P<0.05, **P<0.01 vs ctrl. §P<0.05, §§P<0.01 vs MNNG (ANOVA+Tukey's post hoc test). (b) Phase-contrast micrograph visualization of cells exposed with or without 100 μM MNNG for 1 h in the presence or absence of 100 μM gallotannin (1) or 100 μM 3-galloyl-1,2-O-isopropylidene glucose (4). (c) Cell counting of cultures exposed to 100 μM gallotannin (1) or 100 μM 3-galloyl-1,2-O-isopropylidene glucose (4) in the presence or absence of MNNG. **P<0.01 vs ctrl (ANOVA+Tukey's post hoc test). (d) Cell nuclei visualized with the comet assay of cells exposed for 30 min to MNNG or 3-galloyl-1,2-O-isopropylidene glucose (4) both at 100 μM. (e) Measurement of DNA damage (% of DNA damage in tail) in cells treated as in (d). Bars represent the mean±s.e.mean of three (a, c) and two (e) experiments. Images are representative of three (b) and two (d) experiments. Bar=20 μm.
Figure 7.
Effect of gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) on mitochondrial apoptosis-inducing factor (AIF) release and nuclear morphology of cells exposed to 1-methyl-3-nitro-1-nitrosoguanidine (MNNG). (a) AIF immunostaining of cells under control conditions or 4 h after exposure to 100 μM MNNG for 1 h in the absence or presence of 100 μM gallotannin (1) or 100 μM 3-galloyl-1,2-O-isopropylidene glucose (4). The effect of the two compounds alone is shown. Note the loss of punctate staining and AIF redistribution into the cytoplasm and nucleus in MNNG-exposed cells. (b) Hoechst 33258 staining of nuclei of cells under control conditions or 8 h after exposure to 100 μM MNNG for 1 h in the absence or presence of gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) both at 100 μM. The effect of gallotannin (1) or 3-galloyl-1,2-O-isopropylidene glucose (4) alone is also shown. Note nuclear shrinkage, pyknosis and chromatin condensation (arrows) in MNNG-exposed cells, and partial prevention of these nuclear alterations by 3-galloyl-1,2-O-isopropylidene glucose (4). Arrowheads point to partially shrunken nuclei with irregular surfaces. Images are representative of three experiments. Bar=20 μm (a) and 10 μm (b).
Discussion and conclusions
In this study, we investigated the effects of different hydrolysable tannins with increasing complexity on PARG activity. By means of a novel assay for PARG activity, we report that mono galloyl glucose derivatives inhibit the enzyme with IC50 values in the same range of that of ADP-HPD. It must be pointed out that ADP-HPD is reported to have an IC50 of 0.12 μM (Slama et al., 1995b) or 0.33 μM (Koh et al., 2003a), approximately 10- to 25-fold lower than that reported in this study. At present, we do not know the reason for this apparent discrepancy. It is worth noting, however, that the efficiency of the hydrolytic activity of PARG depends on the molecular weight and structure of the polymer. In particular, PARG targets the terminal ADP-ribose residue as well as the endoglycosidic bond and branching points of PAR. Also, degradation of the polymers starts with a rapid hydrolysis until approximately 50% of the chains are consumed, and then turns into a slow distributive degradation leading to the release of monomers of ADP-ribose. It is noted that the degradation of large polymers (>20 ADP-ribose units) is higher than that of short ones (Hatakeyama et al., 1986), and ADP-HPD more efficiently inhibits PARG when processing high molecular weight polymers (Ramsinghani et al., 1998). Given that we do not know the length of the polymers synthesized in our in vitro assay, we consider that the higher value of the IC50 for ADP-HPD reported here could be ascribed to structural differences of the substrate when compared with that used in previous reports (Slama et al., 1995b; Koh et al., 2003a). These interpretations could also help reconcile the discrepancies reported for the potency of the PARG inhibitor GPI 16552 (Lu et al., 2003; Falsig et al., 2004). Nevertheless, evidence that ADP-HPD is considered the most potent PARG inhibitor currently developed (Zhang and Li, 2002; Gagne et al., 2006) underscores the relevance of our findings, and points to 3- and 2-galloyl glucose as tools useful for pharmacologically modulating PARG activity. The presence of mono-galloyl glucose in gallotannin (Figure 2a) also suggests that these compounds represent the most potent inhibitory components of this natural extract.
Our findings suggest that mono-galloyl glucose derivatives, although characterized by low plasma membrane permeability (see Falsig et al., 2004), can inhibit PAR degradation in intact cells when designed as more lipophilic pro-drugs. Structure–activity relationship analysis of tannins as PARG inhibitors has not been conducted yet. Our finding that mono-galloyl glucose inhibits PARG suggests that this simple structure is, or represents for the most part, the pharmacophore of the complex hydrolysable tannins such as nobotannin (Ying et al., 2001) or sanguinin H-6 (this study). Also, evidence that the methylation of the anomeric carbon of glucose does not affect the inhibitory potency of the compounds suggests that an easy glucose ring opening is not essential for PARG inhibition. These findings, taken together, further our understanding of the mechanisms of inhibition of PARG by tannins, and allow development of more potent PARG inhibitors. In addition, we report the interesting finding that mono-, tri- and pentagalloyl-glucose moieties inhibit PARG, whereas the digalloyl-glucose derivatives are very weak inhibitors. Even though we do not have an explanation for this result, it is worth noting that the binding site(s) of tannins on PARG is (are) unknown. In this regard, studies from Jacobson's group suggest that interaction of PARG with PAR occurs through multiple binding sites that can be differently occupied by chemical inhibitors (Slama et al., 1995a, 1995b; Ramsinghani et al., 1998; Koh et al., 2003a, 2003b). Selective occupancy of one of these putative sites has been postulated to underlie the unusual, non-competitive inhibition of PARG by the product analogue ADP-HPD (Slama et al., 1995b). On this basis, tannins might interact with the extended PAR-binding domain of PARG in a complex manner, and their binding ability to the multiple ADP-ribosyl-binding sites might not be directly proportional to their gallic acid residues. In other words, digalloyl glucose could occupy the enzyme pocket in a manner still permitting binding and hydrolysis of PAR, whereas mono-, tri-, penta- and complex tannin could interact with a number of binding sites which, because of their spatial distribution, more efficiently prevent binding of the polymer to PARG. An additional hypothesis is that tannins interact with PAR rather than PARG. Indeed, both polymers and tannins are highly reactive molecules and their mutual interaction might lead to non-hydrolysable ADP-ribose chains.
Pharmacological inhibition of PARG reportedly affords cytoprotection (Ying et al., 2001; Lu et al., 2003; Cuzzocrea et al., 2005, 2007; Patel et al., 2005; Blenn et al., 2006). Accordingly, we show that both gallotannin (1) and 3-galloyl-1,2-O-isopropylidene glucose (4) reduce MNNG-triggered cell death evaluated 24 h later when present in the incubating media for the first 3 or 6 h, but not 12 or 24 h. Yet a 12 h exposure to the PARG inhibitors partially protects from MNNG toxicity, if cell viability is evaluated at this time point. These findings, together with evidence that both compounds are endowed with intrinsic cytotoxicity, suggest that cytoprotection by PARG inhibition was reduced and, after certain exposure times, was actually abolished, because of the direct toxicity of the compounds. Theoretically, partial protection and intrinsic cytotoxicity might be due to intracellular accumulation of PAR (Figure 4b), a toxic polymer (Andrabi et al., 2006) potentially hindering the cytoprotective effects of PARG inhibition. Overall, this study indicates that acute PARG inhibition is beneficial, reducing or delaying the apoptotic events triggered by PARP-1 (Figure 7), but becomes detrimental when prolonged. This interpretation is in line with prior study showing cell death originating from genetic suppression of PARG (Cortes et al., 2004; Hanai et al., 2004; Koh et al., 2004; Andrabi et al., 2006; Cozzi et al., 2006; Yu et al., 2006). Finally, evidence that radical scavengers do not alter MNNG-dependent cell death (Figure 6a) is at odds with the hypothesis that cytoprotection by tannins is due to their antioxidant properties (Falsig et al., 2004). Conversely, our data strengthen the claims that, under certain experimental settings, the pharmacodynamic effects of tannins are due to PARG inhibition (Tsai et al., 1992; Ying et al., 2001; Dumitriu et al., 2004; Keil et al., 2004; Rapizzi et al., 2004; Cuzzocrea et al., 2005, 2007; Patel et al., 2005; Blenn et al., 2006).
In conclusion, this study identifies two mono-galloyl glucose derivatives as the simplest tannins able to inhibit PARG. Knowledge of the kinetic mechanisms of the enzyme, as well as deeper structural insights into its binding domain(s), is needed to better comprehend its mode of action and develop potent and selective inhibitors. The latter will help clarify the enzyme's function under different pathophysiological conditions as well as the therapeutic relevance of its pharmacological inhibition.
Acknowledgments
This study was supported by grants from the University of Florence, the Italian Ministry of University and Scientific and Technological Research, Associazione Italiana Sclerosi Multipla and Ente Cassa di Risparmio di Firenze.
Abbreviations
- ADP-HPD
ADP-(hydroxymethyl)pyrrolidinediol
- AIF
apoptosis-inducing factor
- MNNG
1-methyl-3-nitro-1-nitrosoguanidine
- PAR
poly(ADP-ribose)
- PARG
poly(ADP-ribose) glycohydrolase
- PARP-1
poly(ADP-ribose) polymerase
- PHE
6(5H)-phenanthridinone
Conflict of interest
The authors state no conflict of interest.
References
- Affar EB, Germain M, Winstall E, Vodenicharov M, Shah RG, Salvesen GS, et al. Caspase-3-mediated processing of poly(ADP-ribose) glycohydrolase during apoptosis. J Biol Chem. 2001;276:2935–2942. doi: 10.1074/jbc.M007269200. [DOI] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, Althaus FR. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat Res. 1989;218:67–74. doi: 10.1016/0921-8777(89)90012-8. [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arapitsas P, Menichetti S, Vincieri FF, Romani A. Hydrolyzable tannins with the hexahydroxydiphenoyl unit and the m-depsidic link: HPLC-DAD-MS identification and model synthesis. J Agric Food Chem. 2007;55:48–55. doi: 10.1021/jf0622329. [DOI] [PubMed] [Google Scholar]
- Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- Blenn C, Althaus FR, Malanga M. Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. Biochem J. 2006;396:419–429. doi: 10.1042/BJ20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonicalzi ME, Vodenicharov M, Coulombe M, Gagne JP, Poirier GG. Alteration of poly(ADP-ribose) glycohydrolase nucleocytoplasmic shuttling characteristics upon cleavage by apoptotic proteases. Biol Cell. 2003;95:635–644. doi: 10.1016/j.biolcel.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Chang P, Jacobson MK, Mitchinson TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Cipriani G, Rapizzi E, Vannacci A, Rizzuto R, Moroni F, Chiarugi A. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J Biol Chem. 2005;280:17227–17234. doi: 10.1074/jbc.M414526200. [DOI] [PubMed] [Google Scholar]
- Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, et al. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi A, Cipriani G, Fossati S, Faraco G, Formentini L, Min W, et al. Poly(ADP-ribose) accumulation and enhancement of postischemic brain damage in 110-kDa poly(ADP-ribose) glycohydrolase null mice. J Cereb Blood Flow Metab. 2006;26:684–695. doi: 10.1038/sj.jcbfm.9600222. [DOI] [PubMed] [Google Scholar]
- Cuzzocrea S, Di Paola R, Mazzon E, Cortes U, Genovese T, Muia C, et al. PARG activity mediates intestinal injury induced by splanchnic artery occlusion and reperfusion. FASEB J. 2005;19:558–566. doi: 10.1096/fj.04-3117com. [DOI] [PubMed] [Google Scholar]
- Cuzzocrea S, Mazzon E, Genovese T, Crisafulli C, Min WK, Di Paola R, et al. Role of poly(ADP-ribose) glycohydrolase in the development of inflammatory bowel disease in mice. Free Radic Biol Med. 2007;42:90–105. doi: 10.1016/j.freeradbiomed.2006.09.025. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- Davidovich L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- Di Meglio S, Denegri M, Vallefuoco S, Tramontano F, Scovassi AI, Quesada P. Poly(ADPR) polymerase-1 and poly(ADPR) glycohydrolase level and distribution in differentiating rat germinal cells. Mol Cell Biochem. 2003;248:85–91. doi: 10.1023/a:1024136927637. [DOI] [PubMed] [Google Scholar]
- Dumitriu IE, Voll RE, Kolowos W, Gaipl US, Heyder P, Kalden JR, et al. UV irradiation inhibits ABC transporters via generation of ADP-ribose by concerted action of poly(ADP-ribose) polymerase-1 and glycohydrolase. Cell Death Differ. 2004;11:314–320. doi: 10.1038/sj.cdd.4401348. [DOI] [PubMed] [Google Scholar]
- Falsig J, Christiansen SH, Feuerhahn S, Burkle A, Oei SL, Keil C, et al. Poly(ADP-ribose) glycohydrolase as a target for neuroprotective intervention: assessment of currently available pharmacological tools. Eur J Pharmacol. 2004;497:7–16. doi: 10.1016/j.ejphar.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Fossati S, Cipriani G, Moroni F, Chiarugi A. Neither energy collapse nor transcription underlie in vitro neurotoxicity of poly(ADP-ribose) polymerase hyper-activation. Neurochem Int. 2007;50:203–210. doi: 10.1016/j.neuint.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol. 2006;18:145–151. doi: 10.1016/j.ceb.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Giovannelli L, Pitozzi V, Riolo S, Dolara P. Measurement of DNA breaks and oxidative damage in polymorphonuclear and mononuclear white blood cells: a novel approach using the comet assay. Mutat Res. 2003;538:71–80. doi: 10.1016/s1383-5718(03)00090-1. [DOI] [PubMed] [Google Scholar]
- Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam E. Natural polyphenols (vegetable tannins) as drugs: possible modes of action. J Nat Prod. 1996;59:205–215. doi: 10.1021/np960040+. [DOI] [PubMed] [Google Scholar]
- Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) J Biol Chem. 1986;261:14902–14911. [PubMed] [Google Scholar]
- Keil C, Petermann E, Oei SL. Tannins elevate the level of poly(ADP-ribose) in HeLa cell extracts. Arch Biochem Biophys. 2004;425:115–121. doi: 10.1016/j.abb.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Khan NS, Ahmad A, Hadi SM. Anti-oxidant, pro-oxidant properties of tannic acid and its binding to DNA. Chem Biol Interact. 2000;125:177–189. doi: 10.1016/s0009-2797(00)00143-5. [DOI] [PubMed] [Google Scholar]
- Koh DW, Coyle DL, Mehta N, Ramsinghani S, Kim H, Slama JT, et al. SAR analysis of adenosine diphosphate (hydroxymethyl)pyrrolidinediol inhibition of poly(ADP-ribose) glycohydrolase. J Med Chem. 2003a;46:4322–4332. doi: 10.1021/jm020541u. [DOI] [PubMed] [Google Scholar]
- Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DW, Patel CN, Ramsinghani S, Slama JT, Oliveira MA, Jacobson MK. Identification of an inhibitor binding site of poly(ADP-ribose) glycohydrolase. Biochemistry. 2003b;42:4855–4863. doi: 10.1021/bi0272048. [DOI] [PubMed] [Google Scholar]
- Labieniec M, Gabryelak T. Effects of tannins on Chinese hamster cell line B14. Mutat Res. 2003;539:127–135. doi: 10.1016/s1383-5718(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Lu XC, Massuda E, Lin Q, Li W, Li JH, Zhang J. Post-treatment with a novel PARG inhibitor reduces infarct in cerebral ischemia in the rat. Brain Res. 2003;978:99–103. doi: 10.1016/s0006-8993(03)02774-4. [DOI] [PubMed] [Google Scholar]
- Maatta-Riihinen KR, Kamal-Eldin A, Torronen AR. Identification and quantification of phenolic compounds in berries of Fragaria and Rubus species (family Rosaceae) J Agric Food Chem. 2004;52:6178–6187. doi: 10.1021/jf049450r. [DOI] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res. 2007;313:2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Kanai M, Hanai S, Uchiumi F, Maruta H, Tanuma S, et al. Subcellular localization of poly(ADP-ribose) glycohydrolase in mammalian cells. Biochem Biophys Res Commun. 2003;307:915–921. doi: 10.1016/s0006-291x(03)01272-5. [DOI] [PubMed] [Google Scholar]
- Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- Ono K, Hasegawa K, Naiki H, Yamada M. Anti-amyloidogenic activity of tannic acid and its activity to destabilize Alzheimer's beta-amyloid fibrils in vitro. Biochim Biophys Acta. 2004;1690:193–202. doi: 10.1016/j.bbadis.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, et al. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolase are protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 2005;16:712–719. doi: 10.1681/ASN.2004080677. [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Putt KS, Hergenrother PJ. A nonradiometric, high-throughput assay for poly(ADP-ribose) glycohydrolase (PARG): application to inhibitor identification and evaluation. Anal Biochem. 2004;333:256–264. doi: 10.1016/j.ab.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Ramsinghani S, Koh DW, Ame JC, Strohm M, Jacobson MK, Slama JT. Syntheses of photoactive analogues of adenosine diphosphate (hydroxymethyl)pyrrolidinediol and photoaffinity labeling of poly(ADP-ribose) glycohydrolase. Biochemistry. 1998;37:7801–7812. doi: 10.1021/bi9730386. [DOI] [PubMed] [Google Scholar]
- Rapizzi E, Fossati S, Moroni F, Chiarugi A. Inhibition of poly(ADP-ribose) glycohydrolase by gallotannin selectively up-regulates expression of proinflammatory genes. Mol Pharmacol. 2004;66:890–898. doi: 10.1124/mol.104.000968. [DOI] [PubMed] [Google Scholar]
- Romani A, Ieri F, Turchetti B, Mulinacci N, Vincieri FF, Buzzini P. Analysis of condensed and hydrolysable tannins from commercial plant extracts. J Pharm Biomed Anal. 2006;41:415–420. doi: 10.1016/j.jpba.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Romani A, Menichetti S, Arapitsas P, Nativi C, Turchetti B, Buzzini P. O-Methylglucogalloyl esters: synthesis and evaluation of their antimycotic activity. Bioorg Med Chem Lett. 2005;15:4000–4003. doi: 10.1016/j.bmcl.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Rossi L, Denegri M, Torti M, Poirier GG, Ivana SA. Poly(ADP-ribose) degradation by post-nuclear extracts from human cells. Biochimie. 2002;84:1229–1235. doi: 10.1016/s0300-9084(02)00017-2. [DOI] [PubMed] [Google Scholar]
- Shieh VM, Ame JC, Wilson MV, Wang ZQ, Kho DW, Jacobson MK, et al. Poly(ADP-ribose)polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- Shirato M, Tozawa S, Maeda D, Watanabe M, Nakagama H, Masutani M. Poly(etheno ADP-ribose) blocks poly(ADP-ribose) glycohydrolase activity. Biochem Biophys Res Commun. 2007;355:451–456. doi: 10.1016/j.bbrc.2007.01.171. [DOI] [PubMed] [Google Scholar]
- Slama JT, Aboul-Ela N, Goli DM, Cheesman BV, Simmons AM, Jacobson MK. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995a;38:389–393. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- Slama JT, Aboul-Ela N, Jacobson MK. Mechanism of inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995b;38:4332–4336. doi: 10.1021/jm00021a023. [DOI] [PubMed] [Google Scholar]
- Tavassoli M, Tavassoli MH, Shall S. Effect of DNA intercalators on poly(ADP-ribose) glycohydrolase activity. Biochim Biophys Acta. 1985;827:228–234. doi: 10.1016/0167-4838(85)90207-9. [DOI] [PubMed] [Google Scholar]
- Tsai YJ, Aoki T, Maruta H, Abe H, Sakagami H, Hatano T, et al. Mouse mammary tumor virus gene expression is suppressed by oligomeric ellagitannins, novel inhibitors of poly(ADP-ribose) glycohydrolase. J Biol Chem. 1992;267:14436–14442. [PubMed] [Google Scholar]
- Ying W, Sevigny MB, Chen Y, Swanson RA. Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci USA. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li J-H. PARP and PARG as novel therapeutic agents. Drugs Future. 2002;27:371–383. [Google Scholar]