Abstract
Many studies have implicated nuclear factor E2-related factor 2 (Nrf2) and nuclear factor-κB1 (Nfkb1) in inflammation and cancer. However, the regulatory potential for crosstalk between these two important transcription factors in inflammation and carcinogenesis has not been explored. To delineate conserved transcription factor-binding site signatures, we performed bioinformatic analyses on the promoter regions of human and murine Nrf2 and Nfkb1. We performed multiple sequence alignment of Nrf2 and Nfkb1 genes in five mammalian species – human, chimpanzee, dog, mouse and rat – to explore conserved biological features. We constructed a canonical regulatory network for concerted modulation of Nrf2 and Nfkb1 involving several members of the mitogen-activated protein kinase (MAPK) family and present a putative model for concerted modulation of Nrf2 and Nfkb1 in inflammation/carcinogenesis. Our results reflect potential for putative crosstalk between Nrf2 and Nfkb1 modulated through the MAPK cascade that may influence inflammation-associated etiopathogenesis of cancer. Taken together, the elucidation of potential relationships between Nrf2 and Nfkb1 may help to better understand transcriptional regulation, as well as transcription factor networks, associated with the etiopathogenesis of inflammation and cancer.
Keywords: Nrf2, Nfkb1, inflammation, carcinogenesis
The National Cancer Institute Inflammation and Cancer Think Tank in Cancer Biology (NCI, 2008) has recognised that epidemiological and clinical research corroborates an increased risk of certain cancers in the setting of chronic inflammation. Indeed, chronic inflammation, because of both infectious and non-infectious etiologies, has been associated with an increased risk of cancer development at a number of organ sites, with infectious agents estimated to be responsible for the development of 18% of all new cancer cases worldwide (Osburn et al, 2007). Infectious agents linked to cancer include hepatitis B virus and liver cancer, Helicobacter pylori and stomach cancer, and liver fluke infection and cholangiocarcinoma (Osburn et al, 2007). In addition, a number of inflammatory conditions without an infectious aetiology result in a significantly increased cancer risk as exemplified by chronic gastroesophageal reflux-induced oesophageal cancer, proliferative inflammatory atrophy-induced prostate cancer and chronic ulcerative colitis-associated colorectal cancer (Schottenfeld and Beebe-Dimmer, 2006; Osburn et al, 2007). Activation of inflammatory cells is accompanied by an increase in the release of reactive oxygen species (ROS) at the site of inflammation. Excess levels of ROS, due to chronic inflammation, may contribute to carcinogenesis by reacting with DNA to form oxidative DNA adducts possibly leading to mutagenesis and impaired regulation of cellular growth (Osburn et al, 2007). Many of the processes involved in inflammation (e.g., leukocyte migration, dilatation of local vasculature with increased permeability and blood flow and angiogenesis), when found in association with tumours, are more likely to contribute to tumour growth, progression and metastasis than to elicit an effective host antitumour response (NCI, 2008). Recently, it has been shown (Dougan and Dranoff, 2008) that signals downstream of the receptor for advanced glycation end products can fuel chronic inflammation, creating a microenvironment that is ideal for tumour formation in a mouse model of skin cancer.
Nuclear factor E2-related factor 2 (Nrf2 or Nfe2l2) is indispensable to cellular defense against many chemical insults of endogenous and exogenous origin, which play major roles in the etiopathogenesis of many cancers and inflammation-related diseases such as inflammatory bowel disease and Parkinson’s disease (Nair et al, 2007a). Under basal conditions, Nrf2 – a member of the Cap-N-Collar family of transcription factors – is sequestered in the cytoplasm by Keap1 resulting in enhanced proteasomal degradation of Nrf2. In conditions of oxidative stress, Nrf2 is released from Keap1 either by direct oxidative modification of Keap1 or after phosphorylation by redox-sensitive protein kinases, translocates to the nucleus and, in combination with other transcription factors, activates transcription of genes containing an antioxidant response element (ARE) in their promoter regions resulting in a cytoprotective adaptive response. This adaptive response is characterised by upregulation of a battery of antioxidative enzymes and decreased sensitivity to oxidative damage and cytotoxicity. These antioxidative enzymes have also been shown to attenuate inflammatory damage and neutralise ROS implicated in inflammatory signalling pathways. We have also reported (Khor et al, 2006) that Nrf2 could play an important role in protecting intestinal integrity, through the regulation of pro-inflammatory cytokines and induction of phase II detoxifying enzymes. Besides, we have demonstrated (Yuan et al, 2006) that mitogen-activated protein kinase (MAPK) pathways such as extracellular signal-regulated kinase (ERK) and c-jun N-terminal kinase (JNK) signalling pathways played important and positive roles in chemopreventive agent butylated hydroxyanisole-induced and Nrf2-dependent regulation of ARE-mediated gene expression, as well as the nuclear translocation of Nrf2 in HepG2 cells. We have observed earlier (Yu et al, 2000) that the activation of MAPK pathways induces ARE-mediated gene expression through a Nrf2-dependent mechanism. In addition, we have also shown (Shen et al, 2004) that different segments of the Nrf2 transactivation domain have different transactivation potentials and that different MAPKs have differential effects on Nrf2 transcriptional activity, with ERK and JNK pathways playing an unequivocal role in the positive regulation of Nrf2 transactivation domain activity (Shen et al, 2004; Nair et al, 2007a).
Importantly, recent mouse studies provide strong and direct genetic evidence that the classical IKK-β (inhibitor of nuclear factor-κB (NF-κB) kinase-β)-dependent NF-κB activation pathway, which was proposed several years ago to be the molecular link between inflammation and carcinogenesis, is a crucial mediator of tumour promotion (Karin and Greten, 2005). Indeed, several pro-inflammatory cytokines and chemokines – such as tumour necrosis factor (TNF), IL-1, IL-6 and CXC-chemokine ligand 8 (CXCL8; also known as IL-8), all of which are encoded by the target genes of the IKK-β-dependent NF-κB activation pathway – are associated with tumour development and progression in humans and mice. It has, thus, been hypothesised that activation of NF-κB by the classical IKK-dependent pathway is a crucial mediator of inflammation-induced tumour growth and progression, as well as an important modulator of tumour surveillance and rejection (Karin and Greten, 2005). We have also demonstrated (Xu et al, 2005) that the suppression of NF-κB and NF-κB-regulated gene expression (VEGF, cyclin D1 and Bcl-XL) by chemopreventive isothiocyanates sulphoraphane and phenethyl isothiocyanate (PEITC) is mainly mediated through the inhibition of IKK phosphorylation, particularly IKK-β, and the inhibition of IκB-α phosphorylation and degradation, as well as the decrease of nuclear translocation of p65 in human prostate cancer PC-3 cells. Recently, it has been observed (Murakami et al, 2007) that PEITC suppresses receptor activator of NF-κB ligand-induced osteoclastogenesis by blocking the activation of ERK1/2 and p38 MAPK in RAW264.7 macrophages. Besides, HL60 cells treated with fisetin presented high expression of NF-κB, activation of p38 MAPK and an increase of phosphoprotein levels (de Sousa et al, 2007).
Pro-inflammatory biomarkers such as IL-1β, IL-6, TNF-α, inducible nitric oxide synthase and cycloxygenase-2, which are all effector genes regulated by the NF-κB pathway, have been noted (Li et al, 2008) to exhibit greater induction in Nrf2-deficient mice as compared with wild-type mice, indicating that ablation of Nrf2 seems to accelerate NF-κB-mediated pro-inflammatory reactions. Besides, it has been shown recently (Liu et al, 2008) that NF-κB competes with Nrf2 for binding to transcriptional coactivator CREB-binding protein and also promotes the recruitment of the corepressor histone deacetylase 3 to MafK leading to local histone hypoacetylation, thus, serving as a negative regulator of Nrf2–ARE signalling. Further, as constitutively active NF-κB occurs in many inflammatory and tumour tissues (Liu et al, 2008), and as Nrf2 is implicated in the etiopathogenesis of many cancers and inflammation-associated conditions (Nair et al, 2007a), we elected to select these two important transcriptional regulators in this study to explore the potential for putative crosstalk between Nrf2 and NF-κB signalling pathways in inflammation/injury and carcinogenesis. We performed in silico bioinformatic analyses to delineate conserved transcription factor-binding sites (TFBSs) or regulatory motifs in the promoter regions of human and murine Nrf2 and Nfkb1, as well as coregulated genes. We performed multiple sequence alignment of Nrf2 and Nfkb1 genes in five mammalian species and studied conserved biological features. We also looked at microarray data from public repositories such as Oncomine (Rhodes et al, 2005), Gene Expression Omnibus (GEO), Public Expression Profiling Resource (PEPR), as well as data sets from the Kong Laboratory, to dissect the role(s) of key regulatory genes in these selected inflammation/cancer signatures and constructed a regulatory network for concerted modulation of Nrf2 and Nfkb1 involving several members of the MAPK family. Our in silico analyses show that concerted modulation of Nrf2 and NF-κB signalling pathways, and putative crosstalk involving multiple members of the MAPK family, may be potential molecular events governing inflammation and carcinogenesis.
Materials and methods
Identification of microarray data sets bearing inflammation/injury or cancer signatures
We perused several microarray data sets from public repositories such as Oncomine, GEO, PEPR, as well as data sets from the Kong Laboratory (Nair et al, 2006, 2007b, 2008). We selected 13 data sets that presented distinct signatures of inflammation or injury or carcinogenesis. Specifically, these studies reflected data on prostate cancer, spinal trauma, inflammatory response to injury and genes modulated by chemopreventive agents/toxicants in Nrf2-deficient animal models. These studies encompassed three mammalian species – human, mouse and rat – and exhibited modulation of both Nrf2 (Nfe2l2) and Nfkb1 genes as well as coregulated genes.
Promoter analyses for transcription factor-binding sites
The promoter analyses were performed in the Cai Laboratory using Genomatix MatInspector (Quandt et al, 1995; Cartharius et al, 2005). Briefly, human promoter sequences of NFE2L2 and NFKB1, or corresponding murine promoter sequences, were retrieved from Gene2Promoter (Genomatix). Comparative promoter analyses were then performed by input of these sequences in FASTA format into MatInspector using optimised default matrix similarity thresholds. The similar and/or functionally related TFBSs were grouped into ‘matrix families,’ and graphical representations of common TFBS were generated. The ‘V$’ prefixes to the individual matrices are representative of the Vertebrate MatInspector matrix library. Similarly, we also elucidated common TFBS among the three topmost conserved human regulatory sequences after multiple alignment (as described below) of NRF2 and NFKB1 sequences.
Multiple species alignment of Nrf2 (Nfe2l2) and Nfkb1 sequences
Non-coding sequences of Nfe2l2 and Nfkb1 genes in five mammalian species – human, chimpanzee, dog, mouse and rat – were retrieved using the Non-Coding Sequence Retrieval System (NCSRS) for comparative genomic analysis of gene regulatory elements that has been previously developed and published (Doh et al, 2007) by the Cai Laboratory, and is readily available at http://cell.rutgers.edu/ncsrs/. Multiple sequence alignment was performed by submitting the non-coding sequences to MLAGAN (Multi-LAGAN, Multi-Limited Area Global Alignment of Nucleotides) (Brudno et al, 2003), which is compatible with the VISTA visualisation tool. The MLAGAN alignments were, thus, visualised using VISTA by projecting them to pairwise alignments with respect to one reference sequence (human) as baseline. The common TFBSs between Nfe2l2 and Nfkb1 between the top biological features that were conserved in multiple species were then determined using Genomatix MatInspector as described earlier in Materials and Methods under Promoter Analyses for TFBS.
Construction and validation of canonical first-generation regulatory network involving Nrf2 (Nfe2l2) and Nfkb1
A putative regulatory network for Nrf2 (Nfe2l2) and Nfkb1, representing 59 nodes and 253 potential interactions, was constructed using Cytoscape 2.5.2 software (Shannon et al, 2003). Further, we validated our network using PubGene (Jenssen et al, 2001), a literature network where connections are strong indicators of biological interaction. Additional validation was achieved by the generation of a biological network with these gene identifiers through the use of Ingenuity Pathways Analysis (Ingenuity Systems®; www.ingenuity.com). For this purpose, a data set containing gene identifiers was uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity. In addition, we used dChip application (Li and Hung Wong, 2001; Li and Wong, 2001) to assess differential expression of MAPKs in cancer vs developmental or non-cancerous tissue/cell lines. Briefly, the CEL files created from each data set were first imported into dChip software for further data characterisation. A gene information file with current annotations and functional gene ontology was generated and the Affymetrix Chip Description File (CDF) was specified. The data were then normalised in dChip, and the expression value for each gene was determined by calculating the average of differences in intensity (perfect match intensity minus mismatch intensity) between its probe pairs. Finally, clustering and enrichment analysis was performed.
A putative model for Nrf2–Nfkb1 interactions in inflammation and carcinogenesis
A pictorial model for Nrf2–Nfkb1 interactions was generated using Pathway Builder Tool 2.0 available from Protein Lounge, San Diego, CA, USA.
Results
Identification of microarray data sets bearing inflammation/injury or cancer signatures
To investigate distinct signatures of inflammation/injury or carcinogenesis, we perused several microarray data sets from public repositories such as Oncomine, GEO, PEPR, as well as microarray data sets from the Kong Laboratory. As summarised in Table 1, we selected 13 data sets that presented distinct signatures of inflammation/injury or carcinogenesis. Specifically, these studies reflected data on prostate cancer, spinal trauma, inflammatory response to injury and genes modulated by chemopreventive agents/toxicants in Nrf2-deficient animal models. These studies encompassed three mammalian species – human, mouse and rat – and exhibited modulation of both Nrf2 (or Nrf2-dependent) and Nfkb1 genes as well as coregulated genes. In other words, all these studies presented modulation of both Nrf2 (Nfe2l2) and Nfkb1 genes in concert, except for the studies with Nrf2-deficient animal models where Nrf2-dependent genes were elucidated. Interestingly, these data sets exhibited modulation of several key members of the MAPK family as well as cofactors of Nrf2 and Nfkb1. This literature pre-screen encouraged us to investigate further the regulatory potential for concerted modulation of Nrf2- and Nfkb1-mediated gene expression in inflammation and carcinogenesis using an in silico bioinformatic approach.
Table 1. Microarray data sets bearing inflammation/injury or cancer signatures.
| Species | Tissue | Study | Data source | Descriptor | Affymetrix platform |
|---|---|---|---|---|---|
| Human | Prostate | Lapointe | Oncomine | Prostate cancer | Non-Affymetrix |
| Human | Prostate | Luo | Oncomine | Prostate cancer and benign prostatic hyperplasia | Non-Affymetrix |
| Mouse | Lung | Kleeberger | GEO | Hyperoxic lung injury | MG U74Av2 |
| Mouse | Lung | Papaiahgari | GEO | Lung injury and inflammatory response | MG 430A 2.0 |
| Mouse | Spleen and liver | Li | GEO | Autoimmune disease and Nrf2 | MG U74Av2 |
| Mouse | Type II cells | Machireddy | GEO | Nrf2 wild-type and knockout cells | MG 430 2.0 |
| Mouse | Prostate | Nair 1 | Kong Laboratory | EGCG+SFN combination treatment | MG 430 2.0 |
| Mouse | Small intestine and liver | Nair 2 | Kong Laboratory | BHA treatment | MG 430 2.0 |
| Mouse | Small intestine and liver | Nair 3 | Kong Laboratory | Induction of ER stress with TM | MG 430 2.0 |
| Rat | Spinal cord | Faden 1 | PEPR | Supraspinal tracts | RG_U34A |
| Rat | Spinal cord | Faden 2 | PEPR | Trauma above T9 | RG_U34A |
| Rat | Spinal cord | Faden 3 | PEPR | Trauma below T9 | RG_U34A |
| Rat | Spinal cord | Faden 4 | PEPR | Trauma T9 | RG_U34A |
BHA=butylated hydroxyanisole; EGCG=epigallocatechin-3-gallate; ER=endoplasmic reticulum; SFN=sulphoraphane; TM=tunicamycin.
All Nair data sets are from the Kong Laboratory as discussed in Materials and Methods; all Faden data sets are as defined by descriptors detailed above at the PEPR resource; all other data sets are as defined by descriptors detailed above at the Oncomine or GEO resources.
Comparative promoter analyses of Nrf2 (Nfe2l2) and Nfkb1 for conserved transcription factor-binding sites
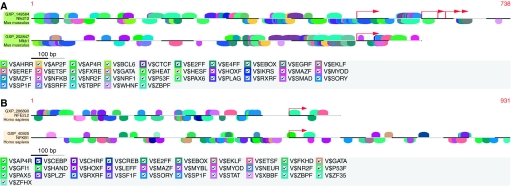
To identify conserved TFBS signatures, we performed comparative analyses of Nrf2 and Nfkb1 murine promoter sequences (Figure 1A) using Genomatix MatInspector (Quandt et al, 1995; Cartharius et al, 2005) as described in Materials and Methods. We also studied NRF2 and NFKB1 human promoter sequences (Figure 1B) similarly. Table 2 includes, as indicated, an alphabetical listing of the conserved vertebrate (V$) matrix families between these two transcription factors. The major human matrix families included activator protein 4 and related proteins, Ccaat/enhancer-binding protein, Camp-responsive element-binding proteins, E2F-myc activator/cell cycle regulator, E-box-binding factors, basic and erythroid krueppel-like factors, fork head domain factors, Myc-associated zinc fingers, nuclear receptor subfamily 2 factors, p53 tumour suppressor, RXR heterodimer-binding sites, SOX/SRY-sex/testis-determining and related HMG box factors, signal transducer and activator of transcription (STAT), X-box-binding factors, Zinc-binding protein factors and two-handed zinc finger homoeodomain transcription factors, among others. Some key conserved murine matrix families included AHR-arnt heterodimers and AHR-related factors, activator protein 2, activator protein 4 and related proteins, E2F-myc activator/cell cycle regulator, E-box-binding factors, basic and erythroid krueppel-like factors, farnesoid X-activated receptor response elements, heat-shock factors, Ikaros zinc finger family, Myc-associated zinc fingers, NF-κB/c-rel, nuclear receptor subfamily 2 factors, nuclear respiratory factor 1, p53 tumour suppressor, pleomorphic adenoma gene, RXR heterodimer-binding sites, SOX/SRY-sex/testis-determining and related HMG box factors, serum response element-binding factor, Tata-binding protein factor, zinc-binding protein factors, among others. Indeed, as evident from Table 2, several matrix families were conserved between Nrf2 and Nfkb1 in both human and murine promoters.
Figure 1.
(A) A conserved TFBS between mouse Nfe2l2 and Nfkb1. Vertebrate (V$) matrix families conserved between murine Nfe2l2 and Nfkb1 promoter regions were identified using Genomatix MatInspector. (B) A conserved TFBS between human NFE2L2 and NFKB1. Vertebrate (V$) matrix families conserved between human NFE2L2 and NFKB1 promoter regions were identified using Genomatix MatInspector.
Table 2. Human and murine matrix families conserved between Nrf2 and Nfkb1.
| Matrix family (Human NRF2 vs NFKB1) | Matrix family (Murine Nrf2 vs Nfkb1) | Matrix family conserved after multiple species alignment (NRF2 vs NFKB1) | Family information |
|---|---|---|---|
| – | V$AHRR | – | AHR-arnt heterodimers and AHR-related factors |
| – | – | V$AIRE | Autoimmune regulatory element-binding factor |
| – | – | V$AP1R | MAF- and AP1-related factors |
| – | V$AP2F | – | Activator protein 2 |
| V$AP4R | V$AP4R | – | Activator protein 4 and related proteins |
| – | – | V$ATBF | AT-binding transcription factor |
| – | V$BCL6 | V$BCL6 | POZ domain zinc finger expressed in B-cells |
| – | – | V$BRAC | Brachyury gene, mesoderm developmental factor |
| – | – | V$BRNF | Brn POU domain factors |
| – | – | V$CAAT | CCAAT-binding factors |
| – | – | V$CART | Cart-1 (cartilage homoeoprotein 1) |
| – | – | V$CDXF | Vertebrate caudal related homoeodomain protein |
| V$CEBP | – | – | Ccaat/enhancer-binding protein |
| V$CHRF | – | V$CHRF | Cell cycle regulators: cell cycle homology element |
| – | – | V$CIZF | CAS-interating zinc finger protein |
| – | – | V$CLOX | CLOX and CLOX homology (CDP) factors |
| – | – | V$COMP | Factors that cooperate with myogenic proteins |
| V$CREB | – | V$CREB | Camp-responsive element-binding proteins |
| – | V$CTCF | – | CTCF and BORIS gene family, transcriptional regulators with 11 highly conserved zinc finger domains |
| V$E2FF | V$E2FF | – | E2F-myc activator/cell cycle regulator |
| – | V$E4FF | – | Ubiquitous GLI-krueppel-like zinc finger involved in cell cycle regulation |
| V$EBOX | V$EBOX | – | E-box-binding factors |
| – | V$EGRF | – | EGR/nerve growth factor-induced protein C and related factors |
| V$EKLF | V$EKLF | V$EKLF | Basic and erythroid krueppel-like factors |
| – | V$EREF | – | Estrogen response elements |
| V$ETSF | V$ETSF | – | Human and murine ETS1 factors |
| – | – | V$EVI1 | EVI1 myeloid-transforming protein |
| V$FKHD | – | V$FKHD | Fork head domain factors |
| – | V$FXRE | – | Farnesoid X-activated receptor response elements |
| V$GATA | V$GATA | V$GATA | GATA-binding factors |
| V$GFI1 | – | V$GFI1 | Growth factor independence transcriptional repressor |
| – | – | V$GREF | Glucocorticoid responsive and related elements |
| – | – | V$GRHL | Grainyhead-like transcription factors |
| V$HAND | – | – | bHLH transcription factor dimer of HAND2 and E12 |
| – | V$HEAT | V$HEAT | Heat-shock factors |
| – | V$HESF | – | Vertebrate homologues of enhancer of split complex |
| – | – | V$HNF1 | Hepatic nuclear factor 1 |
| – | – | V$HNF6 | Onecut homoeodomain factor HNF6 |
| – | – | V$HOMF | Homoeodomain transcription factors |
| – | – | V$HOXC | HOX–PBX complexes |
| V$HOXF | V$HOXF | V$HOXF | Factors with moderate activity to homoeodomain consensus sequence |
| – | V$IKRS | – | Ikaros zinc finger family |
| – | – | V$IRFF | Interferon regulatory factors |
| V$LEFF | – | V$LEFF | LEF1/TCF, involved in the Wnt signal transduction pathway |
| – | – | V$LHXF | Lim homoeodomain factors |
| V$MAZF | V$MAZF | – | Myc-associated zinc fingers |
| – | – | V$MEF2 | MEF2, myocyte-specific enhancer-binding factor |
| V$MYBL | – | – | Cellular and viral myb-like transcriptional regulators |
| V$MYOD | V$MYOD | – | Myoblast-determining factors |
| – | – | V$MYT1 | MYT1 C2HC zinc finger protein |
| – | V$MZF1 | – | Myeloid zinc finger 1 factors |
| V$NEUR | – | V$NEUR | NeuroD, β2, HLH domain |
| – | – | V$NF1F | Nuclear factor 1 |
| – | – | V$NFAT | Nuclear factor of activated T cells |
| – | V$NFKB | – | Nuclear factor κ B/c-rel |
| – | – | V$NKX6 | NK6 homoeobox transcription factors |
| – | – | V$NKXH | NKX homoeodomain factors |
| V$NR2F | V$NR2F | V$NR2F | Nuclear receptor subfamily 2 factors |
| – | V$NRF1 | – | Nuclear respiratory factor 1 |
| – | – | V$OCT1 | Octamer-binding protein |
| – | – | V$OCTP | OCT1-binding factor (POU-specific domain) |
| V$P53F | V$P53F | – | p53 tumour suppressor |
| – | – | V$PARF | PAR/bZIP family |
| V$PAX5 | – | – | PAX-5 B cell-specific activator protein |
| – | V$PAX6 | V$PAX6 | PAX-4/PAX-6 paired domain-binding sites |
| – | – | V$PBXC | PBX1–MEIS1 complexes |
| – | – | V$PDX1 | Pancreatic and intestinal homoeodomain transcription factor |
| – | – | V$PERO | Peroxisome proliferator-activated receptor |
| – | – | V$PIT1 | GHF-1 pituitary-specific pou domain transcription factor |
| – | V$PLAG | – | Pleomorphic adenoma gene |
| V$PLZF | – | V$PLZF | C2H2 zinc finger protein PLZF |
| – | – | V$PRDF | Positive regulatory domain I-binding factor |
| – | – | V$PTF1 | Pancreas transcription factor 1, heterotrimeric transcription factor |
| – | – | V$RBIT | Regulator of B-cell IgH transcription |
| – | – | V$RUSH | SWI/SNF-related nucleophosphoproteins with a RING finger DNA-binding motif |
| V$RXRF | V$RXRF | V$RXRF | RXR heterodimer-binding sites |
| – | – | V$SATB | Special AT-rich sequence-binding protein |
| V$SF1F | – | V$SF1F | Vertebrate steroidogenic factor |
| – | V$SMAD | – | Vertebrate SMAD family of transcription factors |
| – | – | V$SNAP | snRNA-activating protein complex |
| V$SORY | V$SORY | V$SORY | SOX/SRY-sex/testis-determining and related HMG box factors |
| V$SP1F | V$SP1F | V$SP1F | GC-Box factors SP1/GC |
| – | V$SRFF | V$SRFF | Serum response element-binding factor |
| V$STAT | – | V$STAT | Signal transducer and activator of transcription |
| – | – | V$TALE | TALE homoeodomain class-recognising TG motifs |
| – | V$TBPF | V$TBPF | Tata-binding protein factor |
| – | V$WHNF | – | Winged helix-binding sites |
| V$XBBF | – | – | X-box-binding factors |
| V$ZBPF | V$ZBPF | – | Zinc-binding protein factors |
| V$ZF35 | – | – | Zinc finger protein ZNF35 |
| V$ZFHX | – | V$ZFHX | Two-handed zinc finger homoeodomain transcription factors |
Multiple species alignment of Nrf2 (Nfe2l2) and Nfkb1 sequences
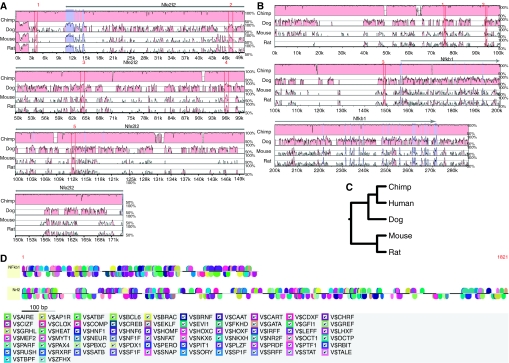
With the objective of investigating conserved biological features across different mammalian species, we performed multiple species alignment of Nrf2 (Nfe2l2) and Nfkb1 sequences as described in Materials and Methods. We used NCSRS for comparative genomic analysis of gene regulatory elements that has been previously developed and published (Doh et al, 2007), and is readily available at http://cell.rutgers.edu/ncsrs/ and retrieved non-coding sequences of Nfe2l2 and Nfkb1 genes in five mammalian species – human, chimpanzee, dog, mouse and rat. As shown in Figures 2A and B, we performed multiple sequence alignment using MLAGAN (Brudno et al, 2003) for Nfe2l2 and Nfkb1 genes, respectively, with respect to one reference sequence (human) as baseline. The phylogenetic tree for Nfe2l2 and Nfkb1 in the five species under consideration was constructed (Figure 2C). The conserved biological features across species for each of Nfe2l2 and Nfkb1 genes were perused, and the top five features for Nfe2l2 and the top three features for Nfkb1, as numbered in Figures 2A and B, are listed in Tables 3A and B, respectively. Sequence 4 for Nfe2l2 and sequence 1 for Nfkb1 exhibited the highest degree of conservation across species at 98.86 and 86.58%, respectively. In addition, the top three conserved sequences in the human sequences of both these genes (sequences 4, 3 and 2 for Nfe2l2 in that order and sequences 1, 2 and 3 for Nfkb1 in that order) as evident from Tables 3A and B were submitted to Genomatix MatInspector. The common TFBSs between Nfe2l2 and Nfkb1 between these biological features that were conserved in multiple species were then determined (Figure 2D) and tabulated along with the other TFBS results for comparative promoter analyses in Table 2 discussed earlier.
Figure 2.
Multiple species alignment. Non-coding sequences of Nfe2l2 and Nfkb1 genes in five mammalian species – human, chimpanzee, dog, mouse and rat – were retrieved using the Non-Coding Sequence Retrieval System (NCSRS) for comparative genomic analysis of gene regulatory elements. Multiple sequence alignment was performed by submitting the non-coding sequences to MLAGAN and visualised by projecting them to pairwise alignments with respect to one reference sequence (human) as baseline. Pink regions, conserved non-coding sequences (CNS); dark blue regions, exons. The numbers indicate CNS that were identified across species. (A) Multiple species alignment for Nfe2l2; (B) multiple species alignment for Nfkb1; (C) phylogenetic tree for Nfe2l2 and Nfkb1; (D) a conserved TFBS between NFE2L2 and NFKB1 among top matching human sequences.
Table 3. (A) Multiple species alignment for Nfe2l2. (B) Multiple species alignment for Nfkb1.
| Nfe2l2 | CNS start | CNS end | % id | Location | Length | Score | Chr | Strand | Start | End | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) | |||||||||||
| 1 | 84.2 | 200.50 | 87.57 | ||||||||
| Human | 4424 | 4568 | – | Intergenic | 144 | 2 | − | 177796698 | 177796842 | ||
| Chimpanzee | 1559 | 12467 | 98.4 | Intergenic | 2b | − | 182248910 | 182259818 | |||
| Dog | 4037 | 4426 | 78.9 | Intergenic | 389 | 36 | − | 24010936 | 24011325 | ||
| Mouse | 3993 | 4131 | 79.6 | Intergenic | 138 | 2 | − | 75470119 | 75470257 | ||
| Rat | 4002 | 4133 | 80.0 | Intergenic | 131 | 3 | − | 58360804 | 58360935 | ||
| 2 | 79.9 | 801.25 | 93.25 | ||||||||
| Human | 46618 | 47576 | – | Intronic | 958 | 2 | − | 177838892 | 177839850 | ||
| Chimpanzee | 16433 | 72741 | 98.0 | Intronic | 2b | − | 182263784 | 182320092 | |||
| Dog | 44082 | 45048 | 76.3 | Intronic | 966 | 36 | − | 24050981 | 24051947 | ||
| Mouse | 49275 | 49911 | 73.0 | Intronic | 636 | 2 | − | 75515401 | 75516037 | ||
| Rat | 48247 | 48892 | 72.3 | Intronic | 645 | 3 | − | 58405049 | 58405694 | ||
| 3 | 81.6 | 746.50 | 94.04 | ||||||||
| Human | 63951 | 64978 | – | Intronic | 1027 | 2 | − | 177856225 | 177857252 | ||
| Chimpanzee | 16433 | 72741 | 98.0 | Intronic | 2b | − | 182263784 | 182320092 | |||
| Dog | 63914 | 64953 | 78.0 | Intronic | 1039 | 36 | − | 24070813 | 24071852 | ||
| Mouse | 69040 | 69553 | 74.9 | Intronic | 513 | 2 | − | 75535166 | 75535679 | ||
| Rat | 68447 | 68854 | 75.5 | Intronic | 407 | 3 | − | 58425249 | 58425656 | ||
| 4 | 82.8 | 962.25 | 98.86 | ||||||||
| Human | 95699 | 97201 | – | Intronic | 1502 | 2 | − | 177887973 | 177889475 | ||
| Chimpanzee | 93603 | 105622 | 98.3 | Intronic | 2b | − | 182340954 | 182352973 | |||
| Dog | 94052 | 95563 | 80.9 | Intronic | 1511 | 36 | − | 24100951 | 24102462 | ||
| Mouse | 99967 | 100384 | 76.4 | Intronic | 417 | 2 | − | 75566093 | 75566510 | ||
| Rat | 103210 | 103629 | 75.7 | Intronic | 419 | 3 | − | 58460012 | 58460431 | ||
| 5 | 82.8 | 418.75 | 89.74 | ||||||||
| Human | 112361 | 112533 | – | Intronic | 172 | 2 | − | 177904635 | 177904807 | ||
| Chimpanzee | 113121 | 117879 | 98.1 | Intronic | 2b | − | 182360472 | 182365230 | |||
| Dog | 105905 | 106887 | 82.1 | Intronic | 517 | 36 | − | 24112804 | 24113786 | ||
| Mouse | 115077 | 115617 | 74.8 | Intronic | 540 | 2 | − | 75581203 | 75581743 | ||
| Rat | 119448 | 119894 | 76.1 | Intronic | 446 | 3 | − | 58476250 | 58476696 | ||
|
(B)
| |||||||||||
| 1 | 78 | 491 | 86.58 | ||||||||
| Human | 75272 | 75516 | – | Intergenic | 244 | 4 | + | 103675774 | 103675545 | ||
| Rat | 190119 | 189890 | 77 | Intergenic | 229 | 2 | − | 233663290 | 233663525 | ||
| Mouse | 585736 | 585501 | 80 | Intergenic | 235 | 3 | − | 135287512 | 135286504 | ||
| Dog | 52140 | 53148 | 78 | Intergenic | 1008 | 32 | + | 26791395 | 26841008 | ||
| Chimpanzee | 67709 | 117322 | 98 | Intergenic | 49613 | 4 | + | 105654198 | 105654198 | ||
| 2 | 80 | 319 | 84.82 | ||||||||
| Human | 93740 | 94016 | – | Intergenic | 276 | 4 | + | 103659473 | 103659195 | ||
| Rat | 173818 | 173540 | 79 | Intergenic | 278 | 2 | − | 233651850 | 233652129 | ||
| Mouse | 574340 | 574061 | 78 | Intergenic | 279 | 3 | − | 135307030 | 135306630 | ||
| Dog | 72266 | 72666 | 81 | Intergenic | 400 | 32 | + | 26791395 | 26841008 | ||
| Chimpanzee | 67709 | 117322 | 98 | Intergenic | 49613 | 4 | + | 105654198 | 105654198 | ||
| 3 | 74 | 469 | 81.92 | ||||||||
| Human | 148427 | 148852 | – | Intergenic | 425 | 4 | + | 103626900 | 103626486 | ||
| Rat | 141245 | 140831 | 73 | Intergenic | 414 | 2 | − | 233620618 | 233620924 | ||
| Mouse | 543135 | 542829 | 73 | Intergenic | 306 | 3 | − | 135342413 | 135341726 | ||
| Dog | 107362 | 108049 | 76 | Intergenic | 687 | 32 | + | 26841253 | 26877988 | ||
| Chimpanzee | 117567 | 154302 | 99 | Intergenic | 36735 | 4 | + | 105654198 | 105654198 | ||
Chr=chromosome; CNS=conserved non-coding sequences.
Construction and validation of a canonical first-generation regulatory network involving Nrf2 (Nfe2l2) and Nfkb1
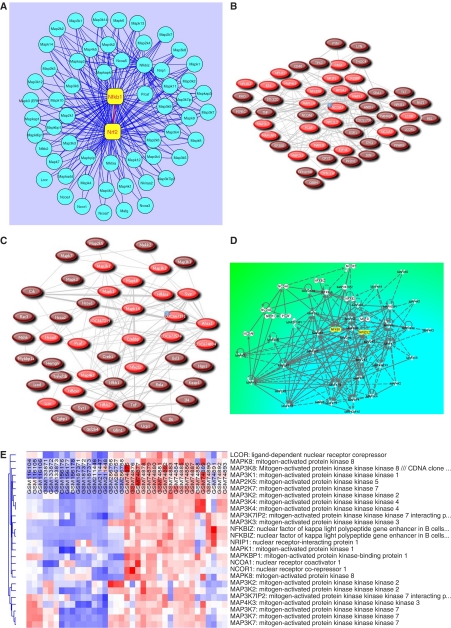
To construct a canonical first-generation biological network for Nrf2–Nfkb1 interactions, we streamlined our study to five data sets summarised in Table 4, which were representative of the most distinct inflammation/injury and cancer signatures from the 13 data sets perused earlier. We obtained gene expression values for 59 genes, as shown in Table 4, including Nrf2 (Nfe2l2), Nfkb1, several cofactors and many members of the MAPK family from these data sets. As shown in Figure 3A, we constructed a putative first-generation regulatory network for Nrf2 (Nfe2l2) and Nfkb1, representing 59 nodes and 253 potential interactions using Cytoscape 2.5.2 software (Shannon et al, 2003). Further, we validated our network by submitting 20 representative genes from Table 4 to PubGene (Jenssen et al, 2001), a literature network where connections are strong indicators of biological interaction, and retrieved biological networks in human (Figure 3B) and mouse (Figure 3C), thus delineating gene signatures that validated the putative biological role(s) of the genes elucidated in this in silico study that served as the source and target nodes in our regulatory network. We further validated our network by querying Ingenuity Pathways Analysis (Ingenuity Systems; www.ingenuity.com) application with the 59 gene identifiers forming the basis of our network and obtained a biological network (Figure 3D) with a high degree of functional crosstalk based on the Ingenuity Knowledge Base reiterating the potential for crosstalk between multiple members of the MAPK family that might modulate the Nrf2–Nfkb1 interactions as indicated in our canonical network (Figure 3A). Furthermore, we used dChip application (Li and Hung Wong, 2001; Li and Wong, 2001) to assess the differential expression of MAPKs in cancerous vs developmental or non-cancerous tissue/cell lines by using several unrelated microarray data sets from the GEO resource at the NCBI including GSM116104–116106: bronchial smooth muscle cells; GSM133871-133873: retinal pigment epithelial cell line; GSM156176–156178: skeletal muscle; GSM187371–187373: untreated LNCaP cells; GSM211446–211448: normal adrenal gland; GSM286756–286758: untreated MCF7 cells; GSM74875–74880: benign prostate tissue; GSM74881–74887: clinically localised primary prostate cancer; and GSM74888–74893: metastatic prostate cancer. Results from this analysis (Figure 3E) show the differential expression of several MAPKs in cancer vs non-cancerous tissue.
Table 4. Canonical first-generation regulatory network members representing putative crosstalk between Nrf2 (Nfe2l2) and Nfkb1 in inflammation-associated carcinogenesisa.
| Sr. no. | GenBank accession no. | Gene name | Gene symbol | Papaiahgari lung injury and inflammation | SFN+EGCG Nrf2-dependent genes | BHA Nrf2- dependent genes | Tunicamycin Nrf2-dependent genes | Faden supraspinal tracts |
|---|---|---|---|---|---|---|---|---|
| 1 | NM_172154 | Ligand-dependent nuclear receptor corepressor | Lcor | – | −3.25 | – | – | – |
| 2 | NM_010756 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) | Mafg | – | – | 2.45 | – | – |
| 3 | NM_008927 | Mitogen-activated protein kinase kinase 1 | Map2k1 | −2.7 | −4.12 | 2.28 | – | 1.59 |
| 4 | NM_023138 | Mitogen-activated protein kinase kinase 2 | Map2k2 | – | – | 2.75 | 1.15 | 1.1 |
| 5 | NM_009157 | Mitogen-activated protein kinase kinase 4 | Map2k4 | −1.55 | −3.31 | – | −1.68 | – |
| 6 | NM_011840 | Mitogen-activated protein kinase kinase 5 | Map2k5 | −1.56 | – | −1.45 | – | 1.59 |
| 7 | NM_011943 | Mitogen-activated protein kinase kinase 6 | Map2k6 | – | −3.98 | – | – | 1.19 |
| 8 | NM_011944 | Mitogen-activated protein kinase kinase 7 | Map2k7 | −1.32 | – | −0.37 | −0.16 | – |
| 9 | NM_011945 | Mitogen-activated protein kinase kinase kinase 1 | Map3k1 | – | – | −0.28 | −0.18 | 1.11 |
| 10 | NM_011946 | Mitogen-activated protein kinase kinase kinase 2 | Map3k2 | – | −5.02 | – | 2.25 | – |
| 11 | NM_011947 | Mitogen-activated protein kinase kinase kinase 3 | Map3k3 | −2.08 | – | – | – | – |
| 12 | NM_011948 | Mitogen-activated protein kinase kinase kinase 4 | Map3k4 | −1.87 | −3.52 | −1.98 | 1.51 | – |
| 13 | NM_008580 | Mitogen-activated protein kinase kinase kinase 5 | Map3k5 | −1.71 | – | – | – | |
| 14 | NM_016693 | Mitogen-activated protein kinase kinase kinase 6 | Map3k6 | 1.58 | −5.25 | 3.25 | – | – |
| 15 | NM_172688 | Mitogen-activated protein kinase kinase kinase 7 | Map3k7 | 1.51 | −3.07 | 2.52 | 3.29 | – |
| 16 | NM_025609 | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 | Map3k7ip1 | – | −3.93 | – | −1.91 | – |
| 17 | NM_138667 | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 | Map3k7ip2 | −1.56 | – | – | – | – |
| 18 | NM_007746 | Mitogen-activated protein kinase kinase kinase 8 | Map3k8 | −2.04 | – | −0.58 | 2.76 | 1.55 |
| 19 | NM_177395 | Mitogen-activated protein kinase kinase kinase 9 | Map3k9 | – | −6.48 | 3.06 | 4.77 | – |
| 20 | NM_009582 | Mitogen-activated protein kinase kinase kinase 12 | Map3k12 | −7.07 | −3.51 | – | – | – |
| 21 | NM_016896 | Mitogen-activated protein kinase kinase kinase 14 | Map3k14 | 1.38 | – | 2.12 | 1.51 | – |
| 22 | NM_008279 | Mitogen-activated protein kinase kinase kinase kinase 1 | Map4k1 | – | −3.35 | – | – | 1.4 |
| 23 | NM_009006 | Mitogen-activated protein kinase kinase kinase kinase 2 | Map4k2 | −6.72 | 2.35 | −1.35 | – | |
| 24 | NM_001081357 | Mitogen-activated protein kinase kinase kinase kinase 3 | Map4k3 | −1.73 | – | – | – | |
| 25 | NM_008696 | Mitogen-activated protein kinase kinase kinase kinase 4 | Map4k4 | −1.58 | −3.54 | 2.46 | – | −0.74 |
| 26 | NM_201519 | Mitogen-activated protein kinase kinase kinase kinase 5 | Map4k5 | −2.45 | −3.28 | 3.94 | −2.52 | – |
| 27 | NM_031248 | Mitogen-activated protein-binding protein-interacting protein | Mapbpip | 2.29 | – | – | – | |
| 28 | NM_001038663 | Mitogen-activated protein kinase 1 | Mapk1 | – | – | – | 1.08 | 1.51 |
| 29 | NM_011952 | Mitogen-activated protein kinase 3 | Mapk3 (ERK1) | −1.4 | −7.48 | – | – | 1.09 |
| 30 | NM_172632 | Mitogen-activated protein kinase 4 | Mapk4 | – | −3.51 | – | – | 1.11 |
| 31 | NM_015806 | Mitogen-activated protein kinase 6 | Mapk6 | – | −3.53 | 2.11 | 1.76 | 1.07 |
| 32 | NM_011841 | Mitogen-activated protein kinase 7 | Mapk7 | −16.44 | – | – | – | −0.99 |
| 33 | NM_016700 | Mitogen-activated protein kinase 8 | Mapk8 | – | – | 10.39 | 12.43 | 1.21 |
| 34 | NM_011162 | Mitogen-activated protein kinase 8-interacting protein 1 | Mapk8ip1 | 2.22 | −10.62 | – | – | −0.84 |
| 35 | NM_016961 | Mitogen-activated protein kinase 9 | Mapk9 | −1.88 | −4.77 | 1.68 | 2.33 | 1.4 |
| 36 | NM_009158 | Mitogen-activated protein kinase 10 | Mapk10 | 1.47 | – | 2.25 | – | 1.29 |
| 37 | NM_011161 | Mitogen-activated protein kinase 11 | Mapk11 | – | −10.9 | 1.78 | 2.32 | – |
| 38 | NM_013871 | Mitogen-activated protein kinase 12 | Mapk12 | – | – | −0.24 | – | −0.88 |
| 39 | NM_011950 | Mitogen-activated protein kinase 13 | Mapk13 | 1.44 | – | – | −0.21 | – |
| 40 | NM_011951 | Mitogen-activated protein kinase 14 | Mapk14 | −1.62 | – | 1.14 | −0.46 | 1.38 |
| 41 | NM_145527 | Mitogen-activated protein kinase-activating death domain | Mapkadd(Madd) | – | 1.03 | |||
| 42 | NM_177345 | Mitogen-activated protein kinase-associated protein 1 | Mapkap1 | −1.62 | −6.56 | −0.34 | – | – |
| 43 | NM_008551 | Mitogen-activated protein kinase-activated protein kinase 2 | Mapkapk2 | – | – | – | 4.31 | 1.34 |
| 44 | NM_178907 | Mitogen-activated protein kinase-activated protein kinase 3 | Mapkapk3 | – | −3.39 | – | 2.38 | 1.19 |
| 45 | NM_010765 | Mitogen-activated protein kinase-activated protein kinase 5 | Mapkapk5 | – | −4.58 | −0.42 | −0.47 | – |
| 46 | NM_011941 | Mitogen-activated protein kinase binding protein 1 | Mapkbp1 | 1.42 | – | – | – | – |
| 47 | NM_010881 | Nuclear receptor coactivator 1 | Ncoa1 | – | −3.55 | – | – | – |
| 48 | NM_008679 | Nuclear receptor coactivator 3 | Ncoa3 | – | – | −0.12 | – | – |
| 49 | NM_144892 | Nuclear receptor coactivator 5 | Ncoa5 | – | – | – | 14.65 | – |
| 50 | NM_172495 | Nuclear receptor coactivator 7 | Ncoa7 | – | −4.46 | – | – | – |
| 51 | NM_011308 | Nuclear receptor corepressor 1 | Ncor1 | – | −5.37 | 3.39 | – | – |
| 52 | NM_008689 | Nuclear factor of kappa light chain gene enhancer in B-cells 1, p105 | Nfkb1 | −1.52 | – | – | – | 1.21 |
| 53 | NM_019408 | Nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 | Nfkb2 | −2.56 | – | – | – | – |
| 54 | NM_010907 | Nuclear factor of kappa light-chain gene enhancer in B-cell inhibitor, alpha | Nfkbia | – | – | – | – | −0.77 |
| 55 | NM_030612 | Nuclear factor of kappa light polypeptide gene enhancer in B cell inhibitor, zeta | Nfkbiz | – | – | – | 2.67 | – |
| 56 | NM_028024 | NFKB inhibitor-interacting Ras-like protein 2 | Nkiras2 | −1.33 | – | – | – | −0.94 |
| 57 | NM_010902 | Nuclear factor, erythroid-derived 2, like 2 | Nrf2 | 1.54 | – | – | – | 1.26 |
| 58 | NM_173440 | Nuclear receptor-interacting protein 1 | Nrip1 | – | – | 2.63 | 2.87 | – |
| 59 | NM_020005 | P300/CBP-associated factor | P/caf | – | – | – | 2.33 | – |
BHA=butylated hydroxyanisole; EGCG=epigallocatechin-3-gallate; ER=endoplasmic reticulum; SFN=sulphoraphane.
Fold-change values are listed.
Figure 3.
A canonical regulatory network for Nrf2–Nfkb1 interactions in inflammation-associated carcinogenesis. (A) A putative regulatory network for Nrf2 (Nfe2l2) and Nfkb1 representing 59 nodes and 253 potential interactions implicating several members of the MAPK family; (B) literature network in humans; (C) literature network in mice; (D) functional crosstalk in biological network of Nfe2l2, Nfkb1 and various members of the MAPK cascade; (E) differential expression of MAPKs in cancer vs developmental or non-cancerous tissue/cell lines (GSM116104–116106: bronchial smooth muscle cells; GSM133871–133873: retinal pigment epithelial cell line; GSM156176–156178: skeletal muscle; GSM187371–187373: untreated LNCaP cells; GSM211446–211448: normal adrenal gland; GSM286756–286758: untreated MCF7 cells; GSM74875–74880: benign prostate tissue; GSM74881–74887: clinically localised primary prostate cancer; GSM74888–74893: metastatic prostate cancer).
A putative model for Nrf2–Nfkb1 interactions in inflammation and carcinogenesis
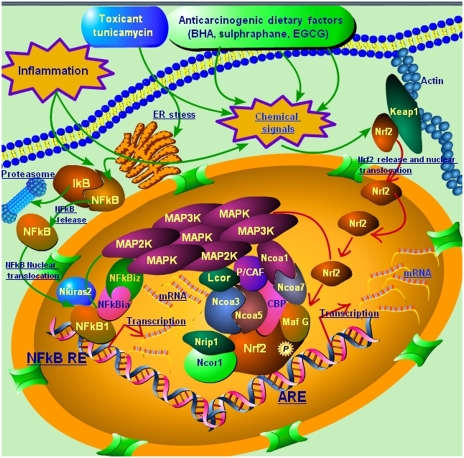
On the basis of the extensive experience (Yu et al, 2000; Li et al, 2005, 2006; Yuan et al, 2006; Nair et al, 2007a; Prawan et al, 2008) of the Kong Laboratory with Nrf2–Keap1 pathway and role(s) of MAPK/dietary chemopreventives/toxicants, our many microarray studies (Shen et al, 2005, 2006; Keum et al, 2006; Nair et al, 2006, 2007b, 2008; Hu et al, 2006a, 2006b; Barve et al, 2008) in Nrf2-deficient mice, our studies (Jeong et al, 2004; Xu et al, 2005) on NF-κB pathway and chemopreventive agents, and the gene signatures elicited in inflammation and carcinogenesis in this in silico study using our data as well as publicly available data from other research groups as indicated earlier, we generated a pictorial model (Figure 4) for Nrf2–Nfkb1 interactions using Pathway Builder Tool 2.0. available from Protein Lounge, San Diego, CA. In essence, chemical signals generated by dietary chemopreventive agents or toxicants, or inflammatory signals, may cause Nrf2 nuclear translocation that sets in motion a dynamic machinery of coactivators and corepressors that may form a multi-molecular complex with Nrf2 to modulate transcriptional response through the ARE. Inflammation may also cause release of NF-κb1 from IκB and stimulate NF-κb1 nuclear translocation to modulate transcriptional response through the NF-κb1 response element, NF-κb-RE, along with cofactors of NF-κb1. Several members of the MAPK family may act in concert with Nrf2 and Nfkb1 with multiple interactions between the members of the putative complex to elicit the chemopreventive and pharmacotoxicological events in inflammation and carcinogenesis.
Figure 4.
A putative model for Nrf2-–Nfkb1 interactions in inflammation and carcinogenesis. Chemical signals generated by dietary chemopreventive agents or toxicants, or inflammatory signals, may cause Nrf2 nuclear translocation that sets in motion a dynamic machinery of coactivators and corepressors that may form a multimolecular complex with Nrf2 for modulating transcriptional response through the antioxidant response element, ARE. Inflammation may also cause release of NF-κB from IκB and stimulate NF-κB nuclear translocation to modulate transcriptional response through the NF-κB response element, NF-κB-RE, along with the cofactors of NF-κB. Several members of the MAPK family may act in concert with Nrf2 and Nfkb1 with multiple interactions between the members of the putative complex to elicit the chemopreventive and pharmacotoxicological events in inflammation and carcinogenesis.
Discussion
Karin and Greten (2005) succinctly noted that carcinogenesis may be divided into three mechanistic phases: initiation (which involves stable genomic alterations), promotion (which involves the proliferation of genetically altered cells) and progression (which involves an increase in the size of the tumour, the spreading of the tumour and the acquisition of additional genetic changes). In 1863, Rudolf Virchow observed leukocytes in neoplastic tissues and suggested (Balkwill and Mantovani, 2001) that the ‘lymphoreticular infiltrate’ reflected the origin of cancer at sites of chronic inflammation. Besides, persistent and recurrent episodes of inflammation (McCulloch et al, 2006), mediated by aberrant activation of innate and acquired immunity, characterise a wide spectrum of idiopathic and infectious chronic inflammatory disorders. In response to tissue injury (Coussens and Werb, 2002), a multifactorial network of chemical signals initiate and maintain a host response designed to ‘heal’ the afflicted tissue involving activation and directed migration of leukocytes (neutrophils, monocytes and eosinophils) from the venous system to sites of damage, and tissue mast cells also play a significant role. Interestingly, inflammation and innate immunity most commonly exert pro-tumorigenic effects (Karin and Greten, 2005) mediated through different types of leukocytes, including normal tissue macrophages, tumour-associated macrophages, dendritic cells, neutrophils, mast cells and T cells, which are recruited to the tumour microenvironment through interactions with local stromal cells and malignant cells. These leukocytes produce cytokines, and growth and angiogenic factors, as well as matrix-degrading proteases (such as the matrix metalloproteinases MMP1, MMP3 and MMP9) and their inhibitors, which allow tumour cells to proliferate, invade and metastasise (Karin and Greten, 2005). Although the causal relationship between inflammation, innate immunity and cancer is more widely accepted, many of the molecular and cellular mechanisms mediating this relationship remain unresolved (Coussens and Werb, 2002). Many studies have implicated Nrf2 (Nfe2l2) in cancer (Ramos-Gomez et al, 2001; Katsuoka et al, 2005; Sporn and Liby, 2005; Nair et al, 2007a; Pearson et al, 2008) or inflammation-associated diseases such as colitis (Khor et al, 2006; Osburn et al, 2007), and Parkinson's disease (Clements et al, 2006), and NF-κB in inflammation (Muller-Ladner et al, 2002; Profita et al, 2008; Puthia et al, 2008) and cancer (Xu et al, 2005; Cilloni et al, 2006; Sun and Zhang, 2007; McDonnell et al, 2008). Indeed, the identification of combinatorial, or synergistic, transcription factors and the elucidation of relationships among them are of great importance for understanding transcriptional regulation as well as transcription factor networks (Hu et al, 2007). However, despite a growing recognition of the important role(s) played by these two pivotal transcription factors, the regulatory potential for crosstalk between these two important transcription factors in inflammation and carcinogenesis has not been explored.
The perusal of several microarray data sets from public repositories such as Oncomine, GEO, PEPR, as well as data sets from the Kong Laboratory, facilitated the identification of 13 data sets (Table 1) presenting distinct signatures of inflammation/injury or carcinogenesis that served as our literature pre-screen for concerted modulation of Nrf2 and Nfkb1 genes. The comparative analyses of TFBS in these two gene promoters revealed that many matrix families were conserved between human NRF2 and NFKB1 promoters, and between murine Nrf2 and Nfkb1 promoter regions (Figure 1 and Table 2). Furthermore, as elucidated in Table 2, several functionally important matrix families were also found to be common across human and murine species, including activator protein 4, E2F-myc activator/cell cycle regulators (V$E2FF), E-box-binding factors, basic and erythroid krueppel-like factors, p53 tumour suppressor and RXR heterodimer-binding sites (V$RXRF), among others. The identification of V$E2FF is significant because disruption of retinoblastoma protein, a key controller of E2F activity and G1/S transition in the cell cycle, can alter the growth-inhibitory potential of TGF-β in the inflammatory milieu of chronic liver disease and contribute to cancer development (Sheahan et al, 2007). Inflammatory conditions can enhance the genotoxic effects of carcinogenic polycyclic aromatic hydrocarbons such as benzo[a]pyrene (BaP) through the upregulation of CYP1B1 expression associated with increased phosphorylation of p53 tumour suppressor at Ser-15 residue, enhanced accumulation of cells in the S-phase of the cell cycle and potentiation of BaP-induced apoptosis (Umannova et al, 2008). Thus, the presence of conserved p53 TFBS in Nrf2/Nfkb1 promoters may point to a critical role for inflammation in the etiopathogenesis of cancer and underscore the relevance of crosstalk between these two transcription factors. The identification of V$RXRF is important as RXR physically interacts with peroxisome proliferator-activated receptor (PPAR-α), a major player in lipid metabolism and inflammation, and PPAR-α agonists such as fenofibrate inhibit NF-κB DNA-binding activity (Xu et al, 2006). In addition, a conserved TFBS for NF-κB itself (Table 2) was found to be present in murine promoter regions of Nrf2 and Nfkb1, strengthening the potential for crosstalk between these two transcription factors. Our multiple sequence alignments (Figures 2A and B and Tables 3A and B) enabled the study of conserved biological features for each gene across five mammalian species and the construction of a phylogenetic tree (Figure 2C). Interestingly, several key biological features were elicited on subjecting the top three conserved human sequences of each gene to comparative promoter analyses (Figure 2D and Table 2). Notably, autoimmune regulatory element-binding factors (V$AIRE) and PPAR (or V$PERO) were conserved in these promoters. Recently, a cyclopentenonic prostaglandin 15-deoxy-delta(12,14)-prostaglandin J(2) has been shown to inhibit TNF-related apoptosis-inducing ligand mRNA expression by downregulating the activity of its promoter in T lymphocytes, with NF-κB being identified as a direct target of this prostanoid that is also regulated by the activation of PPAR-γ (Fionda et al, 2007). The identification of PPAR in the promoter regions of Nrf2 and Nfkb1 in this study, thus, reinforces the significance of these transcription factors and provides a possible mechanistic pathway for crosstalk in inflammation and cancer. Interestingly, a recent study (Kim et al, 2008) has reported that treatment of human brain astrocytes with double-stranded RNA induced interferon regulatory factor 3 (IRF3) phosphorylation and nuclear translocation followed by activation of STAT1 along with a concomitant activation of NF-κB and MAPK cascade members (p38, JNK and ERK). In this study, we identified interferon regulatory factors (IRFF) and STAT as being conserved in Nrf2 and Nfkb1 promoters, as well as MAPK members in our regulatory network (Figure 3A), which agrees with the mechanistic evidence from the brain astrocyte study. It has been observed (Tliba et al, 2006) that stimulation with pro-inflammatory cytokines of CD38, known to be responsible for lung airway inflammation, rendered it insensitive to treatment with glucocorticoids such as fluticasone, dexamethasone or budesonide, by inhibiting steroid-induced glucocorticoid-responsive element (GRE)-dependent gene transcription. We also identified conserved GRE in the Nrf2/Nfkb1 promoters that further validated our results as biologically relevant. In addition, cell cycle regulators, heat-shock factors and several other matrices were found to be conserved between the two genes, thus, underscoring the biological relevance, and the intrinsic complexity, of Nrf2/Nfkb1 crosstalk from a functional standpoint.
Further, we streamlined our study to five data sets (Table 4), which were representative of the most distinct inflammation/injury and cancer signatures of interest and constructed a canonical first-generation regulatory network (Figure 3A) for Nrf2 (Nfe2l2) and Nfkb1, representing 59 nodes and 253 potential interactions. We generated functionally relevant PubGene literature networks in human (Figure 3B) and mouse (Figure 3C), using the Ingenuity Knowledge Base (Figure 3D), thus delineating gene signatures that validated the biological role(s), and potential for crosstalk, of the genes elucidated in this in silico study. We also assessed the expression of MAPKs in several randomly picked, unrelated microarray data sets of cancerous and non-cancerous origin and showed that various MAPKs are differentially expressed in cancer vs developmental or non-cancerous tissue/cell lines (Figure 3E). Our future study includes the expansion of our study objectives to generate more detailed second-generation or third-generation regulatory networks for Nrf2 and Nfkb1 as more functional data emerge on these gene targets of interest and their interactions with coactivator/corepressor modules that associate with them. Interestingly, as shown in our current first-generation network (Figure 3A), several MAPKs play a central role in mediating the transcriptional effects of Nrf2 and Nfkb1. This is, indeed, in consonance with the known role of MAPKs in potentiating Nrf2-mediated ARE activation (Yu et al, 2000; Shen et al, 2004; Yuan et al, 2006) and their role in modulating NF-κB (Murakami et al, 2007; de Sousa et al, 2007), thus, further underscoring the biological applicability of our results. Finally, we present a gestalt pictorial overview (Figure 4) of our current knowledge of concerted modulation of Nrf2 and Nfkb1 based on the data from this study and our extensive experience in cancer chemoprevention.
The results from our current in silico study may strengthen the possibility that scientists could, in the future, consider pursuing Nrf2, Nfkb1 and MAPKs as potential targets in early drug discovery screens for the management of inflammation and cancer. In contemporary times, systems biology has interfaced with the drug discovery process to enable high-throughput screening of multiple drug targets and target-based leads. Indeed, a combination of high-throughput screening, kinase-specific libraries and structure-based drug design has facilitated the discovery of selective kinase inhibitors (Manning and Davis, 2003). Needless to add, the benefits of applying molecular profiling to drug discovery and development include much lower failure rates at all stages of the drug development pipeline, faster progression from discovery through to clinical trials and more successful therapies for patient subgroups (Stoughton and Friend, 2005). Thus, the development of specific inhibitors that might regulate the specific crosstalk between the two central pleiotropic transcription factors Nrf2 and Nfkb1, and with the associated family of kinases, may be one of many strategies that might aid in the drug discovery process. Taken together, our study provides a canonical framework to understand the regulatory potential for concerted modulation of Nrf2 and Nfkb1 in inflammation and cancer. Further studies addressing this question with specific emphasis on cofactor modules binding to these transcription factors and coregulation with upstream signalling molecules in the MAPK cascade will enable a better appreciation of the emerging key role(s) of, and the crosstalk between, these two transcription factors in inflammation and carcinogenesis.
Acknowledgments
This study was supported in part by RO1 CA094828 to Ah-Ng Tony Kong and CA133675 to Li Cai from the National Institutes of Health (NIH).
References
- Balkwill F, Mantovani A (2001) Inflammation and cancer: back to Virchow? Lancet 357: 539–545 [DOI] [PubMed] [Google Scholar]
- Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, Chan JY, Kong AN (2008) Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J Pharm Sci 97(10): 4528–4545 [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Cilloni D, Messa F, Arruga F, Defilippi I, Morotti A, Messa E, Carturan S, Giugliano E, Pautasso M, Bracco E, Rosso V, Sen A, Martinelli G, Baccarani M, Saglio G (2006) The NF-kappaB pathway blockade by the IKK inhibitor PS1145 can overcome imatinib resistance. Leukemia 20: 61–67 [DOI] [PubMed] [Google Scholar]
- Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP (2006) DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci USA 103: 15091–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420: 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa RR, Queiroz KC, Souza AC, Gurgueira SA, Augusto AC, Miranda MA, Peppelenbosch MP, Ferreira CV, Aoyama H (2007) Phosphoprotein levels, MAPK activities and NFkappaB expression are affected by fisetin. J Enzyme Inhib Med Chem 22: 439–444 [DOI] [PubMed] [Google Scholar]
- Doh ST, Zhang Y, Temple MH, Cai L (2007) Non-coding sequence retrieval system for comparative genomic analysis of gene regulatory elements. BMC Bioinformatics 8: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan M, Dranoff G (2008) Inciting inflammation: the RAGE about tumor promotion. J Exp Med 205(2): 267–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fionda C, Nappi F, Piccoli M, Frati L, Santoni A, Cippitelli M (2007) Inhibition of trail gene expression by cyclopentenonic prostaglandin 15-deoxy-delta12,14-prostaglandin J2 in T lymphocytes. Mol Pharmacol 72: 1246–1257 [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN (2006a) Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett 243: 170–192 [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN (2006b) Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci 79: 1944–1955 [DOI] [PubMed] [Google Scholar]
- Hu Z, Hu B, Collins JF (2007) Prediction of synergistic transcription factors by function conservation. Genome Biol 8: R257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenssen TK, Laegreid A, Komorowski J, Hovig E (2001) A literature network of human genes for high-throughput analysis of gene expression. Nat Genet 28: 21–28 [DOI] [PubMed] [Google Scholar]
- Jeong WS, Kim IW, Hu R, Kong AN (2004) Modulatory properties of various natural chemopreventive agents on the activation of NF-kappaB signaling pathway. Pharm Res 21: 661–670 [DOI] [PubMed] [Google Scholar]
- Karin M, Greten FR (2005) NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 5: 749–759 [DOI] [PubMed] [Google Scholar]
- Katsuoka F, Motohashi H, Engel JD, Yamamoto M (2005) Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J Biol Chem 280: 4483–4490 [DOI] [PubMed] [Google Scholar]
- Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, Shakarjian MP, Chong S, Kong AN (2006) Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm Res 23: 2586–2594 [DOI] [PubMed] [Google Scholar]
- Khor TO, Huang MT, Kwon KH, Chan JY, Reddy BS, Kong AN (2006) Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res 66: 11580–11584 [DOI] [PubMed] [Google Scholar]
- Kim H, Yang E, Lee J, Kim SH, Shin JS, Park JY, Choi SJ, Kim SJ, Choi IH (2008) Double-stranded RNA mediates interferon regulatory factor 3 activation and interleukin-6 production by engaging Toll-like receptor 3 in human brain astrocytes. Immunology 124(4): 480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hung Wong W (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2(8): RESEARCH0032.1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jain MR, Chen C, Yue X, Hebbar V, Zhou R, Kong AN (2005) Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J Biol Chem 280: 28430–28438 [DOI] [PubMed] [Google Scholar]
- Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, Kong AN (2008) Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem Pharmacol July 23 [E-pub ahead of print] [DOI] [PMC free article] [PubMed]
- Li W, Yu SW, Kong AN (2006) Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J Biol Chem 281: 27251–27263 [DOI] [PubMed] [Google Scholar]
- Liu GH, Qu J, Shen X (2008) NF-kappaB/p65 antagonizes Nrf2–ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta 1783: 713–727 [DOI] [PubMed] [Google Scholar]
- Manning AM, Davis RJ (2003) Targeting JNK for therapeutic benefit: from junk to gold? Nat Rev Drug Discov 2: 554–565 [DOI] [PubMed] [Google Scholar]
- McCulloch CA, Downey GP, El-Gabalawy H (2006) Signalling platforms that modulate the inflammatory response: new targets for drug development. Nat Rev Drug Discov 5: 864–876 [DOI] [PubMed] [Google Scholar]
- McDonnell TJ, Chari NS, Cho-Vega JH, Troncoso P, Wang X, Bueso-Ramos CE, Coombes K, Brisbay S, Lopez R, Prendergast G, Logothetis C, Do KA (2008) Biomarker expression patterns that correlate with high grade features in treatment naive, organ-confined prostate cancer. BMC Med Genomics 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Ladner U, Gay RE, Gay S (2002) Role of nuclear factor kappaB in synovial inflammation. Curr Rheumatol Rep 4: 201–207 [DOI] [PubMed] [Google Scholar]
- Murakami A, Song M, Ohigashi H (2007) Phenethyl isothiocyanate suppresses receptor activator of NF-kappaB ligand (RANKL)-induced osteoclastogenesis by blocking activation of ERK1/2 and p38 MAPK in RAW264.7 macrophages. Biofactors 30: 1–11 [DOI] [PubMed] [Google Scholar]
- Nair S, Barve A, Khor TO, Shen G, Lin W, Chan JY, Cai L, Kong AN (2008) Regulation of gene expression by a combination of dietary factors sulforaphane (−) epigallocatechin-3-gallate in PC-3 AP-1 human prostate adenocarcinoma cells and Nrf2-deficient murine prostate, Unpublished data, manuscript in preparation
- Nair S, Li W, Kong AN (2007a) Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharmacol Sin 28: 459–472 [DOI] [PubMed] [Google Scholar]
- Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Liew C, Chan JY, Kong AN (2007b) Toxicogenomics of endoplasmic reticulum stress inducer tunicamycin in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Toxicol Lett 168: 21–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Lin W, Keum YS, Liew C, Chan JY, Kong AN (2006) Pharmacogenomics of phenolic antioxidant butylated hydroxyanisole (BHA) in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Pharm Res 23: 2621–2637 [DOI] [PubMed] [Google Scholar]
- NCI (2008) Executive summary of the National Cancer Institute (NCI) inflammation and cancer think tank in cancer biology. Available at http://www.cancer.gov/think-tanks-cancer-biology/page8
- Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW (2007) Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer 121: 1883–1891 [DOI] [PubMed] [Google Scholar]
- Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, Tamashiro KL, Poosala S, Csiszar A, Ungvari Z, Kensler TW, Yamamoto M, Egan JM, Longo DL, Ingram DK, Navas P, de Cabo R (2008) Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci USA 105: 2325–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawan A, Keum YS, Khor TO, Yu S, Nair S, Li W, Hu L, Kong AN (2008) Structural influence of isothiocyanates on the antioxidant response element (ARE)-mediated heme oxygenase-1 (HO-1) expression. Pharm Res 25(4): 836–844 [DOI] [PubMed] [Google Scholar]
- Profita M, Bonanno A, Siena L, Ferraro M, Montalbano AM, Pompeo F, Riccobono L, Pieper MP, Gjomarkaj M (2008) Acetylcholine mediates the release of IL-8 in human bronchial epithelial cells by a NFkB/ERK-dependent mechanism. Eur J Pharmacol 582: 145–153 [DOI] [PubMed] [Google Scholar]
- Puthia MK, Lu J, Tan KS (2008) Blastocystis contains cysteine proteases that mediate interleukin-8 response from human intestinal epithelial cells in a NFkB-dependent manner. Eukaryot Cell 7(3): 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA 98: 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM (2005) Mining for regulatory programs in the cancer transcriptome. Nat Genet 37: 579–583 [DOI] [PubMed] [Google Scholar]
- Schottenfeld D, Beebe-Dimmer J (2006) Chronic inflammation: a common and important factor in the pathogenesis of neoplasia. CA Cancer J Clin 56: 69–83 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan S, Bellamy CO, Dunbar DR, Harrison DJ, Prost S (2007) Deficiency of G1 regulators P53, P21Cip1 and/or pRb decreases hepatocyte sensitivity to TGFbeta cell cycle arrest. BMC Cancer 7: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G, Hebbar V, Nair S, Xu C, Li W, Lin W, Keum YS, Han J, Gallo MA, Kong AN (2004) Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J Biol Chem 279: 23052–23060 [DOI] [PubMed] [Google Scholar]
- Shen G, Xu C, Hu R, Jain MR, Gopalkrishnan A, Nair S, Huang MT, Chan JY, Kong AN (2006) Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther 5: 39–51 [DOI] [PubMed] [Google Scholar]
- Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, Yang CS, Chan JY, Kong AN (2005) Comparison of (−)-epigallocatechin-3-gallate elicited liver and small intestine gene expression profiles between C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Pharm Res 22: 1805–1820 [DOI] [PubMed] [Google Scholar]
- Sporn MB, Liby KT (2005) Cancer chemoprevention: scientific promise, clinical uncertainty. Nat Clin Pract Oncol 2: 518–525 [DOI] [PubMed] [Google Scholar]
- Stoughton RB, Friend SH (2005) How molecular profiling could revolutionize drug discovery. Nat Rev Drug Discov 4: 345–350 [DOI] [PubMed] [Google Scholar]
- Sun XF, Zhang H (2007) NFKB and NFKBI polymorphisms in relation to susceptibility of tumour and other diseases. Histol Histopathol 22: 1387–1398 [DOI] [PubMed] [Google Scholar]
- Tliba O, Cidlowski JA, Amrani Y (2006) CD38 expression is insensitive to steroid action in cells treated with tumor necrosis factor-alpha and interferon-gamma by a mechanism involving the up-regulation of the glucocorticoid receptor beta isoform. Mol Pharmacol 69: 588–596 [DOI] [PubMed] [Google Scholar]
- Umannova L, Machala M, Topinka J, Novakova Z, Milcova A, Kozubik A, Vondracek J (2008) Tumor necrosis factor-alpha potentiates genotoxic effects of benzo[a]pyrene in rat liver epithelial cells through upregulation of cytochrome P450 1B1 expression. Mutat Res 640: 162–169 [DOI] [PubMed] [Google Scholar]
- Xu C, Shen G, Chen C, Gelinas C, Kong AN (2005) Suppression of NF-kappaB and NF-kappaB-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene 24: 4486–4495 [DOI] [PubMed] [Google Scholar]
- Xu J, Chavis JA, Racke MK, Drew PD (2006) Peroxisome proliferator-activated receptor-alpha and retinoid X receptor agonists inhibit inflammatory responses of astrocytes. J Neuroimmunol 176: 95–105 [DOI] [PubMed] [Google Scholar]
- Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN (2000) Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem 275: 39907–39913 [DOI] [PubMed] [Google Scholar]
- Yuan X, Xu C, Pan Z, Keum YS, Kim JH, Shen G, Yu S, Oo KT, Ma J, Kong AN (2006) Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog 45: 841–850 [DOI] [PubMed] [Google Scholar]