Summary
A compelling tool for functional genetics is to silence the expression of multiple related genes concomitantly and reversibly. Such a tool will accelerate the understanding on gene interaction in signaling pathway and the development of comprehensive animal models for human diseases. Multiple gene silencing may be achieved by concurrent expression of multiple miRNA from a Pol II promoter. By comparison, Pol III promoters possess greater capacity to synthesize RNA of high yield and are consisted of compact elements and simple terminators to be convenient for handling. The miRNA-induced gene silencing is a dose-dependent event, and thus, Pol III promoter as a miRNA driver increases the chance to induce phenotypes subsequent to the gene silencing. As a Pol III promoter, endogenous U6 promoter synthesizes small nuclear RNA of high yield and is commonly adapted for miRNA synthesis. Whether U6 promoter is effective to synthesize multiple miRNA in tandem remains to be determined. This study exploited a possibility to express multiple miRNA genes from U6 promoter and also tested the inducibility of varying types of Tet-regulatable U6 promoters. With miR-30a backbone, two miRNA genes were functionally and efficiently expressed from a U6 promoter. The transcriptional activity of Tet-regulatable U6 promoter was tightly regulated by Tetracycline system after sufficient repeats of Tetracycline Operator sequence were introduced within the promoter regions and also between U6 promoter and miRNA gene. This newly developed U6 miRNA system would make multi-gene silencing efficient and reversible.
Keywords: Gene silencing, microRNA, miRNA, RNA polymerase III promoter, U6 promoter, RNA interference, RNAi, Tetracycline regulation
1. Introduction
A class of small noncoding RNA, called microRNA or miRNA, post-transcriptionally regulates gene expression in viruses, plants, and animals [1]. In plants, miRNA initiates sequence-specific cleavage of mRNA molecules with a sequence highly complementary to the miRNA, a process that is similar to RNA interference (RNAi) in gene regulation mechanism [2–4]. In animals, however, miRNA regulates gene expression via a different mechanism. Animal miRNA interferes with translation or mRNA stability because these miRNA hybridize to mRNA molecules with partially complementary miRNA-binding sites that are typically located in the 3’ untranslated regions of the targeted mRNA [1, 5, 6]. In cultured cells and in living animals, artificial miRNA have been shown capable to achieve RNAi effects in mRNA cleavage and in gene inhibition when miRNA is designed to gain a completely-matched binding size in the targeted mRNA [7–11]. Artificial miRNA is increasingly used to silence gene expression and thus to induce hypomorphic phenotypes in cellular and animal models [9–12], attracting wide attention in gene function studies.
Functional genetics would be accelerated by concomitantly silencing the expression of two or more related genes. The concept for concomitant multi-gene silencing has been demonstrated by expression of multiple artificial miRNA from Pol II promoters [13–15]. Compared to Pol II promoters, Pol III promoters such as U6 and H1 possess high and universal transcription activity for a broad and considerable gene expression and also are consisted of compact sequence and simple terminator for easy handling [16]; however, Pol III promoters exhibit a limited capacity to yield long RNA transcript [16, 17]. Thereby, artificial miRNA is commonly expressed from Pol II promoters because a sufficient length of miRNA primary transcript is believed essential for efficient miRNA processing [7–11]. In recent studies, the Pol III promoter U6 is shown to yield a high expression of artificial miRNA with a limited flanking sequence [7, 18], but it still remains unknown whether Pol III promoters such as U6 could be adapted for expression of multiple artificial miRNA in tandem to achieve multiple gene silencing. In addition, reversible gene silencing considerably helps understand the gene functions, and this controllable gene silencing can be achieved by expression of artificial miRNA from Tet-regulatable promoters. Tet-regulatable U6 promoters have been examined [19–22], but shown a considerable leakage (unpublished observation). The limited understanding on Pol III miRNA-expressing system hinders its deserved application to multiple and reversible gene silencing.
This study exploited a possibility to express multiple miRNA genes from a commonly used Pol III promoter—U6 promoter and also tested the inducibility of varying types of Tet-regulatable U6 promoters. With miR-30a backbone, two miRNA genes were functionally and efficiently expressed from the Pol III promoter U6. The transcriptional activity of Tet-regulatable U6 promoter was tightly regulated by Tetracycline system after sufficient repeats of Tetracycline Operator (TetO) sequence were introduced within the promoter regions and also between U6 promoter and miRNA gene. This newly developed Pol III miRNA system would be useful to the studies on gene function in vitro and in vivo.
2. Materials and Methods
2.1. Plasmid construction
The targeting sequence for each miRNA was chosen from the coding sequence of either the mouse c-Jun N-terminal kinase 1 or 3 gene (JNK1; JNK3), and these targeting sequences were used as the guide for miRNA construction. The miRNA plasmids were empirically constructed as described previously [12, 23]. For initial selection of effective miRNA, artificial miRNA was embedded in the miR-30a backbone and expressed from the human cytomegalovirus (CMV) promoter [12]. Single-stranded DNA oligonucleotides were chemically synthesized and then annealed to form double-stranded DNA oligonucleotides with two compatible ends for the restriction sites KpnI or EcoRI. The resulting double-stranded DNA oligonucleotides were inserted at the KpnI and EcoRI restriction sites of pcDNA3.1 plasmid (Invitrogen). Similarly, the selected JNK1 or JNK3 miRNA were inserted downstream of U6 promoter. The U6 promoter was modified by incorporating tetracycline operator sequence (TetO) around its TATA box. Six repeats of TetO sequence were also inserted upstream or downstream of the U6 promoter to yield a tightly regulated U6 promoter. The plasmid pTet-tTs was purchased from Clonetech and used to express the regulating protein—tetracycline responsive transcriptional silencer (tTs). The targeting sequence of each miRNA was inserted at the 3’-end noncoding region downstream of the enhanced green fluorescence protein gene (EGFP) in the plasmid pEGFP-N1 (Clonetech) or downstream of the firefly luciferase gene in the pGL2 vector (Promega). All constructs were verified by DNA sequencing.
2.2. Cell culture and transfection
HEK293 cells were grown in DMEM medium supplemented with 10% fetal bovine serum. When cell confluence reached approximate 80%, cells were detached by trypsin digestion and split into 6-well plates for Northern blotting, into 12-well plates for fluorescence detection, or into 48-well plates for luciferase activity assay. The cells were transfected at a density of approximate 75% and in the presence of fetal bovine serum. For transient transfection, liopfectamine-2000 (Invitrogen) was routinely used as a vehicle to carry plasmids into the transfected cells and usually yielded an efficiency of 90% of the cells transfected. The growth medium was changed every 24 hours.
2.3. Florescence measurement
HEK293 cells were grown in 12-well plates and each well of cells was transfected with a plasmid mixture of two micrograms. The molar ratio of miRNA to its GFP-tagged target was set at 1 to 2. The transfected cells were harvested at 28 hours after transfection, and the cell pellet was lyzed in ice-cold reporter buffer (Promega) supplemented with protease inhibitors (complete, EDTA-free, 1 tablet/10 ml buffer; Roche). The cell lysate was cleared by centrifugation (14000 rpm) at 4°C for 10 minutes. Total protein in the supernatant was measured by BCA assay (Pierce; Rockville IL). Protein concentration in each sample was adjusted to 0.5 mg/ml with the reporter buffer. The fluorescence of green fluorescent protein in 140 µl of samples was measured by fluorescence spectroscopy (Photon Technology International) with excitation at 460 nm and recording from 490 to 570 nm. The spectrum peak detected at 505 nm represented the fluorescence intensity of EGFP. Fluorescence in non-transfected cell lysate was measured as background and subtracted from measurements of the transfected cell lysate [23–25].
2.4. Northern blotting
HEK 293 cells were grown in 6-well plates and each well of the cells was transfected with individual plasmid (2µg/well). The transfected cells were harvested at 24 hours after transfection, and total RNA was extracted from the cell pellets with Trizol reagent (Sigma). RNA samples (20 µg each) were separated on 15% polyacrylamide gels and transferred onto Hybond TM-N+ membranes (Amersham). After UV cross-linking, the membrane was cut into two parts: the low part was probed with an RNA probe complementary to the antisense strand of the artificial miRNA; the upper part was probed with an RNA probe complementary to a part of U6 RNA (the sequence of U6 RNA probe: 5’-TTCACGAATTTGCGTGTCATCCTTGCG-3’). RNA probes were synthesized by in vitro transcription with T7 RNA polymerase and labeled by addition of Digoxigenin-11-uridine-5'-triphosphate instead of uridine-5'-triphosphate during RNA synthesis (RNA probe synthesis kit: Roche). After probing with specific RNA probes, the membranes were incubated with anti-Digoxigenin-AP antibodies (Roche), and the signals were detected with CDP-star kit and documented on a Kodak Image Station.
2.5. Luciferase assay
HEK293 cells were grown in 48-well culture plates and transfected with a mixture (1µg/well) of firefly luciferase (Pp-luc) expressing vector pGL2 with miRNA targeting sequence, rellina luciferase (Rr-luc) expressing vector pRL-TK, and individual miRNA plasmid at a molar ratio of 2:1:1). For gene regulation assay, the plasmid pTet-tTs was added to the mixture with equal mole to the plasmid pGL2. To unlock transcriptional activity of Tet-regulatable U6 promoters, the inducer Doxycycline was added to culture medium (2µg/ml) at 4 hours after transfection and maintained in culture until cell harvest. The cells were harvested at 38 hours after transfection. The luciferase activity in the cleared cell lysate was measured with the Dual Luciferase assay kit (Promega, Madison, Wisconsin, United States) using a Mediators Diagnostika (Vienna, Austria) PhL luminometer. The luciferase activity was defined as the ratio of Pp-luc activity from pGL2 derivative to Rr-luc activity from pRL-TK. Thereafter, the relative luciferase activity was normalized to the control transfected with the miRNA-depleted vector plus two luciferase-expressing vectors.
3. Results
To test the capacity of U6 promoter to drive multiple miRNA genes, we chose c-Jun N-terminal kinase (JNK) genes as miRNA targets because three JNK genes (JNK1, 2, 3) show overlapping expression and redundant functions [26–30]. At present, conventional gene knockout could not achieve a concomitant deletion of the three JNK genes. The technical difficulty in multiple gene silencing may be overcome by miRNA-mediated gene silencing. Three JNK genes share highly homologous sequences that provide an opportunity to locate a commonly effective miRNA-targeting site in the two or more JNK mRNA. From the homologous sequences in the JNK1 and JNK2 genes, we selected two miRNA targeting sites and constructed two artificial miRNA genes (Figure 1: JNK1-miR1 and –miR2). In addition, we selected one unique miRNA targeting site (Figure 1: JNK1-miR3) for the JNK1 gene and three unique sites (Figure 1: JNK3-miR1-3) for the JNK3 gene. These miRNA targeting sequences were cloned downstream of EGFP gene under control of CMV promoter. Artificial miRNA genes were embedded in miR-30a backbone and subsequently cloned downstream of CMV promoter in the pcDNA3.1 plasmid. Individual miRNA and the GFP-tagged target plasmids were contransfected into HEK293 cells. As measured for fluorescence intensity, some miRNA substantially knocked down the expression of their GFP-tagged target (Figure 1), but few exhibited a weak activity in gene silencing. The miRNA JNK1-miR1 was highly effective in gene silencing, and this miRNA targets a common sequence in the JNK1 and JNK2 genes. For the JNK3 gene, two miRNA (Figure 1: JNK3-miR2 and –miR3) significantly silenced the expression of their target gene. Theoretically, a combination of JNK1-miR1 and JNK3-miR3 could silence the expression of the three JNK genes simultaneously.
Figure 1. Selection of effective miRNA targeting the mouse JNK genes.

A, B. For each mouse JNK gene (JNK1 or JNK3), three artificial miRNA were constructed with the backbone of miR-30a gene and expressed under control of the CMV promoter. The sequence of each miRNA-targeting region was selected from the mouse JNK1 or JNK3 mRNA and presented as an order from the 5’-end to the 3’-end. Each miRNA and its GFP-tagged target were contransfected into HEK293 cells for evaluation of the miRNA-mediated gene silencing. Representative photos show that the miRNA silenced the expression of their targets to different degrees. C, D. The transfected cells were lyzed 28 hours after transfection, and the green fluorescence intensity in each sample was quantified.
With the effective miRNA, we first tested the efficiency of U6 promoter to drive single miRNA gene with long flanking sequence. The JNK1-miR1 was embedded in miR-30a backbone because previous studies showed that such a design for artificial miRNA produces an efficient processing of the miRNA primary transcript and gives a better outcome of gene silencing [12]. The JNK1-miR1 along with the incorporated miR-30a sequence was cloned downstream of U6 promoter. Irrelevant sequence of varying length was then inserted upstream or downstream of the JNK1-miR1, resulting in the miRNA primary transcript of different length (Figure 2A). In previous studies, we observed that U6-driven miRNA with the minimal flanking sequence of miR-30a produces a better gene silencing as compared to the counterparts without flanking sequence or with alternative loop sequence [12, 18]. These two kinds of miRNA construction were compared with newly designed miRNA constructs. Northern blotting detected an appropriate processing of the miRNA with long flanking sequence (Figure 2B), but detected unprocessed products of the miRNA without flanking sequence (Figure 2B: lane 1–2). The highest amount of final products was detected for the miRNA with sufficient length of flanking sequence at both ends (Figure 2A, B: construct 7). To test gene silencing effect for each miRNA construct, we transfected HEK293 cells with the individual miRNA and their luciferase tagged targets. All kinds of the miRNA designs were effective in gene silencing, but these miRNA silenced target gene expression to different degrees (Figure 2C). The miRNA with the highest yield produced the most significant gene silencing (Figure 2C). Sufficient length of flanking sequence is required for efficient processing of miRNA primary transcript and also required for effective silencing of target gene expression.
Figure 2. An optimal length of flanking sequence required for efficient processing of U6-synthesized miRNA transcript.

A. Schematics show the structures of each artificial miRNA targeting the mouse JNK1 gene. The loop and flanking sequence of each miRNA is written in light letters, but the coding sequence of each miRNA is written in bolded letters with underline. For constructs 3–8, the JNK1-miR1 was embedded in the miR-30a backbone. B. Northern blotting detected expression of the JNK1-miRNA from various miRNA constructs. Each miRNA construct was transfected into HEK293 cells, and the cellular RNA was extracted 24 hours after transfection. Total RNA (20ug/lane) from each sample was resolved on 15% polyacrylamide gels and transferred onto Hybond TM-N+ membranes that were probed with DIG-labeled RNA probe complementary to the JNK1-miRNA1 coding sequence. The upper part of the same membrane was probed with a DIG-labeled RNA probe complementary to the U6 gene. C. Dual luciferase activity assay measured the gene silencing activity of each miRNA construct. The miRNA target sequence was inserted at the 3’-end of firefly luciferase gene (Pp-luc). Renilla luciferase (Rr-luc) plasmid served as a control for transfection efficiency. HEK 293 cells were transfected with a mixture of the Rr-luc plasmid, the JNK1-Pp-luc plasmid, and the JNK1-miRNA plasmid. The control cells were transfected with an empty U6 plasmid instead of the JNK1-miRNA. Luciferase activity was normalized to the value measured in the lysate of cells transfected with the two luciferase plasmids alone. The values are means with standard deviations (n=8). The construct numbers indicated in B and C match with those in the panel A.
With the defined flanking sequence of optimal length (Figure 2), two miRNA in tandem were expressed from the U6 promoter (Figure 3). In a preceding study, we observed that a long spacing sequence of 500 nucleotides between two adjacent miRNA increases gene silencing effect [13]. However, Pol III promoters usually have a limited capacity to transcribe long RNA [16, 17]. Thereby, we varied the length of spacing sequence between two miRNA and tried to find a minimal spacer for efficient processing of tandem miRNA. When the spacing sequence between two adjacent miRNA was limited to 100 nucleotides, a length of only 5 nucleotides yielded high amount of miRNA and produced effective silencing of target gene expression (Figure 3). Two tandem miRNA did not interfere with each other’s processing or function, but instead, the miRNA yield and the gene silencing effect were substantially increased when two identical miRNA genes were expressed in tandem (Figure 3). The U6 promoter efficiently directed RNA transcription up to 800 nucleotides (data not shown). At least, two miRNA genes could be efficiently transcribed from U6 promoter and functionally processed into final products (Figure 3).
Figure 3. Two functional miRNA in tandem synthesized from the Pol III promoter U6.

A. Schematics show the structure of JNK1-miR1 and JNK3-miR3 embedded in the miR-30a backbone. The miRNA coding sequences were written in bold letters with underline; whereas, the miR-30a backbone was written in light letters. B. Varying constructs were built to incorporate either one or two miRNA under control of the Pol III promoter U6. The length of spacing sequence between two miRNA was varied among the miRNA constructs. C. Northern blotting detected JNK1-miR1 expression from the constructs with the JNK1-miR1 alone or with additional miRNA gene. HEK293 cells were transfected with each construct depicted in the panel B and lyzed for extraction of total RNA at 24 hours after transfection. Total RNA (20ug/lane) from each sample was resolved on polyacrylamide gels and transferred onto Hybond TM-N+ membranes that were probed with DIG-labeled RNA probe complementary to the JNK1-miRNA1 coding sequence. The upper part of the same membrane was probed with a DIG-labeled RNA probe complementary to the U6 gene, serving as a control for equal loading. D. Dual luciferase activity assay measured the gene silencing activity of dual-miRNA expressing construct. The miRNA target sequence was inserted at the 3-end of firefly luciferase gene (JNK1-Pp-luc; or, JNK3-Pp-luc). HEK 293 cells were transfected with a mixture of the Rr-luc plasmid, the JNK1-Pp-luc or JNK3-Pp-luc plasmid, and the miRNA plasmid. The control cells were transfected with an empty U6 plasmid. Luciferase activity in the miRNA-transfected cell lysate was normalized to the value measured in the control cell lysate. The values are means with standard deviations (n=8). The construct numbers indicated in the panels C and D match with those in the panel A.
Next we tried to improve the inducibility of Tet-regulatable U6 promoters. When individual Tetracycline-responsive operator (TetO) was incorporated into U6 promoter around its TATA box, the resulting promoter was largely regulated by Tetracycline system (Figure 4). However, a considerable leakage of the promoter activity was observed at least in transient transfection (Figure 4B, construct 2). Since the U6 promoter can synthesize miRNA with long flanking sequence, we replaced part of the miRNA flanking sequence with six repeats of TetO. The additional TetO sites efficiently blocked the transcriptional activity of the U6 promoter and thus improved the inducibility of the promoter (Figure 4). The transcriptional activity of the modified U6 promoter was subjected to the maximal regulation by Tet system in transient transfection. Addition of TetO sites at the upstream of U6 promoter somewhat improved promoter inducibility, but appreciable leakage of transcriptional activity was still observed (Figure 4). The finding suggested that TetO sites at the downstream of U6 promoter are critical to the promoter inducibility by Tet regulatory system. We then expressed two miRNA genes from the improved Tet-regulatable U6 promoter (Figure 5A). The gene silencing effects for two individual miRNA were not compromised by addition of six TetO sites between the miRNA and the U6 promoter, and the miRNA-mediated gene silencing was tightly regulated by Tetracycline system (Figure 5). These findings indicate that temporal and reversible silencing of multiple related genes was achieved by expression of multiple miRNA from a Tet-regulatable U6 promoter.
Figure 4. Transcriptional activity of the Tet-regulatable U6 promoter subjected to the tight regulation by Tet system.
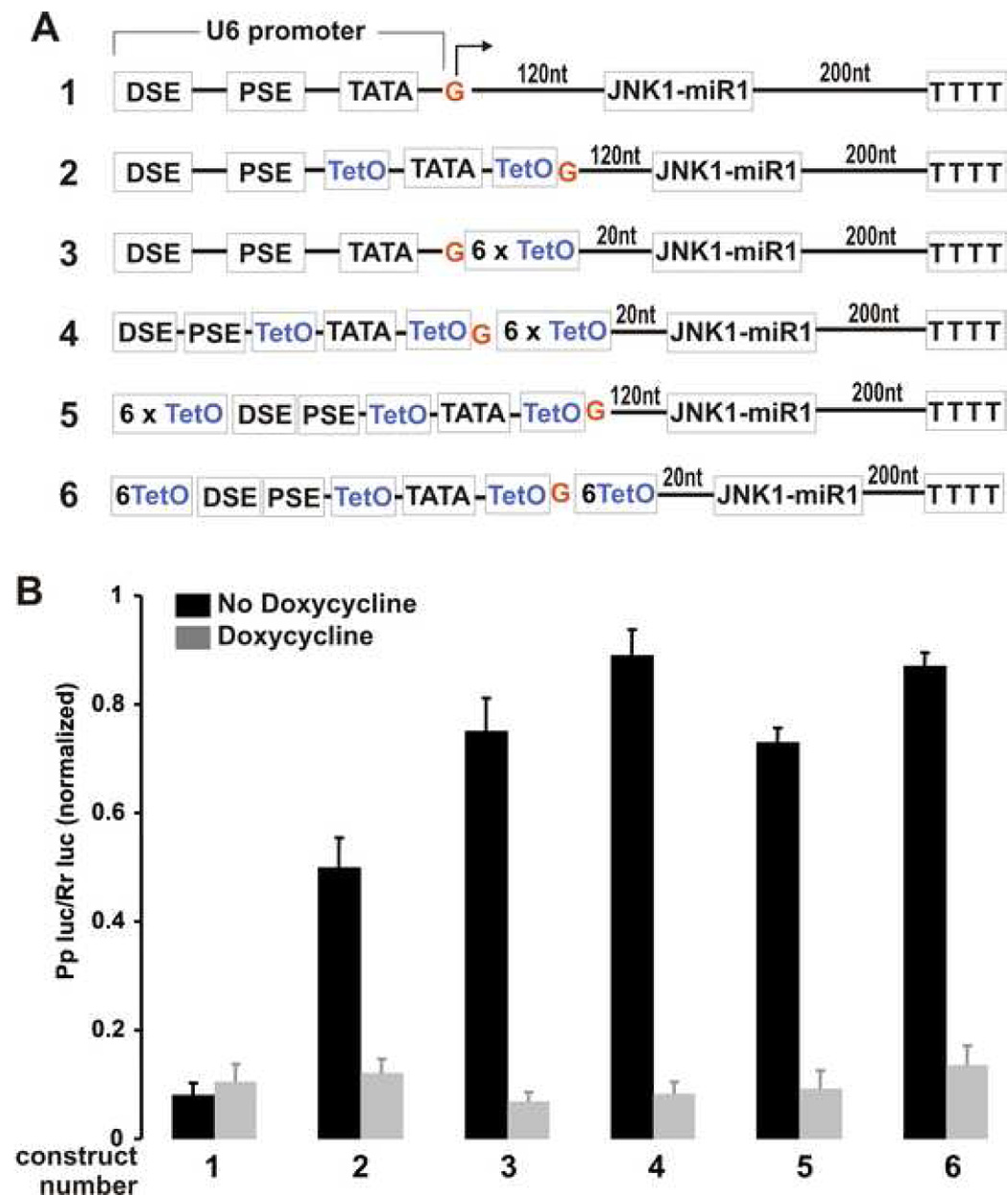
A. Schematics show the structure of Tet-regulatable U6 promoter incorporated with varying combinations of Tetracycline operator sequence (TetO). The U6 promoter consists of three main elements: distance sequence element (DSE), proximate sequence element (PSE), and TATA box. The transcription initiation site for U6 promoter is indicated with the nucleotide G in red. For Tet regulation, the U6 promoter was modified so that the sequence around its TATA box was replaced with one TetO. Six repeats of TetO were inserted at the downstream or the upstream of the native or the modified U6 promoter. B. The transcriptional activity of the modified U6 promoter was regulated by Tet system. HEK293 cells were transfected with a mixture of the JNK1-Pp-luc plasmid, the Rr-luc plasmid, the pTeT-tTs plasmid expressing the Tet-responsive transcriptional silencer, and the JNK1-miR1 plasmid with the native or the modified U6 promoter. In the presence and in the absence of Doxycycline (a derivative of Tetracycline), the transfected cells were allowed to express the transgenes for 38 hours and then were lyzed for measurement of luciferase activity. Luciferase activity in the miRNA-transfected cell lysate was normalized to the value measured in the empty vectortransfected cell lysate. The numerals indicate the constructs depicted in the panel A. The values are means with standard deviations (n=6).
Figure 5. Two miRNA genes expressed from a tightly regulated U6 promoter.
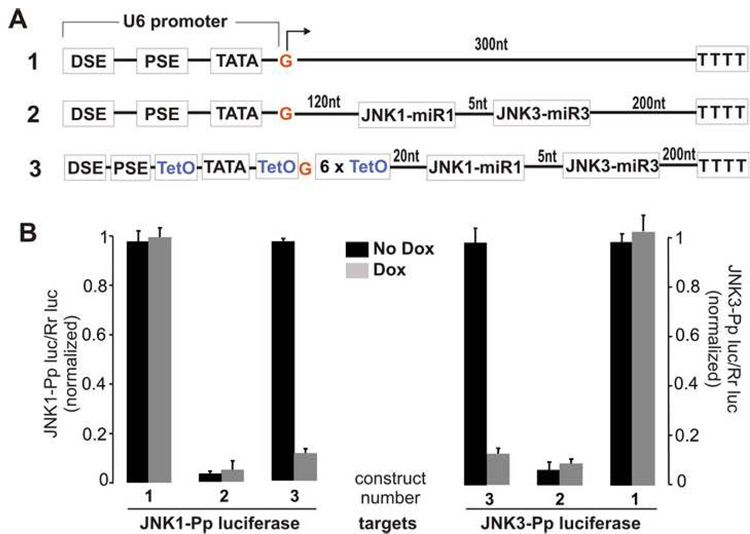
A. Schematics show the construction of the native or the Tet-regulatable U6 promoter for expression of two miRNA genes in tandem. B. Two functional miRNA were expressed from a Tet-regulatable U6 promoter that was tightly regulated by Tet system. HEK293 cells were transfected with a mixture of the JNK1-Pp-luc or the JNK3-Pp-luc plasmid, the Rr-luc plasmid, the pTeT-tTs plasmid, and the miRNA expressing plasmid with the native or the modified U6 promoter. In the absence and in the presence of Doxycycline (Dox), transfected cells were allowed to express the transgenes for 38 hours and then lyzed for measurement of luciferase activity. Luciferase activity in the miRNA-transfected cell lysate was normalized to the value measured in the control cell lysate. The numerals indicate the constructs depicted in the panel A. The values are means with standard deviations (n=8).
4. Discussion
Using Pol II miRNA-expressing system as a template, we developed a Pol III miRNA system for concomitant expression of two or more miRNA genes in tandem. Two tandem miRNA were efficiently expressed from the Pol III promoter U6 and functionally processed from their common primary transcripts. After sufficient TetO sites were integrated, the transcriptional activity of the modified U6 promoter was fully regulated by Tet system. As a result, expression of two tandem miRNA was tightly regulated and their gene silencing effects were reversibly controlled. Using three JNK genes as examples, we were able to silence the expression of two JNK genes simultaneously and conditionally.
In developing this approach, we were concerned that the Pol III promoter U6 was ineffective to transcribe long RNA transcript containing multiple miRNA genes. To produce effective miRNA, we should synthesize miRNA primary transcript long enough to become effective substrate for Drosha-complex. Going through Drosha-processing step, miRNA gains high efficiency in its processing and function [10–12]. As an RNase III enzyme, Drosha requires its substrate of sufficient length and appropriate structure [31, 32]. For efficient processing by Drosha complex, the minimal length of miRNA primary transcript is determined as 110 nucleotides [10–12, 31, 32]. Flanking miRNA minimal transcript, additional sequence significantly enhanced miRNA processing and function (Figure 2). To express two or more miRNA in tandem, U6 promoter is required to be effective in transcribing RNA of more than 500 nucleotides. U6 promoter is an RNA polymerase III promoter and belongs to the type-III subtype [16, 17]. Endogenous U6 promoter directs the synthesis of U6 RNA that is less than 110 nucleotides [17]. Transcriptional efficiency for U6 promoter to transcribe long RNA remains to be determined. By varying the length of flanking sequence around miRNA, we demonstrated that U6 promoter was able to synthesize miRNA-containing transcript of up to 800 nucleotides. Such a length of RNA transcript is sufficient for incorporation of two or three miRNA genes in tandem. To be tightly regulated by Tet system, U6 promoter should be reversibly prevented from transcriptional initiation or elongation. As a pol III promoter, U6 promoter is consisted of highly compact elements that are all located upstream of its transcriptional initiation site [16, 17]. Such a compact structure of U6 promoter provides a full freedom to manipulate the downstream sequence of the promoter, but may not allow for a harsh change in the compacted elements. Indeed, removal of seven nucleotides upstream of TATA box converts U6 promoter into a pol II promoter [33]. Without interrupting U6 promoter elements, we inserted multiple TetO sites immediately downstream of the transcriptional initiation site of U6 promoter. Such a modification subjected U6 promoter to a full regulation by Tet system, and addition of TetO sites did not compromise the capacity of U6 promoter to synthesize multiple miRNA. Compared to previous versions of Tet-regulatable U6 promoters [19–22], this improved version was more responsive to Tet regulation (Figure 4).
Compared to Pol II promoters, Pol III promoters generally possess a greater capacity to synthesize RNA transcripts of a higher yield [16, 34]. In a single cell, U6 promoter-synthesized RNA molecules accumulate more than ten thousand copies [35]. The miRNA-induced gene silencing is a dose-dependent event [24]. Therefore, Pol III miRNA system increases the chance to induce hypomorphic phenotypes subsequent to miRNA-mediated gene silencing [23, 36]. In addition, compact element and simple terminator make U6 promoter convenient for handling, such as assembly of miRNA expression cassette into a viral vector [36, 37]. To study signaling pathway, sometimes we need to block the expression of several related genes simultaneously. For example, three JNK genes perform the same function to phosphorylate c-Jun [26–28]. How these similar genes corporate to maintain cellular functions remains largely unknown; such mysteries may be unraveled by concomitantly silencing the expression of multiple JNK genes. With the developed multi-miRNA expression construct, we are going to test multi-gene silencing in living animals. For multi-gene silencing, multiple U6-miRNA cassettes may be used in a combination [8]. In such a case, the expression of individual miRNA is independent of each other and thus increases the difficulty to control the degrees of individual gene silencing. With an inducible U6 multi-miRNA system, silencing the expression of multiple related genes will become convenient, efficient, and reversible.
Acknowledgement
This work is supported by grants from NIH/NCRR (R21RR024586) to Xu Gang Xia. The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare no conflicts of interest
References
- 1.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 2.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 3.Qi Y, Hannon GJ. Uncovering RNAi mechanisms in plants: biochemistry enters the foray. FEBS Lett. 2005;579:5899–5903. doi: 10.1016/j.febslet.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 4.Tang G, Galili G. Using RNAi to improve plant nutritional value: from mechanism to application. Trends Biotechnol. 2004;22:463–469. doi: 10.1016/j.tibtech.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Fortin K, Mourelatos Z. MicroRNAs: Biogenesis and Molecular Functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fjose A, Drivenes O. RNAi and microRNAs: from animal models to disease therapy. Birth Defects Res C Embryo Today. 2006;78:150–171. doi: 10.1002/bdrc.20069. [DOI] [PubMed] [Google Scholar]
- 7.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez S, Castanotto D, Li H, Olivares S, Jensen MC, Forman SJ, Rossi JJ, Cooper LJ. Amplification of RNAi--targeting HLA mRNAs. Mol Ther. 2005;11:811–818. doi: 10.1016/j.ymthe.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Xia XG, Zhou H, Samper E, Melov S, Xu Z. Pol II-expressed shRNA knocks down Sod2 gene expression and causes phenotypes of the gene knockout in mice. PLoS Genet. 2006;2:27. doi: 10.1371/journal.pgen.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Y, Cai X, Cullen BR. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol. 2005;392:371–380. doi: 10.1016/S0076-6879(04)92022-8. [DOI] [PubMed] [Google Scholar]
- 11.ZENG YAN, CULLEN BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H, Xia XG, Xu Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005;33:e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia XG, Zhou H, Xu Z. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 14.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 16.Medina MF, Joshi S. RNA-polymerase III-driven expression cassettes in human gene therapy. Curr Opin Mol Ther. 1999;1:580–594. [PubMed] [Google Scholar]
- 17.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia XG, Zhou H, Xu Z. Promises and Challenges in Developing RNAi as a Research Tool and Therapy for Neurodegenerative Diseases. Neurodegenerative Diseases. 2005;2:220–231. doi: 10.1159/000089629. [DOI] [PubMed] [Google Scholar]
- 19.Ohkawa J, Taira K. Control of the functional activity of an antisense RNA by a tetracycline-responsive derivative of the human U6 snRNA promoter. Hum Gene Ther. 2000;11:577–585. doi: 10.1089/10430340050015761. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4804. [PubMed] [Google Scholar]
- 21.Matsukura S, Jones PA, Takai D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X, Yang J, Chen J, Gunasekera A, Fesik SW, Shen Y. Development of a tightly regulated U6 promoter for shRNA expression. FEBS Lett. 2004;577:376–380. doi: 10.1016/j.febslet.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Falkenburger BH, Schulz JB, Tieu K, Xu Z, Xia XG. Silencing of the Pink1 Gene Expression by Conditional RNAi Does Not Induce Dopaminergic Neuron Death in Mice. Int J Biol Sci. 2007;3:242–250. doi: 10.7150/ijbs.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia X, Gang , Zhou, Hongxia, Ding, Hongliu, Affar, Bashir E, Shi, Yang, Xu, Zuoshang An enhanced U6 promoter for synthesis of short hairpin RNA. Nucl. Acids. Res. 2003;31:e100. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia XG, Zhou H, Zhou S, Yu Y, Wu R, Xu Z. An RNAi strategy for treatment of amyotrophic lateral sclerosis caused by mutant Cu,Zn superoxide dismutase. J Neurochem. 2005;92:362–367. doi: 10.1111/j.1471-4159.2004.02860.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuan CY, Yang DD, Samanta Roy DR, Davis RJ, Rakic P, Flavell RA. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 27.Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 28.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Genet Dev. 2002;12:14–21. doi: 10.1016/s0959-437x(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 29.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 30.Bogoyevitch MA, Boehm I, Oakley A, Ketterman AJ, Barr RK. Targeting the JNK MAPK cascade for inhibition: basic science and therapeutic potential. Biochim Biophys Acta. 2004;1697:89–101. doi: 10.1016/j.bbapap.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 33.Lobo SM, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 34.Schaub M, Myslinski E, Krol A, Carbon P. Maximization of selenocysteine tRNA and U6 small nuclear RNA transcriptional activation achieved by flexible utilization of a Staf zinc finger. J Biol Chem. 1999;274:25042–25050. doi: 10.1074/jbc.274.35.25042. [DOI] [PubMed] [Google Scholar]
- 35.Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee NS, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, Engelke D, Rossi JJ. The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. Rna. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 36.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluta K, Diehl W, Zhang XY, Kutner R, Bialkowska A, Reiser J. Lentiviral vectors encoding tetracycline-dependent repressors and transactivators for reversible knockdown of gene expression: a comparative study. BMC Biotechnol. 2007;7:41. doi: 10.1186/1472-6750-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]


