Abstract
Photoperiodic control of flowering time is believed to affect latitudinal distribution of plants. The blue light receptor CRY2 regulates photoperiodic flowering in the experimental model plant Arabidopsis thaliana. However, it is unclear whether genetic variations affecting cryptochrome activity or expression is broadly associated with latitudinal distribution of plants. We report here an investigation of the function and expression of two cryptochromes in soybean, GmCRY1a and GmCRY2a. Soybean is a short-day (SD) crop commonly cultivated according to the photoperiodic sensitivity of cultivars. Both cultivated soybean (Glycine max) and its wild relative (G. soja) exhibit a strong latitudinal cline in photoperiodic flowering. Similar to their Arabidopsis counterparts, both GmCRY1a and GmCRY2a affected blue light inhibition of cell elongation, but only GmCRY2a underwent blue light- and 26S proteasome-dependent degradation. However, in contrast to Arabidopsis cryptochromes, soybean GmCRY1a, but not GmCRY2a, exhibited a strong activity promoting floral initiation, and the level of protein expression of GmCRY1a, but not GmCRY2a, oscillated with a circadian rhythm that has different phase characteristics in different photoperiods. Consistent with the hypothesis that GmCRY1a is a major regulator of photoperiodic flowering in soybean, the photoperiod-dependent circadian rhythmic expression of the GmCRY1a protein correlates with photoperiodic flowering and latitudinal distribution of soybean cultivars. We propose that genes affecting protein expression of the GmCRY1a protein play an important role in determining latitudinal distribution of soybeans.
Keywords: blue light, cryptochrome, photoperiodism, photoreceptor
Cryptochromes are blue light receptors that regulate development in plants and the circadian clock in plants and animals (1–3). Plants have at least two types of cryptochromes: cryptochrome 1 (CRY1) and cryptochrome 2 (CRY2) (4, 5). In Arabidopsis, CRY1 mediates mainly blue light control of de-etiolation, whereas CRY2 regulates primarily photoperiodic flowering, defined here as the reaction to change flowering time in response to altered photoperiods (4, 6, 7). In addition to Arabidopsis, cryptochromes have also been studied in other plants, including algae (8), moss (9), fern (10), tomato (11, 12), rapeseed (13), pea (14), and rice (15, 16). Results of these studies indicate that cryptochromes in angiosperms generally regulate developmental aspects in ways that are similar to Arabidopsis.
Light and the circadian clock often regulate gene expression of cryptochromes. For example, the mRNA expression of cryptochrome genes is regulated by the circadian clock in Arabidopsis, tomato, and pea (13, 17, 18), and by blue light in Brassica (19). Most studies of the cryptochrome gene expression are limited to the level of mRNA, which does not necessarily predict the level of protein expression. Blue light regulation of cryptochrome protein expression has been extensively investigated in Arabidopsis. The Arabidopsis CRY2 protein is light labile, whereas the CRY1 protein is light stable; CRY2 is rapidly phosphorylated and degraded in etiolated seedlings exposed to blue light (20–22), by the ubiquitination/26S proteasome apparatus in the nucleus (23). Consistent with CRY2 being a more predominant photoreceptor than CRY1 in the regulation of photoperiodic flowering in Arabidopsis, the protein level of Arabidopsis CRY2, but not CRY1, exhibits a blue light- and photoperiod-dependent diurnal rhythm (24, 25).
As plant species expand their ranges latitudinally, natural selection is likely to favor genetic variations causing the latitudinal clines in flowering time and/or other developmental responses (26, 27). Genetic variations of photoreceptors such as phytochromes and cryptochromes are known to be responsible for some of the natural variations in Arabidopsis (25, 28, 29). For example, a major quantitative trait locus, EDI, which partly accounts for the difference in flowering response to photoperiod between Arabidopsis accessions collected in Northern hemisphere and the Cvi accession collected in the Cape Verde Islands near the equator, encodes a CRY2 variant with the increased protein stability in light (25). However, contrary to the general expectation, a recent study of 150 Arabidopsis accessions appears to show no clear latitudinal cline in flowering time when grown under LD or SD conditions without vernalization (30). Therefore, it remains unclear whether cryptochromes have a broader contribution to the latitudinal distribution of Arabidopsis.
In an attempt to address the question whether the activity or expression of cryptochromes may contribute broadly to the latitudinal distribution of a plant species, we investigated the function and expression of cryptochromes in the facultative SD plant soybean (Glycine max). Soybean was selected for the earlier studies leading to the discovery of photoperiodism in 1920 (31). Most soybean varieties have strong photoperiodic sensitivity, such that soybean is commonly cultivated as different “maturity groups,” each adapted to a narrow latitudinal range (32, 33). The molecular mechanism underlying the “maturity” variation in soybean is almost completely unknown. In this study, we identified six soybean cryptochrome genes that encode four CRY1 (GmCRY1a to GmCRY1d) and two CRY2 (GmCRY2a and GmCRY2b), and investigated in more detail the function, mRNA expression, and protein expression of the GmCRY1a and GmCRY2a genes. Our study demonstrates that, in contrast to Arabidopsis, soybean CRY1 (i.e., GmCRY1a) plays the predominant role in determining flowering time. Consistent with the proposition that soybean GmCRY1a plays a more predominant role regulating photoperiodic flowering, we showed a clear and strong correlation of the circadian rhythmic expression of the GmCRY1a protein with photoperiodic flowering and latitudinal distribution of soybean cultivars.
Results and Discussion
Soybean Cryptochrome Genes.
To investigate possible roles that cryptochromes may play in photoperiodic flowering and its association with the latitudinal distribution of soybean, we searched soybean EST and genome sequence database, identified six soybean cryptochrome-like genes (GmCRY) [supporting information (SI) Fig. S1 and Fig. S2], cloned two representative GmCRY genes (GmCRY1a and GmCRY2a), and prepared antibodies against the more diverged C-terminal domain of GmCRY1a and GmCRY2a (see SI Text). A comparison of the amino acid sequence of GmCRY to that of the Arabidopsis CRY1 and CRY2 indicates that the six GmCRY genes encode 4 CRY1 (GmCRY1a to GmCRY1d) and 2 CRY2 (GmCRY2a and GmCRY2b) apoproteins. As shown in Fig. S1, GmCRY1's have higher sequence similarity to Arabidopsis CRY1 (71–79% identity) than to GmCRY2's (62–65% identity), whereas GmCRY2's are more closely related to Arabidopsis CRY2 (62–65% identity) than to GmCRY1's (52–53% identity). Similar to that found in cryptochromes of other plants, GmCRY's share more extensive sequence similarity in the N-terminal photolyase-like chromophore-binding domains than in the C-terminal domains (Fig. S2). In contrast to the Arabidopsis genome that encodes one CRY1 and one CRY2, the soybean genome encodes twice as many CRY1 as CRY2 (Figs. S1 and S2). Given that CRY1 and CRY2 were most likely derived from gene duplication before the divergence of monocots and dicots ≈150–200 million years ago (2, 34), this phenomenon may be explained by the paleotetraploid nature of soybean. The soybean genome (≈1 Gb) is believed to undergo genome combination, aneuploid loss of chromosomes, and subsequently genome duplication/diploidization (35), which may result in unequal gene duplication or loss of the progenitor cryptochrome genes.
Function of Soybean Cryptochromes.
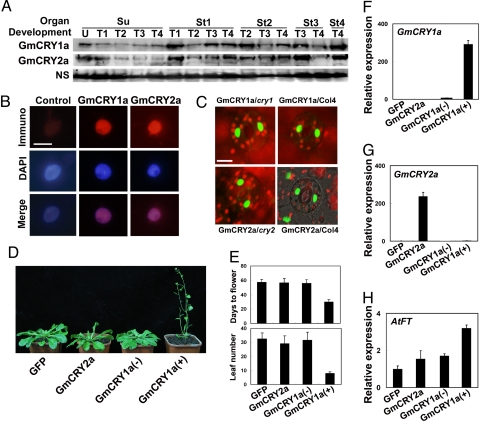
GmCRY1a and GmCRY2a are expressed throughout soybean development, but they appear to express at higher levels in tissues at younger stages of development (Fig. 1A). GmCRY1a and GmCRY2a are nuclear proteins. They were detected in the nuclei of soybean leaf tissues by nuclear immunostaining (Fig. 1B) and in the nuclei of Arabidopsis transgenic plants expressing 35::GFP-GmCRY1a or 35::GFP-CRY2a by GFP fluorescence (Fig. 1C). Similar to previous studies of cryptochromes in other plants (16, 36), GFP-GmCRY1a and GFP-GmCRY2a showed physiological activities mediating blue light inhibition of hypocotyl elongation in transgenic Arabidopsis seedlings (Fig. S3 A–D). Transgenic expression of GFP-GmCRY1a rescued the blue light-specific long hypocotyl phenotype of the Arabidopsis cry1 mutant, and resulted in hypersensitivity to blue light in the wild-type CRY1 background. Similarly, transgenic expression of GFP-GmCRY2 also resulted in hypersensitivity to blue light (Fig. S3 A–D). Given that light inhibition of cell elongation is likely an ancient cellular response, it may not be surprising that this activity of cryptochromes seems universally conserved in different cryptochromes and in different plant species (11–14, 16, 37).
Fig. 1.
Expression, subcellular localization, and function of soybean cryptochromes. (A) Immunoblot showing GmCRY1a and GmCRY2a expression in unifoliolate (Su) and trifoliate leaves (St1, St2, St3, and St4) collected at different developmental stages (U, T1, T2, T3, and T4). (U) T1, T2, T3, and T4 denote the developmental stages, at which the unifoliate leaves, the first, second, third, and the fourth trifoliate leaves fully opened, respectively. (B) Immunostaining showing nuclear localization of GmCRY1a and GmCRY2a. Nuclei isolated from the unifoliate leaves of the 14-day-old etiolated soybean seedlings were probed with anti-GmCRY1a (GmCRY1), anti-GmCRY2a (GmCRY2), or preimmune serum (control), and visualized by DAPI (blue) or fluorescence of rhodamine Red-X conjugated to the goat-anti-rabbit IgG. (Scale bar, 5 μm) (C) GFP fluorescence showing nuclear localization of GFP-GmCRY1a and GFP-GmCRY2a in guard cells of 3-day-old transgenic Arabidopsis seedlings grown under continuous white light. (Scale bar, 10 μm) (D and E) Transgenic expression of 35S::GFP-GmCRY1a, but not 35S::GFP-GmCRY2a, rescued the late-flowering phenotype of the Arabidopsis cry2 mutant. (D) 56-day-old plants of transgenic plants expressing the indicated recombinant proteins in the cry2 mutant background. (E) Flowering time measured by “Days to Flower” and (trifoliate) “Leaf Number” of the indicated genotypes. The phenotype of two independent transgenic lines expressing 35S::GFP-GmCRY1a, one of which [GmCRY1a(+)] expressed high level of GmCRY1a mRNA, but the other line [GmCRY1a(−)] expressed little GmCRY1a mRNA, are shown. Multiple independent lines of each type of transformants exhibited similar phenotypes as the representative lines shown. (F–H) qPCR results showing mRNA expression of the indicated genes in the transgenic lines with the indicated genotype. Note the lack of expression of GmCRY1a in the GmCRY1a(−) and other control lines. AtFT: the Arabidopsis FT gene.
We then examined possible effects of soybean cryptochromes on flowering time, which is apparently a more recent evolutionary “invention” of angiosperm. We first asked whether and which soybean GFP-cryptochrome fusion proteins may rescue the late-flowering phenotype of the Arabidopsis cry2 mutant. Surprisingly, we found that GFP-GmCRY1a, but not GFP-GmCRY2a, rescued the late-flowering phenotype of the cry2 mutant (Fig. 1 D–G). Consistent with this observation, transgenic plants expressing GFP-GmCRY1a, but not GFP-GmCRY2a, also showed accelerated flowering in Arabidopsis of the wild-type CRY2 background. GFP-GmCRY1a promotes flowering by stimulating mRNA expression of the FLOWERING LOCUS T (FT) (Fig. 1H), suggesting a similar mode of action of the soybean GmCRY1a and Arabidopsis CRY2 in the regulation of flowering time (38). Soybean plant transiently transfected by leaf-infiltration with Agrobacterium harboring the Ti plasmid encoding 35S::GFP-GmCRY1a also showed modest but statistically significant acceleration of flowering (Fig. S3 E–G).
Light and Circadian Regulation of the Soybean Cryptochromes.
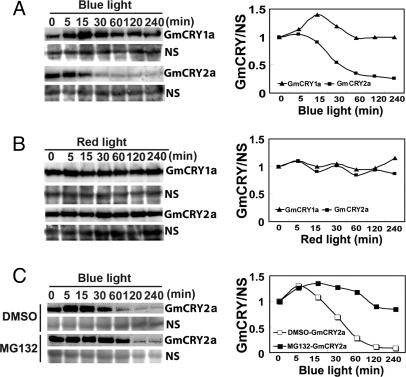
We next tested whether blue light regulation of protein stability of different cryptochromes found in Arabidopsis may be preserved in soybean. In Arabidopsis, CRY2, but not CRY1, undergoes blue light-dependent degradation (22, 23). Similarly, soybean GmCRY2a, but not GmCRY1a, was degraded in blue light (Fig. 2A). In etiolated soybean seedlings exposed to blue light, the level of GmCRY2a decreased rapidly (within 30 min) after blue light treatment, but the GmCRY2a level did not decrease in plants treated with red light for up to 240 min (Fig. 2B). This rapid decline of the GmCRY2a protein in response to blue light was inhibited by the 26S proteasome inhibitor MG132 (Fig. 2C), suggesting that, like Arabidopsis CRY2 (23), soybean GmCRY2a is degraded by the 26S proteasome in response to blue light.
Fig. 2.
GmCRY2a, but not GmCRY1a, undergoes blue light-specific degradation. (A and B) Immunoblots showing GmCRY1a and GmCRY2a in etiolated soybean seedlings exposed to blue light (A) (32 μmol/m2/s) or red light (B) (55 μmol/m2/s) for the indicated time. Protein samples were fractionated by 10% SDS/PAGE, and immunoblots were probed with antibodies against GmCRY1a or GmCRY2a as indicated. NS, a nonspecific band recognized by the antibody that is used to indicate relative loading. Signals of the immunoblot shown on the left were digitized, normalized by the NS signal, and plotted as GmCRY1a/NS on the right. (C) Immunoblots showing inhibition of the blue light-dependent degradation of GmCRY2a by the 26S proteasome inhibitor MG132. Etiolated soybean seedlings were treated with 50 μM MG132, then exposed to blue light for the time indicated, and the immunoblot analyzed by using the anti-GmCRY2a antibody. The relative levels of GmCRY2a proteins were plotted as described in (A and B). Note that different loadings in different lanes shown on the left (NS) were normalized and shown on the right (GmCRY1a/NS).
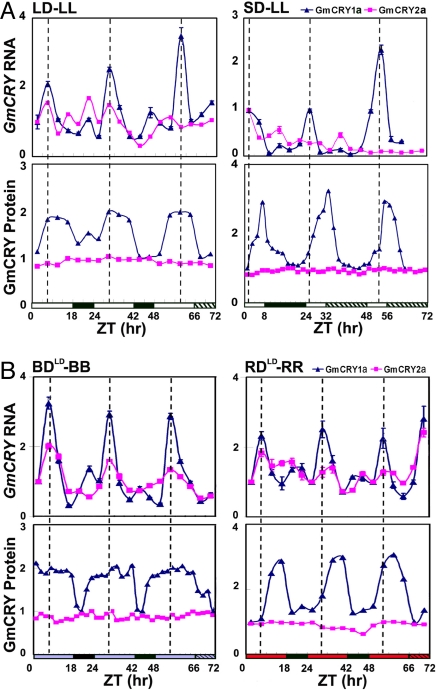
Because blue light-dependent degradation of Arabidopsis CRY2 is thought to be responsible for the photoperiod- and blue light-dependent diurnal rhythm of CRY2 protein expression (24, 25), we tested whether the expression of the blue light-labile GmCRY2a protein would also exhibit a similar diurnal rhythm. We grew soybean in LD (18 hL/6 hD) or SD (8 hL/16 hD) photoperiod (Fig. 3), collected samples every 4 h for 1–2 days, transferred plants to continuous light, collected samples for 1–2 more days, and compared the level of mRNA and protein expression of the GmCRY1a and GmCRY2a genes. Surprisingly, the GmCRY2a protein expression showed neither diurnal rhythm nor circadian rhythms, although its mRNA expression appears to oscillate with a circadian rhythm in LD-entrained conditions, especially when illuminated by blue light (Fig. 3B Upper). This unexpected observation may be explained by that a decrease of the light-labile GmCRY2a protein in the light phase of LD photoperiod is compensated by the increase of the GmCRY2a mRNA expression during this time of the day (Fig. 3B Left). We noted that the GmCRY2a mRNA expression showed no clearly distinguishable circadian rhythm in SD photoperiod (Fig. 3A Right) or in LD photoperiod illuminated by red light, suggesting that a different mechanism may be involved in sustaining a constant cellular level of the light-labile GmCRY2a protein in SD photoperiods.
Fig. 3.
Light and circadian-clock regulation of the GmCRY1a and GmCRY2a genes. (A) Results of qPCR analyses showing the expression of the GmCRY1a and GmCRY2a mRNA (Upper), and immunoblot analyses showing the expression of the GmCRY1a and GmCRY2a protein (Lower) in samples collected at different time from plants treated with different photoperiodic and free-running conditions. LD-LL, samples were collected from unifoliolate leaves of soybean seedlings grown in LD (18 hL/6 hD) for 2 days, and from seedlings transferred to continuous white light for one day, at the time indicated (ZT). SD-LL, samples were collected from seedlings grown in SD (8 hL/16 hD) for one day, and from seedlings transferred to continuous white light for two days, at the time indicated. Black bar: dark phase, white bar: light phase, hatched bar: subjective dark phase but illuminated with light. The triangle and square symbols denote GmCRY1a mRNA (Upper) or protein (Lower), and GmCRY2a mRNA (Upper) or protein (Lower), respectively. Dotted lines indicate the peak time of GmCRY1a mRNA expression. Similar experiments were repeated with similar results, and results of the representative experiment are shown. The last three data points for GmCRY1a in SD-LL were omitted, because of inconsistence in results of those data points in different experiments. (B) Similar to A, but the samples were collected from seedlings treated with different light condition. BDLD-BB, samples were collected for 2 days from unifoliolate leaves of soybean seedlings grown in LD (18hL/6hD) illuminated by blue light, and from seedlings transferred to continuous blue light for two more day, at the time indicated (ZT). RDLD-RR, samples were collected for 2 days from unifoliolate leaves of soybean seedlings grown in LD (18 hL/6 hD) illuminated by red light, and then from seedlings transferred to continuous red light for two additional days.
In contrast to GmCRY2a, both GmCRY1a mRNA and GmCRY1a protein expressions exhibited circadian rhythms (Fig. 3A). The circadian rhythmic expression of the GmCRY1a mRNA partially explains why the level of the light-stable GmCRY1a protein oscillates (Fig. 3). The circadian rhythm of the GmCRY1a mRNA (Fig. 3B Upper) and the GmCRY1a protein expression (Fig. 3B Lower) were similarly observed in photoperiods illuminated by either red light or blue light, suggesting that the circadian clock is regulated redundantly by cryptochromes and phytochromes in not only Arabidopsis (39), but also soybean.
A comparison of the GmCRY1a protein expression in LD and SD revealed two distinct phase characteristics in response to different photoperiods (Fig. 3A). First, the circadian rhythm of the GmCRY1a protein expression in LD and SD had different phase shapes, with the peak level of the GmCRY1a protein expression sustained for the duration that is at least twice as long in LD (>8 h) as that in SD (<4 h) (Fig. 3A Lower). Second, the time that the GmCRY1a protein expression reaches the peak level and its relationship with the time that the level of the GmCRY1a mRNA expression reaches the peak level are different in LD and SD. In LD photoperiods, the protein level of GmCRY1a reached a broad “peak” at approximately noon or subjective noon, which was approximately the same time its mRNA reached the peak level (Fig. 3A Left). In SD photoperiods, the GmCRY1a protein expression reached the peak level at approximately dusk or subjective dusk, which was ≈3 to 5-h lagging behind the time its mRNA reached the peak level (Fig. 3A Right). The differential phase characteristics of the GmCRY1a protein expression in response to different photoperiods are consistent with GmCRY1a being a photoreceptor regulating photoperiodic flowering. Moreover, the photoperiod-dependent deviation in the kinetics of the GmCRY1a protein expression from that of its the mRNA expression indicates that, in addition to the circadian control of the GmCRY1a mRNA expression, other post-transcriptional mechanisms must also be involved in the regulation of the GmCRY1a protein expression. It is intriguing that the kinetics of GmCRY1a protein expression in plants grown in LD illuminated with red light (Fig. 3B, RDLD-RR), which showed a narrow peak lagging behind its mRNA expression, was more similar to that observed in SD illuminated with white light (Fig. 3A, SD-LL) than that found in LD illuminated with white light (Fig. 3A, LD-LL). In contrast, the kinetics of the GmCRY1a protein expression in plants grown in LD illuminated with blue light (Fig. 3B, BDLD-BB) was almost identical to that observed in LD illuminated with white light (Fig. 3A, LD-LL). These results suggest a possible involvement of phytochromes in posttranscriptional regulation of the GmCRY1a protein expression. Regardless of the exact mechanism regulating GmCRY1a expression, the photoperiod-dependent diurnal rhythm of the CRY2 expression in Arabidopsis (24, 25) and the photoperiod-modulated circadian rhythm of GmCRY1a expression in soybean (Fig. 3) appear remarkably consistent with Arabidopsis CRY2 and soybean GmCRY1a being the major cryptochromes that regulate photoperiodic flowering in the respective plant species (5) (Fig. 1 and Fig. S3).
Latitudinal Cline in Photoperiodic Flowering of Soybeans.
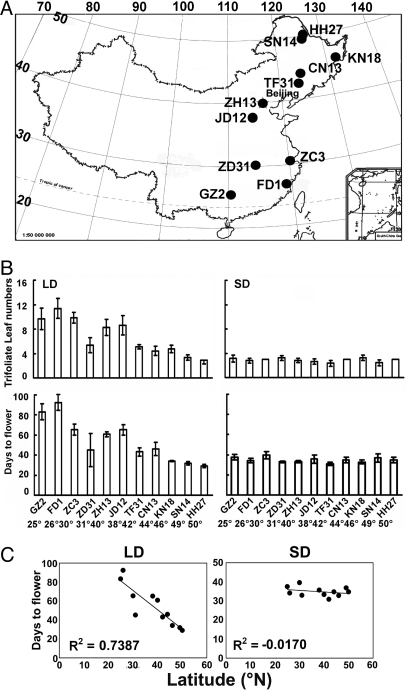
Although photoperiodic control of flowering time in soybean was extensively studied in the early 20th century, there is surprisingly little information concerning latitudinal cline in photoperiodic flowering of soybean examined in defined photoperiod and temperature conditions (7). To further understand the role of cryptochrome in soybean photoperiodic flowering, we analyzed photoperiodic responses of flowering time of soybean cultivars collected from areas in China that range from ≈25°N to ≈50°N (Fig. 4A). When those soybean cultivars were grown in SD photoperiods (8 hL/16 hD), they flowered at approximately the same time, regardless of the latitude of the site of cultivation (Fig. 4B). In contrast, when plants were grown in LD photoperiods (16 hL/8 hD), the cultivars collected from lower latitude flowered later than those collected from higher latitudes (Fig. 4B). A linear regression analysis demonstrated that there is no correlation (R2 = 0.017) between flowering time of cultivars grown in SD photoperiod and latitude of the site of cultivation (Fig. 4C, SD). In contrast, there is a clear and strong correlation (R2 = 0.7387, P < 0.001) between flowering time of those cultivars grown in LD photoperiod and latitude of the site of cultivation of the respective cultivars (Fig. 4C, LD). Soybean (G. max) was domesticated in China from its wild ancestor (G. soja) at least 3,100 year ago (40), therefore, we examined flowering time of 328 wild soybean accessions collected in China. A “common-garden” experiment, performed in a field near Beijing (≈40°N, ≈116°E) in mostly LD photoperiods, demonstrated a latitudinal cline of photoperiodic flowering in wild soybeans (Fig. S4), which is slightly stronger (R2 = 0.8223, P < 0.0001) than that of the domesticated soybean cultivars.
Fig. 4.
A latitudinal cline in flowering time of soybean cultivars (G. max). (A) A diagram showing the geographic centers of cultivation areas of cultivars examined. Numbers on the top or left of the map of China indicate longitude (°E) or latitude (°N), respectively. (B) Flowering time, presented as “Days to Flower” or “Trifoliate Leaf Numbers” at the time of flowering of indicated cultivars grown in LD (18 hL/6 hD) or SD (8 hL/16 hD) with constant temperature (25–28°C). The means and standard deviations (n ≥ 20) were shown. Latitude of the site of cultivation and the cultivar accessions are indicated. (C) Correlation of flowering time and latitude of indicated cultivars grown in LD (R2 = 0.7387, P < 0.004) or SD (R2 = 0.017).
Association of the Circadian Rhythmic Expression of GmCRY1a and Latitudinal Cline in Photoperiodic Flowering of Soybean.
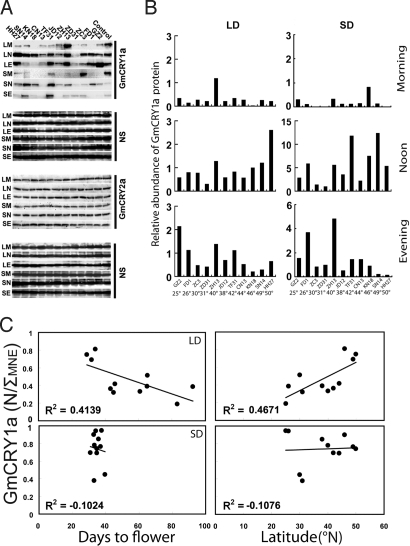
We next analyzed protein expression of cryptochromes in the soybean cultivars grown in LD and SD photoperiods. We collected samples in the morning, noon, and evening, from different cultivars grown in LD or SD photoperiods. The relative abundance of the GmCRY1a protein was analyzed by immunoblot and estimated by two-way normalization, in which the GmCRY1a band signal was normalized for both the relative loading and the variable signal strengths of different immunoblots (see SI Text). Results of this study demonstrated that the GmCRY2a protein expressed constantly throughout the day in different cultivars grown in LD and SD photoperiods (Fig. 5A), whereas the abundance of GmCRY1a oscillated and reached the peak level at noon in most cultivars grown in LD photoperiods (Fig. 5 A and B and Fig. S5). Consistent with GmCRY1a being a positive regulator of floral initiation in soybean that flower earlier in SD than in LD, the relative level of GmCRY1a protein expression was markedly higher in SD than in LD, especially at noon, in all of the cultivars examined (Fig. S6, noon and 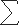 MNE). Importantly, the relative abundance of GmCRY1a at noon (normalized by the sum of the band signals of GmCRY1a of all three time points sampled) in LD-grown plants showed a clear correlation with the flowering time (R2 = 0.414, P < 0.019) and the latitude of the site of cultivation (R2 = 0.467, P < 0.012) (Fig. 5C, LD). No such correlation was detected between GmCRY1a expression in SD photoperiod and flowering time or latitude (Fig. 5C, SD). We conclude that the photoperiod-dependent rhythmic expression of GmCRY1a is associated with the latitudinal cline in photoperiodic flowering of soybean.
MNE). Importantly, the relative abundance of GmCRY1a at noon (normalized by the sum of the band signals of GmCRY1a of all three time points sampled) in LD-grown plants showed a clear correlation with the flowering time (R2 = 0.414, P < 0.019) and the latitude of the site of cultivation (R2 = 0.467, P < 0.012) (Fig. 5C, LD). No such correlation was detected between GmCRY1a expression in SD photoperiod and flowering time or latitude (Fig. 5C, SD). We conclude that the photoperiod-dependent rhythmic expression of GmCRY1a is associated with the latitudinal cline in photoperiodic flowering of soybean.
Fig. 5.
The correlation of the circadian rhythmic expression of the GmCRY1a protein and flowering time or latitude of the site of soybean cultivation. (A) Immunoblots showing GmCRY1a and GmCRY2a expression in samples collected at indicated time from indicated cultivars. LM, LN, and LE: morning (0.5 h after light on), noon (middle of light phase), or evening (0.5 h before light off) in LD photoperiod; SM, SN, and SE: morning (0.5 h after light on), noon (middle of light phase), or evening (0.5 h before light off) in SD photoperiod. Control: the ECL control, an aliquot of the same protein sample prepared at noon in LD from NK18 line was included in each immunoblot. (B) GmCRY1a signals of the immunoblot shown on the left were digitized, treated by the two-way normalization (see SI Text), and plotted as the “Relative abundance of GmCRY1a” of the indicated cultivars and the respective latitude of the site of cultivation. (C) Linear regression analyses showing a strong correlation between the relative abundance of GmCRY1a [GmCRY1a (N/ΣNME)] at noon in LD and flowering time (R2 = 0.4139, P < 0.019) or latitude of the site of cultivation (R2 = 0.4671, P < 0.012), and the lack of a strong correlation between the relative abundance of GmCRY1a [GmCRY1a (N/ΣNME)] at noon in SD and flowering time (R2 = 0.1024) or latitude of the site of cultivation (R2 = 0.1076). 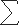 MNE, sum of GmCRY protein abundance in the morning, noon, and evening.
MNE, sum of GmCRY protein abundance in the morning, noon, and evening.
To our knowledge, soybean GmCRY1a is the first plant photoreceptor gene shown to exhibit a latitudinal cline at the level of apoprotein expression. However, two observations argue that the genetic variations affecting the circadian rhythmic expression of the GmCRY1a protein may reside outside of the GmCRY1a gene. First, no clear latitudinal cline of the GmCRY1a mRNA expression was detected in either LD or SD photoperiods (Fig. S7 and data not shown). This is consistent with the notion that, although the circadian rhythmic expression of GmCRY1a mRNA partially explain the circadian oscillation of the level of GmCRY1 protein, additional mechanisms must also be involved to determine the phase changes of GmCRY1a protein expression in response to photoperiods (Fig. 3). Therefore, potential sequence variations in the promoter or other noncoding sequences of the GmCRY1a gene cannot fully explain the natural variations in the GmCRY1a protein expression. Second, no allelic variations detected in the GmCRY1a cDNAs of the 11 soybean cultivars examined in this study showed a clear correlation with the latitudinal cline in the GmCRY1a protein expression or in flowering time (Y. Li, L. Qu, and Q. Zhang, unpublished). This result indicates that, unlike the Arabidopsis CRY2EDI allele (25), genetic variations causing the latitudinal cline in the GmCRY1a protein expression may be better explained by structure variations not readily discernable at the amino acid sequences, at least for the cultivars examined. Consistent with our hypothesis, none of the QTL associated with photoperiodic flowering in soybean has been mapped to the chromosome location near a GmCRY gene (41). Therefore, we are compelled to speculate that the natural variations of genes involved in posttranscriptional regulation of gene expression, such as components of the phytochrome signal transduction, the circadian clock, mRNA export, protein translation, modification, or degradation, are likely involved in determining the latitudinal cline in the circadian rhythmic expression of the GmCRY1a apoprotein and in photoperiodic flowering of soybean. Further studies are needed to identify those genes.
Materials and Methods
Transgenic Arabidopsis “overexpressing” 35S::GFP-GmCRY were prepared in the cry2 mutant (5), cry1 mutant (42), or Col background, respectively. Rabbit antibodies were prepared against the C-terminal domains of GmCRY1a (residues 486 to 681) and GmCRY2a (residue 486 to 634) expressed and purified from E. coli. See SI Text for additional details.
Supplementary Material
Acknowledgments.
The authors thank Drs. Elaine Tobin [University of California, Los Angeles (UCLA)] and Victoria Sork (UCLA) for critical readings of the manuscript, Dr. Tianfu Han [Chinese Academy of Agricultural Sciences (CAAS)] for stimulating discussions, and Drs. Yinhui Li and Lijuan Qiu (CAAS) for communicating their results before publication and providing soybean accessions. This work was supported in part by the Chinese National Key Basic Research “973” Program (2004CB117206), the Chinese National “863” Program (2006AA10Z107, 2006AA10A111, and 2007AA10Z119), the Chinese National Science Foundation (30671245, 30570162), the CAAS Key Technology R&D Program (2007BAD59B02), and the CAAS Special Funding of National Non-Profit Institutes (082060302-8/10). Studies in C.L.'s laboratory at UCLA are supported by the National Institutes of Health (GM56265), UCLA faculty research grants, and the Sol Leshin UCLA-BGU Academic Cooperation program.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ401046 and DQ40104712).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810585105/DCSupplemental.
References
- 1.Cashmore AR. Cryptochromes: Enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 2.Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Yang H, Mockler TC, Lin C. Regulation of Flowering Time by Arabidopsis Photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 6.Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L. ) Heynh. Z Pflanzenphysiol Bd. 1980;100:147–160. [Google Scholar]
- 7.Thomas B, Vince-Prue D. Photoperiodism in plants. New York: Academic; 1997. [Google Scholar]
- 8.Immeln D, Schlesinger R, Heberle J, Kottke T. Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem. 2007;282:21720–21728. doi: 10.1074/jbc.M700849200. [DOI] [PubMed] [Google Scholar]
- 9.Imaizumi T, Kanegae T, Wada M. Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern adiantum capillus-veneris. Plant Cell. 2000;12:81–96. doi: 10.1105/tpc.12.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imaizumi T, Kadota A, Hasebe M, Wada M. Cryptochrome Light Signals Control Development to Suppress Auxin Sensitivity in the Moss Physcomitrella patens. Plant Cell. 2002;14:373–386. doi: 10.1105/tpc.010388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ninu L, et al. Cryptochrome 1 controls tomato development in response to blue light. Plant J. 1999;18:551–556. doi: 10.1046/j.1365-313x.1999.00466.x. [DOI] [PubMed] [Google Scholar]
- 12.Giliberto L, et al. Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 2005;137:199–208. doi: 10.1104/pp.104.051987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee M, Sharma P, Khurana JP. Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 2006;141:61–74. doi: 10.1104/pp.105.076323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platten JD, et al. Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol. 2005;139:1472–1482. doi: 10.1104/pp.105.067462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto N, Hirano T, Iwasaki T, Yamamoto N. Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol. 2003;133:1494–1503. doi: 10.1104/pp.103.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YC, et al. Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J. 2006;46:971–983. doi: 10.1111/j.1365-313X.2006.02753.x. [DOI] [PubMed] [Google Scholar]
- 17.Toth R, et al. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Facella P, et al. Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS ONE. 2008;3:e2798. doi: 10.1371/journal.pone.0002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platten JD, et al. The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol. 2005;59:683–696. doi: 10.1007/s11103-005-0828-z. [DOI] [PubMed] [Google Scholar]
- 20.Shalitin D, et al. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad M, Jarillo JA, Cashmore AR. Chimeric proteins between cry1 and cry2 arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell. 1998;10:197–208. doi: 10.1105/tpc.10.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C, et al. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, et al. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007;19:3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mockler T, et al. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA. 2003;100:2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Assal SE-D, et al. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 26.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Physiol Plant Mol Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 27.Bohlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 28.Maloof JN, et al. Natural variation in light sensitivity of Arabidopsis. Nat Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- 29.Balasubramanian S, et al. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lempe J, et al. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1:109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res. 1920;18:553–606. [Google Scholar]
- 32.Morse W, Cartter J, Williams L. Soybeans: Culture and varieties. US Dep Agric Farmers' Bull. 1947:1–38. No. 1520. [Google Scholar]
- 33.Boerma HR, Specht JE. Soybeans: Improvement, Production, and Uses. Madison, WI: American Society of Agronomy, Inc.; 2004. [Google Scholar]
- 34.Wolfe KH, et al. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoemaker RC, et al. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C, Ahmad M, Gordon D, Cashmore AR. Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc Natl Acad Sci USA. 1995;92:8423–8427. doi: 10.1073/pnas.92.18.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weller JL, et al. Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J. 2001;25:427–440. doi: 10.1046/j.1365-313x.2001.00978.x. [DOI] [PubMed] [Google Scholar]
- 38.Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- 39.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 40.Hymowitz T, Shurtleff WR. Debunking soybean myths and legends in the historical and popular literature. Crop Sci. 2005;45:473–476. [Google Scholar]
- 41.Tasma I, Shoemaker R. Mapping flowering time gene homologs in soybean and their association with maturity (E) loci. Crop Sci. 2003;43:319–328. [Google Scholar]
- 42.Mockler TC, et al. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126:2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.