Abstract
Continental shelf island systems, created by rising sea levels, provide a premier setting for studying the effects of geographical isolation on non-adaptive radiation and allopatric speciation brought about by genetic drift. The Aegean Archipelago forms a highly fragmented complex of mostly continental shelf islands that have become disconnected from each other and the mainland in relatively recent geological times (ca <5.2 Ma). These ecologically fairly homogenous islands thus provide a suitable biogeographic context for assessing the relative influences of past range fragmentation, colonization, gene flow and drift on taxon diversification. Indeed, recent molecular biogeographic studies on the Aegean Nigella arvensis complex, combining phylogenetic, phylogeographic and population level approaches, exemplify the importance of allopatry and genetic drift coupled with restricted gene flow in driving plant speciation in this continental archipelago at different temporal and spatial scales. While the recent (Late Pleistocene) radiation of Aegean Nigella, as well as possible instances of incipient speciation (in the Cyclades), is shown to be strongly conditioned by (palaeo)geographic factors (including changes in sea level), shifts in breeding system (selfing) and associated isolating mechanisms have also contributed to this radiation. By contrast, founder event speciation has probably played only a minor role, perhaps reflecting a migratory situation typical for continental archipelagos characterized by niche pre-emption because of a long established resident flora. Overall, surveys of neutral molecular markers in Aegean Nigella have so far revealed population genetic processes that conform remarkably well to predictions raised by genetic drift theory. The challenge is now to gain more direct insights into the relative importance of the role of genetic drift, as opposed to natural selection, in the phenotypic and reproductive divergence among these Aegean plant species.
Keywords: Aegean palaeogeography, allopatric speciation, continental shelf islands, genetic drift, Nigella, non-adaptive radiation
1. Introduction
The term species radiation, i.e. the divergent evolution of a relatively large, monophyletic group of species within a relatively short time (Stanley 1979; Schluter 2000), is most frequently used in combination with the epithet ‘adaptive’ (Givnish 1997), which in turn implies ecological specialization and the probable co-occurrence of closely related species (MacArthur 1972; Schluter 2000; Savolainen & Forest 2005). In recent years, molecular phylogenetic data have provided a number of spectacular examples of explosive adaptive species radiations for plants in the recent evolutionary past (Late Tertiary/Quaternary), on either oceanic islands (Baldwin & Sanderson 1998; Böhle et al. 1996; see also Whittaker & Fernández-Palacios 2007) or continents (Richardson et al. 2001; Klak et al. 2003; Kay et al. 2005; Hughes & Eastwood 2006; see Linder 2008). In most cases, such radiations have been attributed to ecological opportunities afforded by the emergence of new habitats and the absence of competition on recently formed oceanic islands and continental mountain ranges, rather than to the evolution of key morphological or physiological innovations (but see Klak et al. 2003; Kay et al. 2005).
By contrast, only a very few studies have explicitly addressed the question of whether species proliferation might occur by ‘non-adaptive radiation’ (sensu Cain 1944; Givnish 1997; Savolainen & Forest 2005), i.e. without appreciable ecological divergence and evolution of corresponding adaptations, and simply as a consequence of geographical isolation and allopatric divergence among sibling species maintaining similar ecological niches over evolutionary time scales. Such an allopatric model of non-adaptive species radiation essentially implies that mutation and random genetic drift, rather than habitat-mediated selection, will be the primary factors causing divergence of populations occupying ecologically similar habitats. The best-known examples include species-rich taxa of land snails on either Crete (Albinaria: Gittenberger 1991; Giokas 2000) or the Madeiran island of Porto Santo (Helicidae: Cameron et al. 1996), as well as particular plant lineages in the Aegean Archipelago (e.g. Snogerup 1967; Strid 1970; Stork 1972; see §2). In addition, Savolainen & Forest (2005) claimed that the term non-adaptive radiation might also be an appropriate way of describing the diversification of the temperate legume genus Astragalus (see also Sanderson & Wojciechowski 1996; Sanderson 1998). Similarly, Whittaker & Fernández-Palacios (2007) hypothesized that a non-adaptive component might be responsible for the radiation of several Macaronesian plant genera (i.e. Cheirolophus, Helianthemum, Limonium) that exploit similar habitats in different islands. Although a systematic survey of their prevalence would be valuable, it is perhaps no coincidence that most cases proposed for non-adaptive radiation are from island systems. In such geographical settings, random genetic drift (whether in established populations or during founder events) has long been implicated as contributing to speciation viz. reproductive isolation and phenotypic evolution (e.g. Mayr 1954; Carson 1975; Templeton 1980; see also Grant 1998).
Although patterns of molecular variation in island populations often do correspond to what would be expected if the loci were affected by genetic drift (e.g. Wendel & Percival 1990; Barrett 1996; Berry 1996; Stuessy et al. 2006), the contribution of genetic drift to island speciation remains the subject of keen debate (e.g. Barton 1996; Grant 1998; Whittaker & Fernández-Palacios 2007). However, based on their thorough reassessment of the speciation literature, Coyne & Orr (2004, pp. 383–410) recently concluded that firm empirical evidence for a role of genetic drift in speciation is rare, and they also raised the same objection about drift-induced peripatric or founder-effect speciation. Also, mathematical models indicate that conditions for speciation via drift are fairly restrictive (Barton 1996; Coyne et al. 1997; Turelli et al. 2001; Coyne & Orr 2004). For example, while showing that geographical isolation can lead to complete reproductive isolation (Nei et al. 1983), such models have also revealed that even a small amount of gene flow can retard differentiation by drift (Coyne & Orr 2004, p. 87). Moreover, genetic drift is generally thought to cause speciation and quantitative phenotypic evolution more slowly than does either directional selection alone or in combination with drift (Coyne et al. 1997). On the other hand, there is good evidence for a role of genetic drift in chromosomal speciation, particularly in plants (Rieseberg 2001). Again, especially within plants, a probable important mechanism favouring speciation by drift is selfing, which has recently been invoked in a study of arctic Draba species as providing an efficient non-adaptive means for the accumulation of intraspecific hybrid incompatibilities (Grundt et al. 2006). Another well-known, but fairly unusual, example for speciation by drift involves shell coil reversals in land snails (Gittenberger 1988; Ueshima & Asami 2003).
There can be no doubt that natural (and sexual) selection plays a much larger role in speciation and phenotypic diversification than does drift (e.g. Macnair & Gardner 1998; Rieseberg et al. 2002; Coyne & Orr 2004; Waser & Campbell 2004; Johnson 2006; Noor & Feder 2006; see Lexer & Widmer 2008). Nonetheless, it would seem to be premature to close the book on speciation by drift, no less than dismissing its role in phenotypic evolution at all. Rather, considering both theory and empirical evidence, a potential role of non-adaptive radiation by drift will still apply to all species occurring in effectively isolated (allopatric) populations, especially if these are permanently or temporarily small (Wright 1948; Slatkin 1985). Yet, despite allopatric speciation being widely considered as the most common geographical mode (Barraclough & Vogler 2000; Turelli et al. 2001; Coyne & Orr 2004), and notwithstanding an expanding literature on the joint roles of allopatry and ecology in speciation (Schluter 2000; Ogden & Thorpe 2002; Wiens 2004; Kozak & Wiens 2006; Thorpe et al. 2008), the relationship between geographical isolation, drift, phenotypic differentiation and reproductive isolation remains poorly understood in ‘non-ecological speciation’ (sensu Schluter 2000).
2. The Aegean Archipelago as a laboratory of allopatric speciation and non-adaptive radiation
Perhaps the most suitable natural laboratories for studying the effects of geographical isolation on allopatric speciation are ‘continental shelf islands’ (sensu Whittaker & Fernández-Palacios 2007) that have become disconnected from each other and/or the mainland in relatively recent geological times (e.g. most Aegean islands, the South-East Asian Sundashelf islands (Sumatra, Java, Borneo), and the North Pacific Vancouver and Queen Charlotte islands). Such island systems have many of the advantages of oceanic islands (e.g. Hawaii, Galápagos, Canaries) in that they allow insights into colonization and restricted gene flow. As an important difference, however, continental islands also provide a premier setting for studying the effects of past range fragmentation via geologically dated sea barriers, and thus for testing genetic drift-models of allopatric speciation and non-adaptive radiation (Wright 1940; Mayr 1954; Schluter 2000).
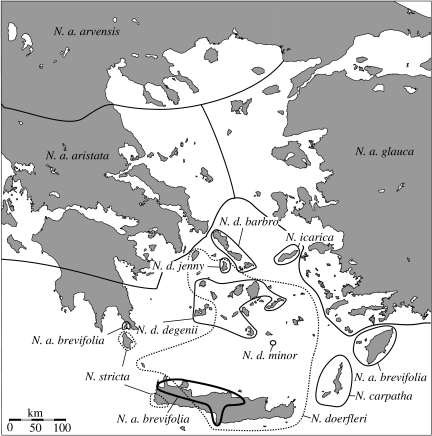
In fact, the most suggestive evidence for non-adaptive radiation comes from plant groups inhabiting the ecologically fairly homogenous continental island system of the Aegean Archipelago (Runemark 1969; Strid 1970; Barrett 1996; Levin 2000). This claim, specifically, relates to the biosystematic work of Strid (1970) conducted on the so-called Nigella arvensis complex (Ranunculaceae, tribe Nigelleae), which contains six species (12 taxa) that are mostly allo-/or parapatrically distributed on numerous islands and adjacent mainland areas of Greece and western Anatolia (figure 1, table 1). Based on crossing experiments, as well as morphological and palaeogeographic evidence, Strid (1970) argued that differentiation in the N. arvensis complex has been determined principally by two factors: (i) range fragmentation triggered by Plio-/Pleistocene changes in sea level; and (ii) subsequent evolution of the fragmented ancestral stock via genetic drift viz. non-adaptive radiation. Strid (1970) further hypothesized that genetic drift in the complex is largely brought about by drastic population bottlenecks in established populations because of (seasonal) fluctuations in population size rather than by repeated interisland (or mainland-island) founding colonizations. His contemporaries working on other endemic components of the Aegean flora favoured similar evolutionary scenarios (e.g. Snogerup 1967; Stork 1972; Bentzer 1973; Von Bothmer 1974, 1987).
Figure 1.
Approximate distribution ranges of the six species (12 taxa) of the Aegean Nigella arvensis complex (after Strid 2002; see also table 1). Solid versus dashed lines delineate outcrossing versus selfing taxa. Note that the range of N. arvensis ssp. brevifolia on Crete comprises only four known populations. N. a., N. arvensis; N. d., N. degenii.
Table 1.
Geographical distribution, habitat preference, flowering period and breeding system of the taxa of the Nigella arvensis complex (after Strid 1970 and modified according to Bittkau & Comes in press). Note that most taxa occur at low altitudes (approx. 0–500 m a.s.l.) except for Nigella arvensis ssp. glauca (0–1000 m a.s.l.) and Nigella icarica (500–1000 m a.s.l.).
| Nigella taxon | geographical distribution | habitat | flowering period | breeding system |
|---|---|---|---|---|
| N. arvensis L. | ||||
| ssp. arvensis | N Greece/Europe and N Africa | cultivated/abandoned fields | VI–IX | outcrosser |
| ssp. aristata (Sibth. and Sm.) Nym. | S and C Greek mainland, Euboea | phrygana, abandoned fields, roadsides | mid-V–VII | outcrosser |
| ssp. glauca (Boiss.) Terracc. | E Aegean islands, W Turkey, Thrakia | phrygana, abandoned fields, olive groves | VI–VIII | outcrosser |
| ssp. brevifolia Strid | Elafonisos (south of Peloponnisos) W Cretea, Rhodes | phrygana, abandoned fields, roadsides | VI–VIII | outcrosser |
| N. carpatha Strid | S Karpathos, Kasos | phrygana | mid-V–mid-VII | outcrosser |
| N. degenii Vierh. | ||||
| ssp. degenii | C and S Cycladesb | phrygana, cultivated fields, seashores | VI–VII | outcrosser |
| ssp. barbro Strid | NW Cyclades (Andros, Tinos, Mikonos) | phrygana, roadsides, seashores | mid-V–mid-VII | outcrosser |
| ssp. jenny Strid | NW Cyclades (Siros) | phrygana, abandoned fields | end-V–mid-VI | outcrosser |
| ssp. minor Strid | S Cyclades (Pakhia, south of Anafi) | phrygana | end-V–VI | outcrosser |
| N. icarica Strid | Ikaria | phrygana | VI–VII | outcrosser |
| N. doerfleri Vierh. | Andikithira, Cyclades, Crete | phrygana | end-IV–mid-V | selfer |
| N. stricta Strid | Kithira, SW Crete | stable sand dunes | mid-IV–mid-V | selfer |
Known from only four locations on Crete (Strid 1970; Z. Kypriotakis 2002, personal communication).
Known from the following islands: Amorgos, Andiparos, Folegandros, Kimolos, Milos, Naxos, Paros, Santorin, Sifnos, Sikinos.
Here, we will synthesize data from our recent and ongoing molecular biogeographic studies of the N. arvensis complex using phylogenetic, phylogeographic and population level approaches in order to dissect the evolutionary history of this plant group at increasingly smaller temporal and spatial scales. We will first introduce the Aegean region, its palaeogeographic history and our study group, and then give a concise overview of the main results, with a focus on the times and orders of evolutionary divergence within the complex, and the population genetic processes underlying its supposedly non-adaptive radiation. In essence, our data indicate that allopatry (often but not exclusively vicariant) and genetic drift (coupled with restricted gene exchange) are the dominant evolutionary processes driving population differentiation and speciation in Aegean Nigella. Because of lack of comparable data, however, we are presently far from being able to evaluate whether this scenario also holds for other plant radiations in the Aegean Archipelago or similarly complex continental island systems. Moreover, as a critical limitation, our surveys of neutral molecular polymorphisms alone provide little direct insight into the potential evolutionary processes (drift versus selection) and genetic changes causing phenotypic differentiation and reproductive isolation in our study group. We conclude, therefore, with some current and possible future approaches for tackling these more difficult issues.
3. The study area and its palaeogeographic history
(a) The Aegean Archipelago
The Aegean Archipelago (figure 1) is located between the Greek peninsula in the west and the Turkish coast of Asia Minor in the east. Approximately 611 km long and 299 km wide, the total area of the Aegean Sea is some 214 000 km2, whereby the total area of the multitude of Aegean islands and islets (>100) is approximately 9012 km2 (Strid 1997, 2002). Geographically, these islands can be roughly arranged into four groups: (i) Euboea and the Sporades, a group lying off mainland Greece; (ii) the east Aegean islands, including Ikaria and Samos, lying off the Turkish coast; (iii) the Cyclades, including 16 major islands such as Andros, Tinos, Mikonos, Siros, Paros, Naxos and the volcanic island of Santorin/Thera (from north to south); and (iv) the southern Aegean (‘Hellenic Arc’) islands, including Kithira, Crete, Karpathos, Kasos and Rhodes, which mark the southern boundary of the archipelago.
Ecologically, the commonest habitat type in these islands is a typical dry Mediterranean maquis-phrygana community of evergreen, mostly degraded (under) shrub vegetation (Blondel & Aronson 1999), which is also one of the most important habitats for the N. arvensis complex. Concomitantly, this vegetation type reflects the influence of humans on all aspects of the environment that dates back to more than 5000 years BP (Broodbank 2000). This may have led to a high degree of human-mediated secondary sympatry of plant populations, and thus may create an obstacle to the interpretation of extant biogeographic and molecular patterns in the Aegean region. In the N. arvensis complex, however, there is only limited sympatry (except for a pair of species in the Cyclades; see §4), as would be predicted under a scenario of non-adaptive radiation.
(b) Aegean palaeogeography
Paleogeographic/climatic changes affecting the entire Mediterranean basin during the Late Tertiary/Quaternary have likely exerted a major influence on the patterns of geographical isolation and allopatric speciation in the N. arvensis complex. Of particular relevance is the fact that several palaeogeographic events provide maximum times of isolation between the Aegean islands from each other and the mainland. In this respect, the ‘Messinian salinity crisis’ (ca 6.0–5.3 Ma; Duggen et al. 2003) clearly sets an upper time boundary for the last, large-scale reconnection of the entire region. During this time period, the Mediterranean completely dried up as a result of the closing of the Strait of Gibraltar, and the Aegean islands then became mountains in a steppe or desert (Blondel & Aronson 1999). At some 5.2±0.1 Ma, the Strait of Gibraltar reopened and the basin was refilled from the Atlantic Ocean within 1000 years (Duggen et al. 2003). As a result, one of the first Aegean sea barriers to become permanently established was the East Aegean Sea, separating the modern Cyclades from the east Aegean islands (ca 4.5 Ma; Chatzimanolis et al. 2003).
The sequence of Plio-/Pleistocene island formation is particularly well documented for ‘Hellenic Arc’ islands. As another consequence of post-Messinian flooding, Crete became isolated, and it has remained isolated since; Kithira, however, was completely submerged and did not re-emerge until the late Pliocene (ca 3.0 Ma; Meulenkamp et al. 1972). At about the same time, Karpathos became permanently disconnected from Rhodes-Anatolia (Daams & Van der Weerd 1980), while the permanent separation of Rhodes from Anatolia occurred at the Plio-/Pleistocene boundary (ca 2.4 Ma; Kuss 1975).
For much of the remainder of the archipelago, the Quaternary sea-level record can be used to approximate times of island separation. During the glacial periods of the Riss (250–125 ka) and Würm (75–10 ka), sea levels were, respectively, 200 and 120 m lower than at present (Sfenthourakis 1996; Perissoratis & Conispoliatis 2003). Thus, because of shallow sea barriers, Euboea and several east Aegean islands (e.g. Samos) have been separated from the nearby mainland only after the last glacial maximum (LGM; 23–18 ka). The seafloor between Samos and Ikaria, however, is much deeper (albeit less than 200 m), so these neighbouring islands were already separated by the sea transgression following the Riss glacial (Beerli et al. 1996).
As regards the Cyclades, most of these islands once formed a landmass that remained connected to mainland Greece/Euboea until the end of the Pliocene (ca 2 Ma; Fattorini 2002; Chatzimanolis et al. 2003). During the glacial/interglacial periods of the Pleistocene, most central islands were alternately joined and disentangled, with the most recent fragmentation dating back to late-glacial/Early Holocene times (Lambeck 1996). Notably, several Cycladic islands further south (e.g. Milos, Santorin/Thera, Anafi) have been formed by volcanic activity that started in the Pliocene, and most of them were temporarily connected to nearby islands during some period of their existence (Sfenthourakis 1996). As will be seen in §7, the Cyclades’ intricate configuration and post-glacial palaeogeography offers an outstanding context in which to explore the times and orders of recent population splitting in a Nigella species endemic to these islands.
4. The model system: the Nigella arvensis complex
(a) Breeding system and experimental crossing barriers
Nigella is a genus of approximately 23 annual, diploid (2n=12) and self-compatible species of mainly Mediterranean/Southwest Asian distribution (Strid 2002). The six species (12 taxa) of the N. arvensis complex (figure 1, table 1) are mainly distinguished by flower and fruit characters and, to a lesser extent, by vegetative characters, always in combination with their geographical distribution (Strid 1970). Four species (N. arvensis, Nigella carpatha, Nigella degenii, Nigella icarica) are summer-flowering and predominantly outbreeding, whereas self-pollination is the normal condition in the spring-flowering, smaller-sized Nigella doerfleri and Nigella stricta, because their styles regularly twist around the dehiscing anthers (Strid 1969). Attempts to produce fertile hybrids between N. doerfleri and N. stricta, or between the selfers and outcrossers, have been unsuccessful, whereas species of the latter group are interfertile, with pollen-fertility values in hybrids ranging between 30 and 100% (Strid 1970).
(b) Geographical distribution and habitat
The 10 taxa of the outcrossing species exhibit a striking allo- or parapatric pattern of geographical distribution (figure 1, table 1). The four outcrossing subspecies of N. arvensis are distributed in the form of a ring around the Cyclades: ssp. aristata occurs in southern Greece, ssp. arvensis in northern Greece, ssp. glauca in the east Aegean islands and Anatolia, and ssp. brevifolia is disjunctly distributed on the ‘Hellenic Arc’ (Elafonisos, Crete, Rhodes). The three remaining outcrossing species are island endemics: N. carpatha is found on Karpathos (and nearby Kasos), N. icarica on Ikaria, and N. degenii is widespread throughout the Cyclades, where four allopatric subspecies are recognized which are endemic to island groups (spp. barbro, degenii) or single islands (spp. jenny, minor). Like the outcrossing taxa, the selfing species are also not sympatric with one another: N. stricta is restricted to Kithira and southwestern Crete, whereas N. doerfleri is more widespread (Andikithira, central/eastern Crete, Cyclades, numerous small islets). The broad range overlap between N. doerfleri and N. degenii in the Cyclades is exceptional for the entire complex (figure 1). It thus appears that only species of the complex that differ in breeding system (and associated traits) are able to live in sympatry on the same islands.
In accordance with one of the major criteria for non-adaptive radiation, the habitats occupied by most taxa of the N. arvensis complex vary little among islands or the mainland (table 1). At least from a macro-habitat perspective, these taxa are ecologically unspecialized with populations occurring in a wide variety of more or less disturbed habitats (0–1000 m a.s.l.) such as stony seashores, phrygana communities, abandoned/cultivated fields or roadsides. In contrast to the outcrossers, however, the two selfers tend to occupy or tolerate more arid habitats (Strid 1969, 1970), including stable sand dunes (N. stricta) or phrygana sites on small, dry and isolated islands (N. doerfleri).
5. Species-level phylogenetics and phylogeography
(a) Phylogenetics
A recent molecular-phylogenetic study (Bittkau & Comes in press), with a complete species representation of Nigella s. lat. (= Nigella and its sister genus Garidella) and based on internal transcribed spacer (ITS) sequences of nuclear ribosomal DNA, revealed that the N. arvensis complex truly qualifies as a ‘radiation’. The clock-calibrated ITS phylogeny generated showed that all the species of the complex began diverging rapidly from a single ancestor in the Late Pleistocene, between ca 0.78 (±0.39) and 0.16 (±0.08) Ma, whereas the origin of this ancestor could only roughly be approximated to between 6.2 and 1.3 Ma (Messinian to mid-Pleistocene). In addition, both log lineages-through-time (LTT) plots (Harvey et al. 1994; Nee et al. 1994) and likelihood survival analyses (Paradis 1998) indicated that the proliferation of Nigella species in the Aegean region caused a significant departure from a stochastic, speciation–extinction process of diversification during the evolution of Nigella s. lat. Otherwise, the genus would have followed a ‘constant-rates birth–death’ (CR-BD) model (Harvey et al. 1994; Barraclough & Nee 2001), a scenario under which speciation rate and extinction rate are assumed to be constant across lineages within a phylogeny and over time (the null hypothesis). Finally, we could show that the observed upturn curve in the LTT-plot towards the present, mainly associated with the Aegean N. arvensis complex, was much more recent than would have been expected under the CR-BD model (Bittkau & Comes in press). This finding indicates that the recent radiation of the Aegean group truly reflects an increase in the rate of speciation and not decreased extinction (Barraclough & Nee 2001). Thus, the major hypothesis emerging from this molecular-phylogenetic study is that the accelerated rate of speciation in the Aegean N. arvensis complex is possibly related to increased opportunities for allopatric speciation afforded by the (palaeo)geographic complexity of the archipelago, combined with Late Pleistocene changes in climate and sea level (Bittkau & Comes in press).
(b) Phylogeography
For plant groups inhabiting continental island systems, the smaller effective population size of their chloroplast (cp)DNA, compared with the nuclear genome (Birky et al. 1989), should be particularly useful for detecting signatures of past range fragmentation (because of rising seas) and its consequences, including restricted dispersal, drift and population bottlenecks/founder events. Initial evidence that present Aegean sea barriers keep Nigella populations isolated emerged from a survey of polymerase chain reaction-restriction fragment length polymorphism haplotype variation of cpDNA (which is maternally inherited in Nigella) in the four outcrossing and cross-compatible species of the complex (Bittkau & Comes 2005), henceforth called the ‘N. arvensis alliance’. Based on the geographical distributions of major haplotype clades, as inferred from genealogical (nested clade) reconstructions, this study identified the Eastern and, to a lesser extent, the Western/Southern Aegean Seas as important isolating barriers to Nigella dispersal, as had long been suspected for other members of the Aegean flora (Rechinger 1950; Runemark 1969, 1980; Greuter 1979; Strid 1996). Notably, there was a sharp break in cpDNA haplotype frequencies observed across the East Aegean Sea, i.e. at the ‘Rechinger's line’ (sensu Strid 1996), the phytogeographic borderline between Europe and Asia Minor. For the first time this testified molecularly to the long-held notion that the East Aegean Sea poses a strong physical barrier to dispersal in many plants (Greuter 1979; Runemark 1980; Strid 1996), as well as for various animals (e.g. Giokas 2000; Fattorini 2002; Poulakakis et al. 2003, 2005). Furthermore, the restricted distribution of a large number of derived cpDNA haplotypes or clades, on both mainland areas and particular islands, identified the N. arvensis alliance as a plant group with low seed dispersal ability and, consequently, high susceptibility to genetic drift (Bittkau & Comes 2005).
The main conclusion drawn from the phylogeographic cpDNA study is that various regional sets of populations of the Aegean N. arvensis alliance are geographically isolated and probably on independent evolutionary trajectories. That said, none of its four constituent species is fixed for a particular cpDNA haplotype, or uniquely characterized by a haplotype clade. The same applies to the two selfers N. doerfleri and N. stricta, which are also nearly fixed for an ancestral haplotype (A) present in all species of the alliance except N. icarica (C. Bittkau & H. P. Comes 2004, unpublished data). In general, such cpDNA haplotype sharing across species boundaries could indicate either the retention of ancestral haplotypes or cytoplasmic gene flow/hybridization following species divergence.
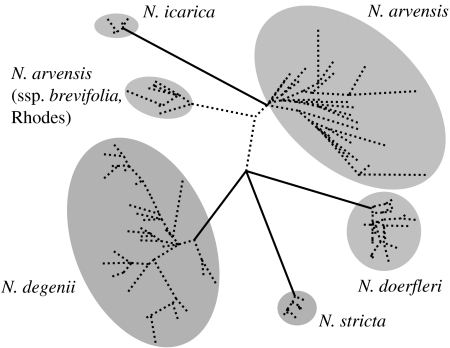
To distinguish between these two possibilities, we have undertaken a preliminary survey of amplified fragment length polymorphisms (AFLPs) in all species of the complex (except N. carpatha), taking advantage of the fact that such fast evolving markers are predominantly of nuclear origin and also potentially suitable for resolving ‘shallow phylogenies’ (Meudt & Clarke 2007). Genetic distance analysis of individual AFLP phenotypes results in an unrooted network (figure 2) in which almost all species of the complex surveyed are separated in clusters, albeit of unresolved relationships. While a more extensive AFLP survey of approximately 500 individuals with three (instead of two) primer combinations is currently underway to obtain better-resolved species and population relationships (U. Jaros, A. Tribsch & H. P. Comes 2008, unpublished data), two important insights are already evident from figure 2. First, most nominal species of the complex are genetically distinct entities, suggesting that erratic instances of cpDNA haplotype sharing across species boundaries reflect incomplete lineage sorting rather than ongoing, interspecific hybridization. Second, N. arvensis ssp. brevifolia sampled from Rhodes forms a separate cluster relative to conspecific material of mainland origin surveyed (spp. aristata/glauca). This pronounced intraspecific divergence of N. arvensis, which is also supported by cpDNA data (Bittkau & Comes 2005), once more indicates a complete lack of gene flow across the East Aegean Sea viz. the Strait of Marmaris (approx. 18 km), which separates Rhodes from the nearby Anatolian coast. It is noteworthy, however, that lack of gene exchange between Nigella populations from either Rhodes (N. arvensis ssp. brevifolia) or the Cyclades (N. degenii) and Anatolia (N. arvensis ssp. glauca) may also be explained, at least in part, by reciprocal cross-incompatibility or reduced hybrid fertility, respectively, as indicated by experimental crosses (Strid 1970, p. 117). Thus, even if seed material from Rhodes or the Cyclades manages to cross the East Aegean Sea, the probability of gene exchange with resident Anatolian populations is probably small.
Figure 2.
Unrooted neighbour-joining phenogram based on Nei & Li's (1979) genetic distances among 106 multilocus AFLP phenotypes observed in 106 plants from 42 populations of the Nigella arvensis complex, and representing all species and subspecies except N. carpatha, N. arvensis ssp. arvensis and N. degenii ssp. minor. A list containing detailed geographical information on the location of all collection sites and the number of individuals surveyed per taxon/site is available upon request. Solid (dashed) branches indicate bootstrap support values above (below) 70% based on 10 000 replicates. AFLP profiles were generated from total genomic DNA samples (Bittkau & Comes 2005) using the AFLP methodology and scoring procedure described in Kropf et al. (2006) with two EcoRI–MseI primer combinations (E38/M53; E37/M56) that resulted in a total of 264 polymorphic fragments.
6. Biogeography of Aegean Nigella speciation
Knowing about the strength of a geographical barrier in currently restricting gene flow does not tell us anything about its importance in speciation (Coyne & Orr 2004). Hence, a crucial question is whether the establishment of Aegean sea barriers actually caused allopatric vicariant speciation within the N. arvensis complex, i.e. via range fragmentation of a formerly widespread (pan-Aegean) ancestor of Messinian to mid-Pleistocene origin (Bittkau & Comes 2005). However, considering the Late Pleistocene diversification of the complex (see §5a) and in the light of our current understanding of post-Messinian sea-level changes in the Aegean (see §3b), there are only two probable instances of sea barrier-induced vicariant speciation, i.e. (i) the origin of N. degenii following the separation of the Cycladic landmass (peninsula) from mainland Greece/Euboea (ca 2 Ma); and (ii) the probably more recent origin of N. icarica following a post-glacial rise in sea level that separated Ikaria from Samos/Anatolia (ca 125–75 ka). By contrast, for the remaining island species (N. doerfleri, N. stricta, N. carpatha), as well as the enigmatic N. arvensis ssp. brevifolia, there is little reason to suspect that rising seas were required for their origin. This is because these taxa mainly (albeit not exclusively) occur on a group of southern ‘Hellenic Arc’ islands that became permanently isolated well before the Late Pleistocene (i.e. Crete, ca 5.5 Ma; Kithira, Karpathos/Kasos, ca 3 Ma; Rhodes, ca 2.4 Ma; see § 3b). In support of this hypothesis, the cpDNA data indicate a peripatric origin of N. carpatha through relatively recent over-sea dispersal from Anatolia (or Ikaria) to Karpathos (Bittkau & Comes 2005). However, the spatial-temporal origins of N. doerfleri, N. stricta and also N. arvensis ssp. brevifolia are not yet fully understood. Nonetheless, our current cpDNA and AFLP data (Bittkau & Comes 2005; figure 2) show no evidence of a founder origin of these island taxa from the mainland. Rather, they may constitute remnants of the formerly widespread ancestral stock from which they originated in situ within islands long in existence, and subsequently dispersed to others to attain their presently disjunct distributions (figure 1).
At this juncture it is worth emphasizing that the change from outcrossing to predominant selfing in the ancestor(s) of N. doerfleri and N. stricta probably had a major effect on the origin of these species. Although ‘selfing’ not necessarily constitutes a reproductive isolating-barrier causing speciation (Coyne & Orr 2004), it nonetheless may contribute to pre-mating barriers between diverging populations and accelerate the development of intrinsic post-zygotic isolation (Fishman & Stratton 2004; Grundt et al. 2006). In fact, the reproductive failure of hybrids in crosses involving these two selfers (see §4a) may reflect genic or chromosomal incompatibilities that evolved as a result of reductions in effective population size (because of selfing) and the fixation of alternate, negatively interacting alleles or chromosomal segments in separated populations, often referred to as Dobzhansky–Muller incompatibilities (Orr & Turelli 2001). Moreover, both species show other possible effects of selfing, including reductions in overall habit and flower size, advanced flowering, as well as successful interisland colonization and production of populations in novel, particularly arid habitats at which the outcrossers are absent (see §3b). Hence, the evolution of selfing may have also led to reproductive isolation as a by-product of local adaptation in response to low water availability, which thus would render the two selfers unlikely candidates for non-adaptive speciation. Given their genetic distinctness (figure 2), it also seems unlikely that selfing evolved under sympatric conditions as a consequence of reinforcement to reduce gene flow from nearby outcrossing populations not adapted to arid habitats. Rather, we presume that selfing and associated isolating mechanisms (phenological, ecological, genetic) evolved more or less simultaneously as by-products of geographical isolation (Coyne & Orr 1999). Experimental crossings and, ideally, sequence analyses of relevant genes (Noor & Feder 2006) will be needed to determine the temporal order in which these multiple isolating mechanisms evolved between outcrossers and selfers. Importantly, while most species of the N. arvensis complex have allopatric distributions, precluding direct observation of reproductive isolation, N. doerfleri and N. degenii coexist in the Cyclades (figure 1) without hybridizing (Strid 1970). Evidently, it is here, in an area of possibly secondary range overlap, that these multiple isolating factors act together to prevent gene flow between these two species, allowing their coexistence.
To summarize so far, the above data indicate that plant speciation in the Aegean Archipelago cannot simply be viewed as mainly resulting from vicariant speciation via historical range fragmentation because of post-Messinian (Plio-/Pleistocene) rises in sea level. This was the view commonly held by earlier biosystematists working on allopatric plant ‘neo-endemics’ of the Aegean region (e.g. Snogerup 1967; Stork 1972; Bentzer 1973; Von Bothmer 1974, 1987). In contrast, the molecular data presented here suggest a remarkably recent (Late Pleistocene) radiation of the Aegean N. arvensis complex that resulted from a combination of vicariant, (infrequent) peripatric and intra-island speciation, with the latter process probably facilitated or even accomplished by a change in pollination system (selfing). That speciation via founder events had only a minor role in the group's history perhaps comes as a surprise, but apart from taxon-specific attributes (e.g. dispersal ability, genetic incompatibilities) this could also reflect a migratory situation more typical for continental (compared with oceanic) island systems, where most successful introductions of plant diaspores are generally thought to fail because of niche pre-emption of a resident flora that has long been well adapted to existing habitat conditions (Runemark 1969; Silvertown 2004; Silvertown et al. 2005).
More generally speaking, and to compound this complexity, it should be recalled that although plant radiations in continental archipelagos are liable to be partly conditioned by (palaeo)environmental (extrinsic) factors, such as changes in sea level and geographical configurations, they may also be profoundly influenced by taxon-specific (intrinsic) attributes that are thought to influence rates of diversification, including types of pollination, mechanisms of dispersal, generation time, population size or hybridization/polyploidization (Coyne & Orr 2004; Ricklefs 2007). Future evaluation of Aegean plant radiations will thus require not only good phylogenetic and phylogeographic data for the existence of sister species and accurate molecular clocks to broaden our understanding of the biogeography of plant speciation in the Aegean Archipelago, but also similar molecular investigations across evolutionarily independent lineages to separate the effects of historical versus taxon-specific factors on diversification. Finally, further detailed intraspecific investigations of the population genetic structure of Aegean plant species together with experimental studies are needed to test the strong hypothesis of Strid (1970) and others (Snogerup 1967; Runemark 1969, 1980; Stork 1972; Bentzer 1973; Von Bothmer 1974, 1987) that allopatric divergence via genetic drift is the rule rather than the exception for plant evolution in the Aegean Archipelago.
7. Incipient speciation in the Cyclades
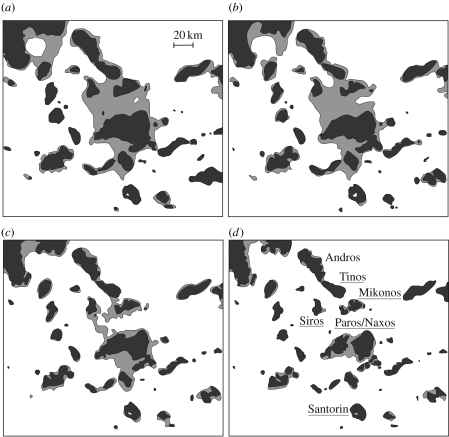
An ideal Aegean plant species with which to test the impact of recent population fragmentation on the potential for incipient allopatric speciation and genetic/phenotypic divergence via drift is the Cycladic endemic N. degenii. The Cycladic region de facto provides an excellent setting for studying the history of population splitting and its evolutionary consequences, given that sea-level changes in the area since the LGM (ca <23–18 ka) are well documented (Van Andel & Shackleton 1982; Lambeck 1996; Perissoratis & Conispoliatis 2003). Starting with the LGM, there was a major drop in sea level that created a large insular landmass of approximately 6000 km2, named ‘Cycladia’ (with several satellite islands), in the place of the modern Cyclades. Moreover, refined calibrations of eustatic changes combined with modelling of crustal rebound enabled Lambeck (1996) to generate remarkably precise reconstructions of the topography of Cycladia and the degrees by which it drowned towards the end of the Pleistocene because of rising seas (ca 14–10 ka; figure 3). By the start of the Holocene, there was a substantial tract of land still present between ‘Greater Paros’ (i.e. the current islands of Paros, Antiparos, Despotikon) and Naxos, but this land bridge also rapidly vanished by ca 9 ka (Van Andel & Shackleton 1982).
Figure 3.
Reconstruction of the topography of the insular landmass of Cycladia at the LGM (ca 23–18 ka; (a) 18 ka, (b) 14 ka, (c) 12.5 ka, (d) 10 ka), and the degrees by which it drowned because of rising sea levels towards the end of the Pleistocene (modified from Lambeck (1996) as illustrated in Broodbank (2000)). Names of islands from where Nigella degenii populations were sampled for AFLP analysis (see figure 4, table 2) are underlined.
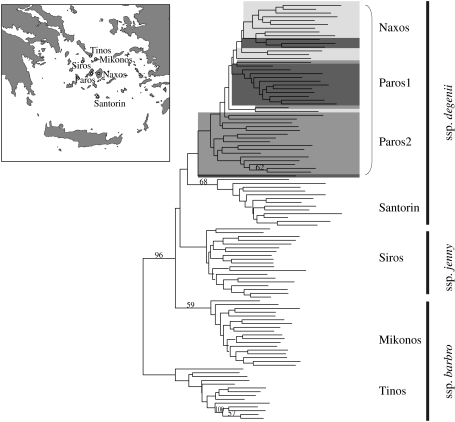
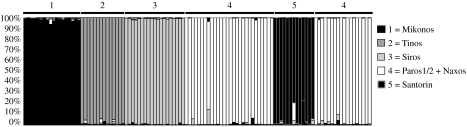
Our cpDNA population-level survey of Cycladic N. degenii (Bittkau & Comes 2005), although suffering from low levels of haplotype variation, has revealed high levels of genetic differentiation (FST) both between sampled islands connected during the LGM (i.e. Tinos, Mikonos, Siros, Paros, Naxos), as well as between populations at the species-wide range (FST=0.782 versus 0.840). Both results indicate that seed flow (Nm=0.139 versus 0.095) is not strong enough to prevent N. degenii populations from diverging purely by drift (i.e. Nm≪1; Wright 1931; Slatkin 1985). To further investigate the species’ history of population splitting and divergence in the Cyclades, we have undertaken a survey of AFLP variation in 110 individuals, representing six Cycladia populations (ssp. barbro: Tinos, Mikonos; ssp. jenny: Siros; ssp. degenii: Paros1/2, Naxos), plus one ssp. degenii population from the volcanic island of Santorin/Thera, which has been isolated from Cycladia at least since the last interglacial (Snogerup 1967; Greuter 1979; Sfenthourakis 1996; see table 2 and inset of figure 4 for sampling sites). Genetic distance analysis of these individuals results in an unrooted neighbour-joining phenogram (figure 4) in which most individuals cluster according to their populations except for those from Paros (1/2) and Naxos, which cannot be clearly distinguished. Bayesian analysis of population structure as implemented in the program Structure v. 2.1 (Pritchard et al. 2000) confirms this pattern by recognizing five population clusters corresponding to Tinos, Mikonos Siros, Santorin and Paros (1/2)/Naxos (figure 5). Moreover, there was no significant association between genetic and geographical distances of these populations (r=0.249, Mantel test, p=0.177). This lack of isolation-by-distance in conjunction with pronounced population subdivision (FST =0.290; figure 5) once more suggests that genetic drift has been of much greater historical importance in these Cycladic populations compared with recurrent gene flow (Hutchison & Templeton 1999).
Table 2.
Location of sites used in the present AFLP study for Nigella degenii distributed across the Cyclades, and summary of AFLP variation for a total of seven populations. Variation is described by the proportion of AFLP markers (‘loci’) polymorphic (PLP) and Nei's (1973) expected heterozygosity He (or ‘gene diversity’, with FIS assumed to be zero) calculated by the program Fdist (Beaumont & Nichols 1996).
| N. degenii taxon | localitya | latitude (N) | longitude (E) | PLP | He | n |
|---|---|---|---|---|---|---|
| ssp. barbro | Tinos, E of Agios Fokas beach | 37° 31′ 54″ | 25° 13′ 11″ | 0.342 | 0.110 | 14 |
| Mikonos, Panormos beach | 37° 28′ 27″ | 25° 21′ 42″ | 0.361 | 0.109 | 18 | |
| ssp. jenny | Siros, near Kokkina beach (Finikas) | 37° 23′ 44″ | 24° 52′ 16″ | 0.390 | 0.152 | 19 |
| ssp. degenii | Paros1, S of Cape Korakas | 37° 08′ 52″ | 25° 13′ 20″ | 0.417 | 0.128 | 15 |
| Paros2, S of Lefkes | 37° 03′ 08″ | 25° 12′ 45″ | 0.426 | 0.155 | 18 | |
| Naxos, W of Himaros | 37° 03′ 36″ | 25° 27′ 39″ | 0.333 | 0.110 | 13 | |
| Santorin, S of Fira | 36° 24′ 40″ | 25° 26′ 15″ | 0.491 | 0.173 | 13 |
All collections by C. Bittkau. See figure 4 (inset) for geographical location.
Figure 4.
Neighbour-joining analysis of 110 multilocus AFLP phenotypes observed in 110 plants from seven populations of Nigella degenii from the Cyclades (inset) based on Nei & Li's (1979) genetic distances (see table 2 for locality information). Intermingled individuals from populations Paros1, Paros2 and Naxos are highlighted (dark, medium and light grey, respectively). Bootstrap values (>50%) based on 10 000 replicates are shown above branches. AFLP profiles were generated from total genomic DNA samples (Bittkau & Comes 2005) according to Schönswetter et al. (2002) with slight modifications, and using two EcoRI–MseI primer combinations (E+ACA/M+CAGC; E+ACC/M+CAGA) that resulted in a total of 108 polymorphic fragments.
Figure 5.
Histogram of Structure assignment test for seven Cycladic Nigella degenii populations (110 individuals) based on 108 polymorphic AFLP fragments. Each vertical bar represents an individual and its assignment proportion into one of five (white to black coloured) population clusters (K). After a burn-in period of 200 000 iterations, 10 independent runs of K=1–7 were performed at 106 Markov chain Monte Carlo samplings. The model with K=5 explained the data as satisfactorily as models with K=6 or 7 (showing similarly high log-likelihood values), whereby the latter did not further improve the resolution among the populations Paros1, Paros2 and Naxos (data not shown).
Despite low internal bootstrap support, the tree topology shown in figure 4 is strikingly congruent with the temporal order in which Cycladia fragmented (figure 3), with Tinos and Mikonos separating first (14–12.5 ka), followed by Siros (12.5–10 ka), and Paros and Naxos last (10–9 ka). Under this scenario, the high genetic similarity viz. admixture observed between populations from these latter islands would reflect recent common ancestry rather than post-divergence gene exchange. While we cannot presently distinguish between these two possibilities (but see §8), a more intriguing issue to be solved is the origin of the Santorin population of N. degenii, which does not conform to the expectations of a recent colonization (e.g. Carstens et al. 2005; Kropf et al. 2006). Instead this population groups next to the Paros/Naxos cluster rather than being nested within this group (figure 4) and shows elevated rather than reduced levels of genetic diversity (table 2). Together, this suggests that an over-sea dispersal event occurred from the Paros/Naxos area to Santorin, and that time was sufficient for genetic diversity to evolve in the colonized area, and to erase any pattern of nestedness because of the accumulation of de novo mutations and/or the effects of continued lineage sorting (Avise 2000). Hence, if our inferred times and orders of Cycladia population splitting are correct, and barring insufficient population sampling, N. degenii must have dispersed to Santorin shortly before the Paros/Naxos mega-island broke up (ca 9 ka). In turn, this would raise the intriguing possibility that the massive eruption of the Thera volcano in the mid-second millennium BC (Heiken & McCoy 1984) did not eradicate N. degenii on Santorin (see Thorpe et al. 2008, for a similar scenario in Lesser Antillean Anolis lizards).
8. Conclusions and future directions
The geographical distribution of intraspecific genetic (cpDNA, AFLP) variation in N. degenii from the Cyclades is broadly consistent with a pure drift/isolation model. In other words, historical, non-equilibrium sampling processes following post-glacial fragmentation of Cycladia or (rarely) long-distance colonization have indeed been the primary determinants of population divergence across these islands. These patterns of intraspecific genetic divergence therefore may render N. degenii at an incipient stage of allopatric speciation. As such, the species reiterates the major evolutionary and biogeographic processes of speciation as invoked for the entire N. arvensis complex, albeit at a time scale probably much shorter than that required for most speciation in this plant group. Clearly, the population demographic history of N. degenii remains to be explored more fully, ideally with multi-locus coalescent methods at the DNA sequence level (Majoram & Tavaré 2006), and with the target of directly estimating times since population isolation, population-size changes at the time of divergence and post-divergence rates of gene flow. Inferring such key demographic parameters in an Aegean plant species such as N. degenii, occupying a patchwork of approximately 20 islands in the Cyclades, would provide an excellent test case for the rapidly expanding field of ‘divergence population genetics’ (Hey & Nielsen 2004, 2007; Dolman & Moritz 2006; Marjoram & Tavaré 2006).
However, whether the magnitude of genetic divergence among populations within a species or between species may be considered an indicator of, respectively, incipient and full speciation is likely to depend on the species concept invoked (Baum 1992; Avise 2000; Hey 2001; Coyne & Orr 2004). In any event, based on our current molecular (AFLP) evidence (figure 2), the two selfers (N. doerfleri and N. stricta) and most species of the outcrossing N. arvensis alliance did evolve as independent and coherent gene pools (except N. arvensis). However, whereas the selfers accumulated sufficient reproductive isolation barriers (inter-se and towards the alliance), species of the latter group remained interfertile (see §4b). That said, closer scrutiny of Strid's (1970) crossing data for the alliance reveals various degrees of partial hybrid sterility both at the inter- and intraspecific levels. While pending better-resolved species and population relationships, it is tempting to speculate that the magnitude of such partial (intrinsic post-zygotic) reproductive isolation between pairs of species or populations will increase with genetic distance, as would be expected for Dobzhansky–Muller incompatibilities (see §6) that evolve because of intragenomic conflicts as by-products of the divergence of genomes in allopatry and via drift (Price & Bouvier 2002; Moyle et al. 2004; Bolnick & Near 2005; Burt & Trivers 2006; see Mallet 2008; Cozzolino & Scopece 2008). Also, it would be worth exploring the genetic basis of such partial hybrid sterility in the N. arvensis alliance by means of quantitative trait locus (QTL) analysis. In view of recent studies of chromosomal speciation (Noor et al. 2001; Rieseberg 2001; Navarro & Barton 2003), the prediction would be that QTLs affecting hybrid fertility are associated with chromosomal rearrangements rather than non-rearranged regions of the genome.
Finally, perhaps the most formidable challenge is to falsify the hypothesis that much of the phenotypic diversity seen in the N. arvensis complex in general, and N. degenii in particular, is largely non-adaptive and the result of random genetic drift (cf. Strid 1970). One approach to address this issue is to use estimates of population structure for quantitative phenotypic traits (QST) to infer natural selection acting on those traits by their departure from patterns (FST) seen at ostensibly neutral molecular markers (Merilä & Crnokrak 2001; McKay & Latta 2002). Using AFLP markers, Jorgensen et al. (2006) recently applied this method separately to island populations of N. degenii from Mikonos (ssp. barbro) and Siros (ssp. jenny). Within each of the two islands, they found little support for diversifying selection being important in structuring variation in a suite of quantitative developmental, vegetative and floral characters (QST≈FST). Essentially, the same results emerge when populations of N. degenii from different islands (i.e. Mikonos, Siros, Paros, Naxos) are subjected to AFLF-based QST/FST analyses for similar quantitative traits (C. Bittkau & H. P. Comes, unpublished data). As a notable exception, Jorgensen et al. (2006) showed that intra-island estimates of FST for dimorphic (light versus dark) pollen colour variation, which is under single, dominant gene control in N. degenii, significantly exceed neutral expectations estimated from AFLP data. The mechanism underlying this signal of diversifying selection may well involve morph-by-environment interactions (because of pleiotropy and/or linkage) in response to local water or nutrient availability, rather than reflecting pollinator-mediated selection acting directly on pollen colour (Jorgensen et al. 2006; see also Strid 1970, p. 142).
On balance, the N. degenii results strongly argue in favour of Strid's (1970) hypothesis that genetic drift rather than selection is shaping the phenotypic population structure of this species, at both the inter- and intra-island levels. This outcome accords well with the biogeographic history of N. degenii and its local population dynamics viz. large, seasonal fluctuations in population size (Strid 1970; C. Bittkau 2004, personal observation). It may also indicate that populations experience too similar ecological conditions for diversifying selection to be of any major importance (Jorgensen et al. 2006). Consequently, there is a need for additional studies on the nature of phenotypic differences between species of the complex that strongly differ in habitat requirements and/or breeding system, and for which, therefore, the diversifying effect of selection should become more apparent (e.g. N. degenii versus N. doerfleri, see §6). This type of species pair, especially if identified as sister, may provide the best model systems for comparative genomics (i) to detect signatures of adaptive molecular variation per se, for example, via FST-outlier tests (Luikhart et al. 2003) or expression-based approaches (Bouck & Vision 2007); and (ii) to isolate and characterize genes that control traits important in adaptation and reproductive isolation (Noor & Feder 2006). These technological approaches may not only provide additional insights into our conception of how new Aegean plant species form, but will also improve our understanding of the genetic changes that make such species and the relative roles of selection versus drift in their evolution.
Acknowledgments
This research was primarily funded by a grant from the Deutsche Forschungsgemeinschaft (DFG Co254/3-1) with additional support from the Department of Organismic Biology, Salzburg University. The authors thank Michaela Eder, Ursula Jaros (Salzburg University), Petra Siegert and Marion Kever (Mainz University) for laboratory assistance. The kind offer by Roger S. Thorpe (Bangor University) of a preprint is gratefully acknowledged as well as the insightful comments of three anonymous reviewers.
Footnotes
One contribution of 12 to a Theme Issue ‘Speciation in plants and animals: pattern and process’.
References
- Avise J.C. Harvard University Press; Cambridge, MA: 2000. Phylogeography: the history and formation of species. [Google Scholar]
- Baldwin B.G, Sanderson M.J. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc. Natl Acad. Sci. USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. doi:10.1073/pnas.95.16.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough T.G, Nee S. Phylogenetics and speciation. Trends Ecol. Evol. 2001;16:391–399. doi: 10.1016/s0169-5347(01)02161-9. doi:10.1016/S0169-5347(01)02161-9 [DOI] [PubMed] [Google Scholar]
- Barraclough T.G, Vogler A.P. Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 2000;15:419–434. doi: 10.1086/303332. doi:10.1086/303332 [DOI] [PubMed] [Google Scholar]
- Barrett S.C.H. The reproductive biology and genetics of island plants. Phil. Trans. R. Soc. B. 1996;351:725–733. doi:10.1098/rstb.1996.0067 [Google Scholar]
- Barton N.H. Natural selection and random genetic drift as causes of evolution on islands. Phil. Trans. R. Soc. B. 1996;351:785–795. doi: 10.1098/rstb.1996.0073. doi:10.1098/rstb.1996.0073 [DOI] [PubMed] [Google Scholar]
- Baum D. Phylogenetic species concepts. Trends Ecol. Evol. 1992;7:1–2. doi: 10.1016/0169-5347(92)90187-G. doi:10.1016/0169-5347(92)90187-G [DOI] [PubMed] [Google Scholar]
- Beaumont M.A, Nichols R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B. 1996;263:1619–1626. doi:10.1098/rspb.1996.0237 [Google Scholar]
- Beerli P, Hotz H, Uzzell T. Geologically dated sea barriers calibrate a protein clock for Aegean water frogs. Evolution. 1996;50:1676–1687. doi: 10.1111/j.1558-5646.1996.tb03939.x. doi:10.2307/2410903 [DOI] [PubMed] [Google Scholar]
- Bentzer B. Taxonomy, variation and evolution in representatives of Leopoldia Parl. (Liliaceae) in the southern and central Aegean. Bot. Notiser. 1973;126:69–132. [Google Scholar]
- Berry R.J. Small mammal differentiation on islands. Phil. Trans. R. Soc. B. 1996;351:753–764. doi: 10.1098/rstb.1996.0070. doi:10.1098/rstb.1996.0070 [DOI] [PubMed] [Google Scholar]
- Birky C.W, Fuerst P, Maruyama T. Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics. 1989;121:613–627. doi: 10.1093/genetics/121.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittkau C, Comes H.P. Evolutionary processes in a continental island system: molecular phylogeography of the Aegean Nigella arvensis alliance (Ranunculaceae) inferred from chloroplast DNA. Mol. Ecol. 2005;14:4065–4083. doi: 10.1111/j.1365-294X.2005.02725.x. doi:10.1111/j.1365-294X.2005.02725.x [DOI] [PubMed] [Google Scholar]
- Bittkau, C. & Comes, H. P. In press. Molecular inference of a Late Pleistocene shift in Nigella s. lat. (Ranunculaceae) resulting from increased speciation in the Aegean archipelago. J. Biogeogr
- Blondel J, Aronson J. Oxford University Press; Oxford, UK: 1999. Biology and wildlife of the Mediterranean region. [Google Scholar]
- Böhle U.-R, Hilger H.H, Martin W.F. Island colonization and evolution of the insular woody habit in Echium L. (Boraginaceae) Proc. Natl Acad. Sci. USA. 1996;93:11 740–11 745. doi: 10.1073/pnas.93.21.11740. doi:10.1073/pnas.93.21.11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick D.I, Near T.J. Tempo of hybrid inviability in centrarchid fishes (Teleostei: Centrarchidae) Evolution. 2005;59:1754–1767. doi:10.1554/04-563.1 [PubMed] [Google Scholar]
- Bouck A, Vision T. The molecular ecologist's guide to expressed sequence tags. Mol. Ecol. 2007;16:1007–1022. doi: 10.1111/j.1365-294X.2006.03195.x. doi:10.1111/j.1365-294X.2006.03195.x [DOI] [PubMed] [Google Scholar]
- Broodbank C. Cambridge University Press; Cambridge, UK: 2000. An island archaeology of the early Cyclades. [Google Scholar]
- Burt A, Trivers R. Harvard University Press; Cambridge, MA: 2006. Genes in conflict: the biology of selfish genetic elements. [Google Scholar]
- Cain S.A. Harper and Row; New York, NY: 1944. Foundations of plant biogeography. [Google Scholar]
- Cameron R.A.D, Cook L.M, Hallows J.D. Land snails on Porto Santo: adaptive and non-adaptive radiation. Phil. Trans. R. Soc. B. 1996;351:309–327. doi:10.1098/rstb.1996.0025 [Google Scholar]
- Carson H.L. The genetics of speciation at the diploid level. Am. Nat. 1975;109:83–92. doi:10.1086/282975 [Google Scholar]
- Carstens B.C, Brunsfeld S.J, Demboski J.R, Good J.M, Sullivan J. Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: hypothesis testing within a comparative phylogeographic framework. Evolution. 2005;59:1639–1652. doi:10.1554/04-661.1 [PubMed] [Google Scholar]
- Chatzimanolis S, Trichas A, Mylonas M. Phylogenetic analysis and biogeography of Aegean taxa of the genus Dendarus (Coleoptera: Tenebrionidae) Insect Syst. Evol. 2003;34:295–312. [Google Scholar]
- Coyne J.A, Orr H.A. The evolutionary genetics of speciation. In: Magurran A.E, May R.M, editors. Evolution of biological diversity. Oxford University Press; Oxford, UK: 1999. pp. 1–36. [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer Associates; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Coyne J.A, Barton N.H, Turelli M. A critique of Sewall Wright's shifting balance theory. Evolution. 1997;51:643–671. doi: 10.1111/j.1558-5646.1997.tb03650.x. doi:10.2307/2411143 [DOI] [PubMed] [Google Scholar]
- Cozzolino S, Scopece G. Specificity in pollination and consequences for postmating reproductive isolation in deceptive Mediterranean orchids. Phil. Trans. R. Soc. B. 2008;363:3037–3046. doi: 10.1098/rstb.2008.0079. doi:10.1098/rstb.2008.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daams R, Van der Weerd A.V. Early Pliocene small mammals from the Aegean island of Karpathos (Greece) and their paleogeographic significance. Geol. Mijnbouw. 1980;59:327–331. [Google Scholar]
- Dolman G, Moritz C. A multilocus perspective on refugial isolation and divergence in rainforest skinks (Carlia) Evolution. 2006;60:573–582. doi:10.1111/j.0014-3820.2006.tb01138.x [PubMed] [Google Scholar]
- Duggen S, Hoernle K, Van den Bogaard P, Rüpke L, Morgan J.P. Deep roots of the Messinian salinity crisis. Nature. 2003;422:602–606. doi: 10.1038/nature01553. doi:10.1038/nature01553 [DOI] [PubMed] [Google Scholar]
- Fattorini S. Biogeography of the tenebrionid beetles (Coleoptera, Tenebrionidae) on the Aegean Islands (Greece) J. Biogeogr. 2002;29:49–67. doi:10.1046/j.1365-2699.2002.00656.x [Google Scholar]
- Fishman L, Stratton D.A. The genetics of floral divergence and postzygotic barriers between outcrossing and selfing populations of Arenaria uniflora (Caryophyllaceae) Evolution. 2004;58:296–307. doi:10.1554/03-069 [PubMed] [Google Scholar]
- Giokas S. Congruence and conflict in Albinaria (Gastropoda, Clausiliidae). A review of morphological and molecular phylogenetic approaches. Belg. J. Zool. 2000;130:93–100. [Google Scholar]
- Gittenberger E. Sympatric speciation in snails: a largely neglected model. Evolution. 1988;42:826–828. doi: 10.1111/j.1558-5646.1988.tb02502.x. doi:10.2307/2408875 [DOI] [PubMed] [Google Scholar]
- Gittenberger E. What about non-adaptive radiation? Biol. J. Linn. Soc. 1991;43:263–272. doi:10.1111/j.1095-8312.1991.tb00598.x [Google Scholar]
- Givnish T.J. Adaptive radiation and molecular studies: aims and conceptual issues. In: Givnish T.J, Sytsma K.J, editors. Molecular evolution and adaptive radiation. Cambridge University Press; Cambridge, UK: 1997. pp. 1–54. [Google Scholar]
- Grant P.R. Speciation. In: Grant P.R, editor. Evolution on islands. Oxford University Press; Oxford, UK: 1998. pp. 83–101. [Google Scholar]
- Greuter W. The origins and evolution of island floras as exemplified by the Aegean Archipelago. In: Bramwell D, editor. Plants and islands. Academic Press; New York, NY: 1979. pp. 87–106. [Google Scholar]
- Grundt H.H, Kjølner S, Borgen L, Rieseberg L.H, Brochmann C. High biological species diversity in the arctic flora. Proc. Natl Acad. Sci. USA. 2006;103:972–975. doi: 10.1073/pnas.0510270103. doi:10.1073/pnas.0510270103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P.H, May R.M, Nee S. Phylogenies without fossils. Evolution. 1994;48:523–529. doi: 10.1111/j.1558-5646.1994.tb01341.x. doi:10.2307/2410466 [DOI] [PubMed] [Google Scholar]
- Heiken G, McCoy F. Caldera development during the Minoan eruption, Thera, Cyclades, Greece. J. Geophys. Res. 1984;89:8441–8462. doi:10.1029/JB089iB10p08441 [Google Scholar]
- Hey J. Oxford University Press; Oxford, UK: 2001. Genes, categories, and species: the evolutionary and cognitive causes of the species problem. [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. doi:10.1534/genetics.103.024182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl Acad. Sci. USA. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. doi:10.1073/pnas.0611164104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl Acad. Sci. USA. 2006;103:10 334–10 339. doi: 10.1073/pnas.0601928103. doi:10.1073/pnas.0601928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison D.W, Templeton A.R. Correlation of pairwise genetic and geographic distance measures: the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. doi:10.2307/2640449 [DOI] [PubMed] [Google Scholar]
- Johnson S.D. Pollinator-driven speciation in plants. In: Harder L.D, Barrett S.C.H, editors. Ecology and evolution of flowers. Oxford University Press; Oxford, UK: 2006. pp. 295–310. [Google Scholar]
- Jorgensen T.H, Richardson D.S, Andersson S. Comparative analysis of population structure in two subspecies of Nigella degenii: evidence for diversifying selection on pollen-color dimorphisms. Evolution. 2006;60:518–528. doi:10.1111/j.0014-3820.2006.tb01133.x [PubMed] [Google Scholar]
- Kay M.K, Reeves P.A, Olmstead R.G, Schemske D.W. Rapid speciation and the evolution of hummingbird pollination in Neotropical Costus subgenus Costus (Costaceae): evidence from nrDNA ITS and ETS sequences. Am. J. Bot. 2005;92:1899–1910. doi: 10.3732/ajb.92.11.1899. doi:10.3732/ajb.92.11.1899 [DOI] [PubMed] [Google Scholar]
- Klak C, Reeves G, Hedderson T. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature. 2003;427:63–65. doi: 10.1038/nature02243. doi:10.1038/nature02243 [DOI] [PubMed] [Google Scholar]
- Kozak K.H, Wiens J.J. Does niche conservatism promote speciation? A case study in North American salamanders. Evolution. 2006;60:2604–2621. doi:10.1111/j.0014-3820.2006.tb01893.x [PubMed] [Google Scholar]
- Kropf M, Comes H.P, Kadereit J.W. Long-distance dispersal vs. vicariance: the origin and genetic diversity of alpine plants in the Spanish Sierra Nevada. New Phytol. 2006;172:169–184. doi: 10.1111/j.1469-8137.2006.01795.x. doi:10.1111/j.1469-8137.2006.01795.x [DOI] [PubMed] [Google Scholar]
- Kuss S.E. Die pleistozänen Hirsche der ost-mediterranen Inseln Kreta, Kasos, Karpathos und Rhodos (Griechenland) Ber. Naturforsch. Ges. Freib. Breisgau. 1975;65:25–79. [Google Scholar]
- Lambeck K. Sea-level change and shore-line evolution in Aegean Greece since Upper Palaeolithic time. Antiquity. 1996;70:588–611. [Google Scholar]
- Levin D.A. Oxford University Press; Oxford, UK: 2000. The origin, expansion, and demise of plant species. [Google Scholar]
- Lexer C, Widmer A. The genic view of plant speciation: recent progress and emerging questions. Phil. Trans. R. Soc. B. 2008;363:3023–3036. doi: 10.1098/rstb.2008.0078. doi:10.1098/rstb.2008.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder H.P. Plant species radiations: where, when, why? Phil. Trans. R. Soc. B. 2008;363:3097–3105. doi: 10.1098/rstb.2008.0075. doi:10.1098/rstb.2008.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England P.R, Tallon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 2003;4:981–994. doi: 10.1038/nrg1226. doi:10.1038/nrg1226 [DOI] [PubMed] [Google Scholar]
- MacArthur R.W. Princeton University Press; Princeton, NJ: 1972. Geographical ecology. [Google Scholar]
- Macnair M.R, Gardner M. The evolution of edaphic endemics. In: Howard D.J, Berlocher S.H, editors. Endless forms: species and speciation. Oxford University Press; Oxford, UK: 1998. pp. 157–171. [Google Scholar]
- Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Phil. Trans. R. Soc. B. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. doi:10.1098/rstb.2008.0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram P, Tavaré S. Modern computational approaches for analysing molecular genetic variation. Nat. Rev. Genet. 2006;7:759–770. doi: 10.1038/nrg1961. doi:10.1038/nrg1961 [DOI] [PubMed] [Google Scholar]
- Mayr E. Change of genetic environment and evolution. In: Huxley J, Hardy A.C, Ford E.B, editors. Evolution as a process. Allen and Unwin; London, UK: 1954. pp. 157–180. [Google Scholar]
- McKay J.K, Latta R.G. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 2002;17:285–291. doi:10.1016/S0169-5347(02)02478-3 [Google Scholar]
- Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 2001;14:892–903. doi:10.1046/j.1420-9101.2001.00348.x [Google Scholar]
- Meudt H.M, Clarke A.C. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;12:106–117. doi: 10.1016/j.tplants.2007.02.001. doi:10.1016/j.tplants.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Meulenkamp J.E, De Mulder E.F.J, Van de Wird A. Sedimentary history and paleogeography of the late Cenozoic of the island of Rhodos. Z. Dtsch. Geol. Ges. 1972;123:541–553. [Google Scholar]
- Moyle L.C, Olson M.S, Tiffin P. Patterns of reproductive isolation in three angiosperm genera. Evolution. 2004;58:1195–1208. doi: 10.1111/j.0014-3820.2004.tb01700.x. doi:10.1111/j.0014-3820.2004.tb01700.x [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton N.H. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution. 2003;57:447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x. doi:10.1554/0014-3820(2003)057[0447:APIGIP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nee S, Holmes E.C, May R.M, Harvey P.H. Extinction rates can be estimated from molecular phylogenies. Phil. Trans. R. Soc. B. 1994;344:77–82. doi: 10.1098/rstb.1994.0054. doi:10.1098/rstb.1994.0054 [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl Acad. Sci. USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. doi:10.1073/pnas.70.12.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. doi:10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Maruyama T, Wu C.-I. Models of evolution of reproductive isolation. Genetics. 1983;103:557–579. doi: 10.1093/genetics/103.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M.A.F, Feder J.L. Speciation genetics: evolving approaches. Nat. Rev. Genet. 2006;7:851–861. doi: 10.1038/nrg1968. doi:10.1038/nrg1968 [DOI] [PubMed] [Google Scholar]
- Noor M.A.F, Grams K.L, Bertucci L.A, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl Acad. Sci. USA. 2001;98:12 084–12 088. doi: 10.1073/pnas.221274498. doi:10.1073/pnas.221274498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R, Thorpe R.S. Molecular evidence for ecological speciation in tropical ecotopes. Proc. Natl Acad. Sci. USA. 2002;99:13 612–13 615. doi: 10.1073/pnas.212248499. doi:10.1073/pnas.212248499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;55:1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. doi:10.1111/j.0014-3820.2001.tb00628.x [DOI] [PubMed] [Google Scholar]
- Paradis E. Detecting shifts in diversification rates without fossils. Am. Nat. 1998;152:176–187. doi: 10.1086/286160. doi:10.1086/286160 [DOI] [PubMed] [Google Scholar]
- Perissoratis C, Conispoliatis N. The impacts of sea-level changes during latest Pleistocene and Holocene times on the morphology of the Ionian and Aegean seas (SE Alpine Europe) Mar. Geol. 2003;196:145–156. doi:10.1016/S0025-3227(03)00047-1 [Google Scholar]
- Poulakakis N, Lymberakis P, Antoniou A, Chalkia D, Zouros E, Mylonas M, Valakos E. Molecular phylogeny and biogeography of the wall-lizard Podarcis erhardii (Squamata: Lacertidae) Mol. Phylogenet. Evol. 2003;28:38–46. doi: 10.1016/s1055-7903(03)00037-x. doi:10.1016/S1055-7903(03)00037-X [DOI] [PubMed] [Google Scholar]
- Poulakakis N, Lymberakis P, Tsigenopoulos C.S, Magoulas A, Mylonas M. Phylogenetic relationships and evolutionary history of snake-eyed skink Ablepharus kitaibelii (Sauria: Scincidae) Mol. Phylogenet. Evol. 2005;34:245–256. doi: 10.1016/j.ympev.2004.10.006. doi:10.1016/j.ympev.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Price T.D, Bouvier M.M. The evolution of F1 postzygotic incompatibilities in birds. Evolution. 2002;56:2083–2089. doi:10.1554/0014-3820(2002)056[2083:TEOFPI]2.0.CO;2 [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donelly P. Inference of population structure from multilocus genotype data. Mol. Ecol. 2000;16:2852–2854. [Google Scholar]
- Rechinger K.H. Grundzüge der Pflanzenverbreitung in der Aegäis I-III. Vegetatio. 1950;2:55–119. doi:10.1007/BF00151672 See also pp. 239–308, 365–386. [Google Scholar]
- Richardson J.E, Weitz F.M, Fay M.F, Cronk Q.C.B, Linder H.P, Reeves G, Chase M.W. Rapid and recent origin of species richness in the Cape flora of South Africa. Nature. 2001;412:181–183. doi: 10.1038/35084067. doi:10.1038/35084067 [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E. Estimating diversification rates from phylogenetic information. Trends Ecol. Evol. 2007;22:601–610. doi: 10.1016/j.tree.2007.06.013. doi:10.1016/j.tree.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. doi:10.1016/S0169-5347(01)02187-5 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, Widmer A, Arntz A.M, Burke J.M. Directional selection is the primary cause of phenotypic diversification. Proc. Natl Acad. Sci. USA. 2002;99:12 242–12 245. doi: 10.1073/pnas.192360899. doi:10.1073/pnas.192360899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runemark H. Reproductive drift, a neglected principle in reproductive biology. Bot. Notiser. 1969;122:90–129. [Google Scholar]
- Runemark H. Studies in the Aegean Flora XXIII. The Dianthus fruticosus complex (Caryophyllaceae) Bot. Notiser. 1980;133:475–490. [Google Scholar]
- Sanderson M.J. Reappraising adaptive radiation. Am. J. Bot. 1998;85:1650–1655. doi:10.2307/2446495 [Google Scholar]
- Sanderson M.J, Wojciechowski M.F. Diversification rates in a temperate legume clade: why are there ‘so many species’ of Astragalus (Fabaceae) Am. J. Bot. 1996;83:1488–1502. doi:10.2307/2446103 [Google Scholar]
- Savolainen V, Forest F. Species-level phylogenetics from continental biodiversity hotspots. In: Bakker F.T, Chatrou L.W, Gravendeel B, Pelser P.B, editors. Plant species-level systematics: patterns, processes and new applications. Koeltz; Koenigstein, Germany: 2005. pp. 17–30. (Regnum Vegetabile 142). [Google Scholar]
- Schluter D. Oxford University Press; Oxford, UK: 2000. The ecology of adaptive radiation. [Google Scholar]
- Schönswetter P, Tribsch A, Barfuss M, Niklfeld H. Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium in the European Alps. Mol. Ecol. 2002;11:2637–2647. doi: 10.1046/j.1365-294x.2002.01651.x. doi:10.1046/j.1365-294X.2002.01651.x [DOI] [PubMed] [Google Scholar]
- Sfenthourakis S. A biogeographic analysis of terrestrial isopods (Isopoda, Oniscidea) from central Aegean islands (Greece) J. Biogeogr. 1996;23:687–698. doi:10.1111/j.1365-2699.1996.tb00029.x [Google Scholar]
- Silvertown J. The ghost of competition past in the phylogeny of island endemic plants. J. Ecol. 2004;92:168–173. doi:10.1111/j.1365-2745.2004.00853.x [Google Scholar]
- Silvertown J, Francisco-Ortega J, Carine M. The monophyly of island radiations: an evaluation of niche pre-emption and some alternative explanations. J. Ecol. 2005;93:653–657. doi:10.1111/j.1365-2745.2005.01038.x [Google Scholar]
- Slatkin M. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 1985;16:393–430. doi:10.1146/annurev.ecolsys.16.1.393 [Google Scholar]
- Snogerup S. Studies in the Aegean flora. IX. Erysimum sect. Cheiranthus. B. Variation and evolution in the small-population system. Opera Bot. 1967;14:1–86. [Google Scholar]
- Stanley S.M. W. H. Freeman; San Francisco, CA: 1979. Macroevolution: pattern and process. [Google Scholar]
- Stork A.L. Studies in the Aegean flora. XX. Biosystematics of the Malcolmia maritima complex. Opera Bot. 1972;33:1–118. [Google Scholar]
- Strid A. Evolutionary trends in the breeding system of Nigella (Ranunculaceae) Bot. Notiser. 1969;122:380–397. [Google Scholar]
- Strid A. Studies in the Aegean flora. XVI. Biosystematics of the Nigella arvensis complex. Opera Bot. 1970;28:1–169. [Google Scholar]
- Strid A. Phytogeographia Aegaea and the Flora Hellenica database. Ann. Naturhist. Mus. Wien. 1996;96B:279–289. [Google Scholar]
- Strid, A. 1997 Introduction. In Flora Hellenica, vol. 1 (eds A. Strid & K. Tan), pp. ix–xxxv. Koenigstein, Germany: Koeltz.
- Strid, A. 2002 Nigella L. In Flora Hellenica, vol. 2 (eds A. Strid & K. Tan), pp. 5–13. Koenigstein, Germany: Koeltz.
- Stuessy T.F, Jakubowsky G, Salguero Gómez R, Pfosser M, Schlüter P.M, Fer T, Sun B.-Y, Kato H. An alternative model for plant evolution in islands. J. Biogeogr. 2006;33:1259–1265. doi:10.1111/j.1365-2699.2006.01504.x [Google Scholar]
- Templeton A.R. The theory of speciation via the founder principle. Genetics. 1980;94:1011–1038. doi: 10.1093/genetics/94.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe R.S, Surget-Groba Y, Johansson H. The relative importance of ecology and geographic isolation for speciation in anoles. Phil. Trans. R. Soc. B. 2008;363:3071–3081. doi: 10.1098/rstb.2008.0077. doi:10.1098/rstb.2008.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Barton N.H, Coyne J.A. Theory and speciation. Trends Ecol. Evol. 2001;16:330–343. doi: 10.1016/s0169-5347(01)02177-2. doi:10.1016/S0169-5347(01)02177-2 [DOI] [PubMed] [Google Scholar]
- Ueshima R, Asami T. Single-gene speciation by left-right reversal. Nature. 2003;425:679. doi: 10.1038/425679a. doi:10.1038/425679a [DOI] [PubMed] [Google Scholar]
- Van Andel T.H, Shackleton J.C. Late Paleolithic and Mesolithic coastlines of Greece and the Aegean. J. Field Archaeol. 1982;9:445–454. doi:10.2307/529681 [Google Scholar]
- Von Bothmer R. Studies in the Aegean Flora XXI. Biosystematic studies in the Allium ampeloprasum complex. Opera Bot. 1974;34:1–104. [Google Scholar]
- Von Bothmer R. Differentiation patterns in the E. Mediterranean Scutellaria rubicunda group (Lamiaceae) Plant Syst. Evol. 1987;155:219–249. doi:10.1007/BF00936301 [Google Scholar]
- Waser N.M, Campbell D.R. Ecological speciation in flowering plants. In: Dieckmann U, Doebeli M, Metz J.A.J, Tautz D, editors. Adaptive speciation. Cambridge University Press; Cambridge, UK: 2004. pp. 264–277. [Google Scholar]
- Wendel J.F, Percival A.E. Molecular divergence in the Galapagos Islands-Baja California species pair, Gossypium klotzschianum and G. davidsonii (Malvaceae) Plant Syst. Evol. 1990;171:99–115. doi:10.1007/BF00940598 [Google Scholar]
- Whittaker R.J, Fernández-Palacios J.M. 2nd edn. Oxford University Press; Oxford, UK: 2007. Island biogeography: ecology, evolution, and conservation. [Google Scholar]
- Wiens J.J. What is speciation and how should we study it? Am. Nat. 2004;163:914–923. doi: 10.1086/386552. doi:10.1086/386552 [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The statistical consequences of Mendelian heredity in relation to speciation. In: Huxley J.S, editor. The new systematics. Clarendon Press; Oxford, UK: 1940. pp. 161–183. [Google Scholar]
- Wright S. On the rôles of directed and random changes in gene frequencies in the genetics of populations. Evolution. 1948;2:279–294. doi: 10.1111/j.1558-5646.1948.tb02746.x. doi:10.2307/2405519 [DOI] [PubMed] [Google Scholar]