Abstract
Homoploid hybrid speciation (HHS) is the establishment of a novel species through introgressive hybridization without a change in chromosome number. We discuss different routes by which this might occur and propose a novel term, ‘hybrid trait speciation’, which combines the idea that hybridization can generate adaptive novelty with the ‘magic trait’ model of ecological speciation. Heliconius butterflies contain many putative examples of hybrid colour patterns, but only recently has the HHS hypothesis been tested explicitly in this group. Molecular data has shown evidence for gene flow between many distinct species. Furthermore, the colour pattern of Heliconius heurippa can be recreated in laboratory crosses between Heliconius melpomene and Heliconius cydno and, crucially, plays a role in assortative mating between the three species. Nonetheless, although the genome of H. heurippa shows evidence for hybridization, it is not a mosaic of the two parental species. Instead, ongoing hybridization has likely blurred any signal of the original speciation event. We argue that where hybridization leads to novel adaptive traits that also cause reproductive isolation, it is likely to trigger speciation.
Keywords: mimicry, homoploid hybrid speciation, biodiversity
1. Introduction
Homoploid hybrid speciation (HHS) is the formation of a novel, reproductively isolated population through hybridization, without a change in chromosome number (Coyne & Orr 2004). The process is distinct from hybrid speciation through polyploidization, which is a well established route to speciation, particularly in plants (Stebbins 1950; Hegarty et al. 2008). HHS has commonly been considered rare or absent in animals (but see Stebbins 1959; Templeton 1981), although there have been several putative examples published recently (Nolte et al. 2005; Schwarz et al. 2005; Gompert et al. 2006; Mavárez et al. 2006; Meye et al. 2006). The reasons for its apparent rarity are probably partly real and partly historical. First, there are theoretical reasons why it is hard to imagine a hybrid lineage becoming established, primarily because such a lineage is likely to be eliminated by its parents either through backcrossing or ecological competition, which might suggest hybrid speciation is genuinely a rare phenomenon (Mallet 2007). Second, there is a logistical problem of detection inherent in the genetic and morphological methodologies available. Specifically, determining that novel adaptive traits (as opposed to neutral genetic variation) are a result of hybridization can be challenging. Finally, however, there has also been an inherent scepticism of the importance of hybrids in the animal literature, perhaps because of the widespread adoption of the biological species concept, which specifically precludes hybridization between ‘good’ species (Mallet 2005, 2008). This scepticism is in marked contrast to the plant literature and is an attitude that is changing rapidly in the light of new evidence. In particular, hybridization has been resurrected as a major force in adaptive animal radiations (Rieseberg et al. 1999; Seehausen 2004).
(a) Models of homoploid hybrid speciation
Hybrid speciation has recently been defined as the ‘process in which natural hybridization results in the production of an evolutionary lineage that is at least partially reproductively isolated from both parental lineages, and which demonstrates a distinct evolutionary and ecological trajectory’ (Arnold 2006). Importantly, hybridization must lead to at least some reproductive isolation from both parental species, allowing the daughter population to diverge independently and this is the definition that we adopt here. HHS has been best studied in the sunflower genus Helianthus, where the stabilization of hybrid species has involved the establishment of a lineage with genomic blocks derived from one or other parent, distributed throughout the genome (Ungerer et al. 1998). The stabilization of such hybrid species takes a number of generations during which incompatible genomic regions are rapidly eliminated by selection. Incompatibility between parental and daughter species is thus rapidly established because the resultant hybrid genome possesses large numbers of genes that cause epistatic hybrid breakdown when crossed back to either parent (Ungerer et al. 1998).
This mode of hybrid speciation is unlikely to apply in more recent adaptive radiations, such as cichlid fish, where hybridizing species have not accumulated genetic incompatibility at multiple genetic loci. In such lineages speciation typically does not result from intrinsic hybrid incompatibility at large numbers of loci, but rather through sexual and ecological isolation at a few traits driven by strong selection. Recent species pairs can be strongly isolated in the wild but produce completely compatible hybrid offspring in laboratory crosses (Seehausen et al. 1997). Commonly, adaptive divergence at a single or a few traits causes reproductive isolation as a side effect of divergence (i.e. magic traits, Gavrilets 2004; Jiggins et al. 2004). Examples include beak size and vocal signature in Darwin's finches (Podos 2001), body size and colour in stickleback fish (Boughman 2001) and colour patterns in Hypoplectrus fish (Puebla et al. 2007). Pleiotropic effects of ecological traits on mating can be divided into three classes (Jiggins et al. 2004): Pleiotropic assortment traits, in which a single trait leads directly to assortative mating, the classic example being host choice in phytophagous insects; Pleiotropic mating cues, in which a cue used in mate choice diverges under ecological selection, such as butterfly colour patterns that are under selection for signalling to predators as well as to potential mates; and finally pleiotropic mating preferences in which adaptation of the sensory system leads to pleiotropic divergence in mate preference because of ecological selection (Jiggins et al. 2004).
Given the long-standing suggestion that hybridization might contribute to adaptively significant variation, either within species or in novel lineages (Stebbins 1959; Lewontin & Birch 1966; Templeton 1981; Arnold 1997, 2006), it seems probable that novel magic traits might also arise through hybridization. Thus, a hybrid lineage might colonize a new habitat by combining one or more ecological traits from two divergent species, and those novel traits would also confer reproductive isolation from the parents. The hybrid characteristics must necessarily be controlled by two or more genes, such that both parents contribute adaptively significant variation to the novel lineage, but could involve just a handful of genetic loci. This process might also be facilitated by ‘transgressive segregation’ of an ecological trait. This is the phenomenon whereby hybrid trait values lie outside the range of the two parents, and is a surprisingly frequent occurrence (Rieseberg et al. 2003; Hegarty et al. 2008).
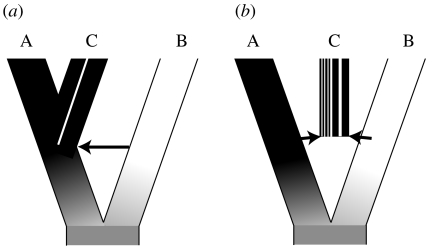
It might therefore be helpful to distinguish different modes of HHS. We propose the terms hybrid trait speciation and mosaic genome hybrid speciation to describe this distinction (figure 1). To summarize the main characteristics, hybrid trait speciation involves colonization of a novel adaptive peak through introgression of genes controlling adaptive traits (Mallet 2007). The novel adaptive trait must also confer a degree of reproductive isolation from the parental lineages to qualify as an example of HHS. In contrast, mosaic genome hybrid speciation involves stabilization of a hybrid genome, where the parental species differ in multiple loci conferring intrinsic hybrid incompatibility. Thus, the distinction is primarily based on the nature of selection acting during the establishment of the hybrid lineage (ecological versus intrinsic incompatibility), but the two models are also likely to differ in the proportion of the genome involved in hybridization. The former might involve just a handful of adaptively important genes introgressing into a related species, whereas the latter will commonly involve establishment of a hybrid lineage comprising similar proportions from two or more parental genomes (figure 1). These two modes of hybrid speciation are not so much mutually exclusive alternatives as opposite ends of a continuum and, indeed, in Helianthus hybrid lineages novel ecological characteristics are important as well as intrinsic incompatibility from their parents (Rieseberg et al. 2003).
Figure 1.
Two possible routes to hybrid speciation. (a) Hybrid trait speciation involves backcrossing of genes from lineage B into the genome of A. If those genes control a trait with adaptive potential that also causes reproductive isolation (similar to a magic trait, sensu Gavrilets 2004), then this might give rise to establishment of the novel species C. (b) In contrast, under mosaic genome hybrid speciation the novel species C is formed by the establishment and stabilization of a recombinant lineage combining compatible regions of the genome in similar proportions from A and B.
Nonetheless, the motivation for highlighting this distinction is twofold. First, it is done to emphasize that hybridization can generate novel adaptive variation that might facilitate both colonization of novel habitats and speciation at a faster rate than through mutation alone, potentially accelerating the rate of adaptive radiation (Seehausen 2004; Mallet 2007). One possible example of this is the establishment of the apple race of Rhagoletis pomonella through introgression of genetic variation from more southerly populations (Feder et al. 2003). The colonization of this novel niche was facilitated by combining existing genetic variation rather than waiting for novel mutation. Second, it is also done to highlight the fact that detection of putative hybrid species will be challenging following hybrid trait speciation, as there will not necessarily be a strong signal of overall hybridity in the genome of the novel species. The difficulty of both defining and identifying HHS when only a small proportion of the genome is involved was also highlighted in a recent review (Mallet 2007). Thus, the study of a handful of neutral molecular markers will not necessarily give a strong signal of hybridization in the putative hybrid species. Instead it will require the study of the genetic basis of traits with a putative hybrid origin and, ideally, genealogical study of the genes controlling such traits—a difficult task but one that is becoming increasingly feasible (Colosimo et al. 2005). Furthermore, the key distinction between hybrid trait speciation and merely adaptive introgression is the role of the novel trait in reproductive isolation, which needs to be demonstrated experimentally. Nonetheless, we strongly feel that there is a need to move away from studying putative hybrid species at neutral markers, and for a stronger emphasis on documenting the hybrid origin of the adaptive traits that allow those lineages to persist.
(b) Hybrid speciation in Heliconius butterflies
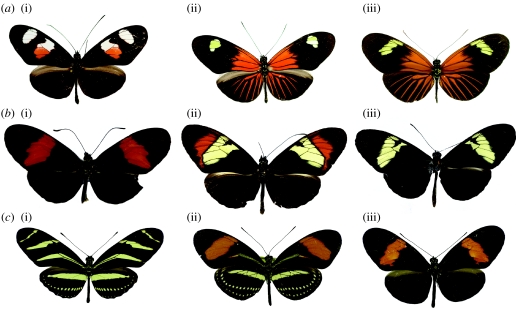
Perhaps because of the abundance of natural hybrids, there has long been an emphasis on the potential importance of hybridization in the evolution of these brightly coloured butterflies (Linares 1989; Gilbert 2003; Mallet 2005, 2007). Heliconius are known for their bright warning colour patterns and mimicry between divergent species, and the genetic basis of these diverse patterns has been widely studied through laboratory hybridization over many years. Furthermore, natural hybrid zones are common and provide a huge natural diversity of phenotypes, which have been avidly sought after by collectors. One of the observations made as a result of field and laboratory studies has been that the phenotypes of extant taxa often show marked similarity to hybrids between related species (figure 2). This has led to the suggestion by several Heliconius biologists that hybridization may play a significant constructive role in the generation of novel pattern diversity.
Figure 2.
Some putative examples of hybrid patterns in Heliconius and their progenitors. (a) (i) Heliconius erato notabilis, (ii) Heliconius erato etylus and (iii) Heliconius erato emma. The position of the forewing spot in H. erato etylus is apparently homologous to that of the most distal spot of H. erato notabilis. (b) (i) Heliconius melpomene melpomene, (ii) Heliconius heurippa and (iii) Heliconius cydno cordula. Crosses have shown that the H. heurippa pattern can be recreated by crossing the other two forms. (c) (i) Heliconius charithonia, (ii) Heliconius hermathena renatae and (iii) Heliconius erato hydara. Heliconius hermathena has yellow bands and spots similar to that of H. charithonia and a red band similar to that of H. erato hydara.
Natural hybridization is known to occur between races and species and shows an exponential decline with genetic distance (Mallet 2007). This demonstrates that speciation is a gradual process of decreasing compatibility and that introgression can potentially occur long after speciation is considered complete (Mallet 2008). Putative hybrid entities occur at all levels of this continuum. At a racial level, the forewing yellow spot of Heliconius erato etylus appears homologous in location to the white spot found slightly further north in the race Heliconius erato notabilis (J. Mallet 1994, personal communication, figure 2). Thus H. erato etylus is putatively a hybrid between H. erato notabilis and the rayed Amazonian form Heliconius erato lativitta (see figure 2). This is supported by recent crossing experiments showing that in both Heliconius erato races this element is controlled by the same genetic locus, known as Sd (Papa et al. submitted). At the other end of the scale, the species Heliconius hermathena has a pattern that shows marked similarity on the hindwing to Heliconius charithonia, but on the forewing it has a red band similar to sympatric H. erato (figure 2). Heliconius charithonia and H. erato are rather distantly related, but a single hybrid specimen is known from Mexico (Mallet 2007); so, it is not implausible that H. hermathena might have arisen through hybridization between these two species or their ancestors. Furthermore, H. hermathena has mixed patterns of relationships in phylogenetic studies, and an unusual pupal morphology for the H. erato group (Brown & Benson 1977). This mixed phylogenetic signal might be further indicative of a hybrid origin (Beltrán et al. 2007), although an alternative hypothesis that has been proposed is that H. hermathena is ancestral to the H. erato and H. charithonia lineages (Brown & Benson 1977).
The best-studied examples, however, are in the Heliconius cydno species complex. Heliconius melpomene and H. cydno are closely related species that hybridize both in the wild and the laboratory (Mallet 2007). They occur sympatrically in Central America and in the Andes, although they are ecologically and altitudinally segregated to some degree (Smiley 1978; Estrada & Jiggins 2002). While female F1 hybrids are sterile, males are fertile and can be used to introgress genes between the two species in the laboratory. Colour pattern genes, such as the yellow hindwing bar controlled by the Yb gene, appear homologous between the two species on the basis of identical gene action and linkage (Linares 1996; Naisbit et al. 2003; Joron et al. 2006). For example, when Heliconius melpomene vulcanus and H. cydno from west Colombia are crossed, the H. melpomene yellow bar behaves as another allelic variant at the same locus in the H. cydno genome (Linares 1989). The H. cydno complex actually consists of several species that are parapatric replacements of one another. All three geographical isolates of H. cydno, namely Heliconius pachinus found on the Pacific slopes of Costa Rica and western Panama, Heliconius heurippa, found on the eastern slopes of the Andes near Villavicencio in Colombia (figure 2), and Heliconius timareta, which occurs in the eastern Andes of Ecuador, have been suggested at some time as hybrid entities (Linares 1989; Mallet 1999; Gilbert 2003; Salazar et al. 2005). All three species are ecologically and genetically similar to H. cydno but share pattern elements with nearby races of H. melpomene, putatively acquired by introgression.
(c) Neutral marker surveys of introgression
The first major line of evidence demonstrating the evolutionary importance of hybridization are surveys of neutral genetic variation, which suggest widespread introgression in the wild even between fairly distantly related species (Bull et al. 2006; Kronforst et al. 2006b; Kronforst 2008). For example, strong positive evidence for introgression was found at one of the four loci sequenced in sympatric H. melpomene and H. cydno populations from Panama (Bull et al. 2006). An allopatric population of H. melpomene from French Guiana provided a natural control, showing that genetic similarity in sympatry was not simply a result of a slow evolutionary rate for a particular locus. A complementary genome-wide approach using large numbers of amplified fragment length polymorphism markers, in addition to sequence data for 16 genes, showed similar evidence for recent and persistent introgression between the same two species in Costa Rica (Kronforst et al. 2006b). These studies provide confirmation that hybrids are not an evolutionary dead end—hybridization is a genuine source of novel genetic variability.
However, such studies of putatively neutral genetic variation do not directly address the hypothesis proposed originally, which is that novel adaptive colour patterns have arisen through hybridization. The basic problem is that genetic markers used are not themselves informative regarding the origin of the colour pattern phenotype.
(d) Laboratory reconstruction of extant patterns
A better approach is to directly study the phenotypes. The easiest way to do this is through laboratory crosses. For example, the broad black band that divides the forewing yellow pattern into two thin bands in H. pachinus can be recreated by introgression of a forewing ‘shutter’ gene from Heliconius melpomene rosina into Heliconius cydno galanthus (Gilbert 2003). Similarly, the unusual all-black forewing phenotype of Heliconius cydno weymeri can be created by introgression of genetic variation from Heliconius melpomene vulcanus into H. cydno races from the northern Cauca valley (Colombia; Linares 1989; Gilbert 2003). In Ecuador the variation observed in the polymorphic Heliconius cydno alithea, which mimics Heliconius sapho and Heliconius eleuchia, has also been recreated in the crosses between H. melpomene and H. cydno from Costa Rica (Gilbert 2003).
Moreover, a recent study has demonstrated a route for the formation of H. heurippa, by reconstructing its pattern through laboratory crosses of H. melpomene and H. cydno (Mavárez et al. 2006). Earlier crossing experiments had suggested that the H. heurippa pattern could arise through hybridization of these species, which led to experiments specifically with the races geographically adjacent to the current range of H. heurippa (Linares 1989). The colour pattern of H. heurippa essentially consists of the NN allele from H. cydno controlling the yellow forewing band, combined with the B and br alleles from H. melpomene controlling the red forewing band (B), hindwing red spots and the absence of forceps-shaped brown marking on the hindwing ventral surface (br). These loci have to be combined on an H. cydno genetic background in order for the full expression of the yellow band; hence, the reconstruction involved backcrossing with H. cydno. The B and br loci are tightly linked, which facilitates their introgression into H. cydno, and led to the surprising result that this reconstruction only required three generations of crosses. Heliconius heurippa shares one other pattern trait with H. melpomene, whose genetics is not well understood, namely the lack of structural blue iridescent colour. The reconstructed pattern is remarkably similar, but not an exact match to wild H. heurippa—the only difference being a thin black line between the red and yellow bands, which is only occasionally faintly seen in laboratory hybrids, perhaps implying additional pattern evolution subsequent to the speciation event.
Although such a reconstruction experiment in itself cannot prove a hybrid origin for H. heurippa, this hypothesis is strongly supported by the geographical distribution of the species involved. Of all the genetic variation present in H. melpomene and H. cydno, natural overlap between the red forewing band of H. melpomene and the yellow forewing of H. cydno only occurs geographically adjacent to the current range of H. heurippa. Nonetheless, there are two alternative hypotheses to hybrid origin that should be considered. First, the H. heurippa pattern might be ancestral and have given rise to H. melpomene and H. cydno lineages that inherited different aspects of the ancestral pattern. This seems unlikely as the H. heurippa pattern is not seen in any other species in the genus, and is not a plausible ancestor in any parsimony or likelihood ancestral state reconstructions based on a recent phylogeny of the genus (Beltrán et al. 2007; C. D. Jiggins 2006, personal observation). Second, the pattern alleles might have arisen through mutation rather than hybridization. In principle it might be possible to estimate the relative probability of a particular pattern arising through mutation versus hybridization. We have never observed novel pattern mutants in many thousands of lab-reared butterflies and the probability of them arising and being convergent to patterns found locally in related species is low. Of course Heliconius do frequently evolve convergent patterns, but this is driven by mimicry selection, which does not apply to the non-mimetic H. heurippa. This contrasts with the ease of generating novel hybrid patterns through laboratory crosses (see above). In the wild the relative probability of hybrids versus novel pattern mutants is harder to estimate, but the great majority of ‘aberrant’ wild phenotypes are most readily explained as hybrids (Mallet et al. 2007). Furthermore, a large proportion of known wild hybrids are between H. melpomene and H. cydno. It seems probable that novel patterns generated through hybridization are an order of magnitude more common than those generated through mutation. On balance, the relative frequency of occurrence of hybrids versus novel mutants in both laboratory and wild populations provides further evidence to support the hybrid origin hypothesis for the formation of H. heurippa.
In summary, therefore, the genetic data show that hybrid introgression is rife between Heliconius species, and provide the most probable scenario for the origin of the H. heurippa pattern. Ultimately, however, the positional cloning of loci controlling pattern diversity offers the potential to reconstruct the genealogical history of colour pattern alleles and flanking markers and, hence, estimate directly the importance of hybridization in pattern evolution.
(e) The origin of reproductive isolation
Importantly, what makes H. heurippa a possible case of hybrid speciation, rather than merely adaptive introgression, is that the hybrid colour pattern itself has a direct influence on reproductive isolation. Previous work has shown that species in this group use colour in mate recognition (Jiggins et al. 2001; Kronforst et al. 2006a). This was confirmed with mate choice experiments on H. heurippa and adjacent races of H. melpomene and H. cydno. Males of all three species are about half as likely to approach the colour pattern of another species. Furthermore, when the H. heurippa pattern was manipulated, both red and yellow elements were necessary to stimulate approach and courtship by H. heurippa males (Mavárez et al. 2006). Thus, the novel colour pattern contributes to partial reproductive isolation from both the parental lineages, as required by our definition of hybrid speciation.
This strong preference for the hybrid pattern in the H. heurippa lineage raises the question of how such a preference would have arisen. There are essentially two scenarios—either the mating preference came about through evolution of mate-finding subsequent to the establishment of the novel colour pattern as a ‘race’ of H. cydno, or it arose as a direct result of the hybridization event. The former is fairly trivial to envisage, but the latter would imply a greater and more direct role for hybridization in speciation and seems less likely a priori. Under the first hypothesis, hybridization plays a role basically equivalent to mutation at multiple loci in generating a novel colour pattern phenotype. Under the second hypothesis there is a more central role for hybridization in producing a genetic association between the novel colour pattern and mate preferences in the novel lineage. In support of the latter hypothesis, earlier work indeed suggests that F1 hybrids between H. melpomene and H. cydno actually prefer to mate with each other rather than their parents (Naisbit et al. 2001). Furthermore, a recent quantitative trait locus analysis of the genetics of mate preference in H. cydno and H. pachinus showed a genetic association between a colour pattern shift (from yellow to white) and the preference for that pattern (Kronforst et al. 2006a). This might imply that preference genes could be introgressed between species alongside patterning genes. Between them, these studies imply that it is plausible to imagine that hybrids in which a novel red pattern element was introgressed into an H. cydno genome might actually also show a mate preference for their novel pattern. This remains to be demonstrated experimentally.
(f) Patterns of post-mating isolation
One of the major barriers to HHS is hybrid breakdown. In the case of H. melpomene and H. cydno, F1 female hybrids are sterile, while males are fully fertile (Naisbit et al. 2002). Sterility segregates in backcross broods and provides a partial barrier to introgression. Nonetheless, as described previously it is possible to introgress colour pattern genes from one species into another and therefore overcome the sterility barrier. In addition, numerous backcross hybrids have been collected in San Cristobal, Venezuela, and shown to be genetically indistinguishable from H. cydno at microsatellite markers. This clearly demonstrates multiple generations of natural backcrossing and shows that this hybrid sterility can be overcome in the wild as well. Heliconius heurippa itself is completely compatible with H. cydno, but shows unidirectional F1 female hybrid sterility with H. melpomene—in other words female offspring of a male H. melpomene crossed with a female H. heurippa are sterile, while offspring of the reciprocal cross are fully fertile (Salazar et al. 2005). This is in contrast to the bi-directional hybrid sterility between H. melpomene and H. cydno (although male H. cydno rarely mate with female H. melpomene, so crosses in this direction are very poorly documented). The pattern is therefore compatible with a hybrid origin, as H. heurippa is somewhat intermediate between the other two species in the expression of hybrid sterility. Nonetheless, the same pattern could equally be generated by an entirely bifurcating speciation scenario, in which additional incompatibility alleles were acquired along the H. cydno lineage subsequent to divergence of H. heurippa. These data do not therefore provide positive support for hybrid speciation, but are consistent with the hypothesis.
(g) Evidence for hybrid genomes?
Unfortunately, the large amount of shared neutral genetic variation between related species actually makes it difficult to identify hybrid taxa. A study of H. pachinus failed to show any positive evidence for a hybrid origin at neutral loci, despite identifying a substantial number of markers diagnostic for H. melpomene and H. cydno. Instead, H. pachinus appears as a divergent form of H. cydno, consistent with a bifurcating origin for this species (Kronforst et al. 2007). The power of such a study is weakened however when the putative parents themselves share such a large amount of genetic variation. The H. melpomene and H. cydno genomes are clearly far less differentiated than, for example, those of Helianthus annus and Helianthus petiolaris (Ungerer et al. 1998).
The pattern in H. heurippa is more unusual, with sequence data for two loci, fragments of the genes Invected and Distalless, showing a striking pattern in which H. melpomene and H. cydno are differentiated, but H. heurippa shares allelic variation with both the species. Of the three further loci sampled since, one showed a similar pattern, the other showed the three species intermixed and the third showed the three species mutually differentiated (Salazar et al. 2008). This data initially led us to state that H. heurippa has a hybrid genome (Mavárez et al. 2006, in which this result was erroneously attributed to Salazar et al. 2005). However it actually seems probable that this allele sharing can also be explained by ongoing introgression between the three species. Thus, H. melpomene and H. cydno sampled from Costa Rica also show evidence for introgression at the Distalless locus, with a strong frequency difference between the species but a few very similar shared alleles (Kronforst et al. 2006b). Heliconius heurippa is likely to show higher levels of introgression with H. melpomene and H. cydno than occurs between those two species, given the intermediate colour pattern and, hence, mating signal.
Distinguishing the alternate hypotheses of a branching speciation event from that of hybrid founding, with the likelihood of ongoing gene flow under either scenario, is a challenge. Previous models of divergence population genetics were designed for the study of two species rather than three and primarily developed to distinguish shared ancestral polymorphism from introgression (Wakeley 2000). Our own attempts to develop a coalescent model incorporating three species and the possibility of either branching or hybrid founding speciation have failed to provide a good estimate of parameter values for H. heurippa based on our sequence data (J. Taylor 2007, personal communication). The only finding was that parameter estimates for introgression between the species were consistently non-zero. In the future we aim to focus our sequencing efforts on regions surrounding colour pattern loci in order to directly test the hypothesis that the B locus has been introgressed from H. melpomene into the H. cydno genome. In summary, the data are probably consistent with either a hybrid founding or a bifurcating origin for H. heurippa, and more than anything provide evidence for ongoing introgression.
The evidence for hybrid origin is based largely on the three Mendelian loci that control colour pattern and their direct role in reproductive isolation through mate choice. Thus, our basic hypothesis for the origin of H. heurippa argues for the role of a hybrid trait in the speciation event, but makes no specific prediction regarding the rest of the genome. In the recently discovered Venezuelan hybrid zone between H. melpomene and H. cydno, the hybrids are all indistinguishable from pure H. cydno at 12 microsatellite loci (Mavárez et al. 2006) and must therefore represent greater than fifth generation backcrosses to this species. If a similar scenario were involved in the origin of H. heurippa, then it would be very difficult to detect a significant signal of the hybrid founding event—the genome would primarily be derived from H. cydno despite the crucial role of introgression of the patterning genes in the formation of the novel lineage. We therefore propose H. heurippa to be an example of hybrid trait speciation.
2. Conclusions
We primarily envisage the role of hybridization as a means of instantaneously generating novel variants at multiple genetic loci, providing the raw material for the establishment of lineages with adaptive potential (Seehausen 2004). While the adaptive traits in question also contribute to reproductive isolation with parental lineages, this can lead to hybrid trait speciation. In the case of H. heurippa, there is evidence from laboratory crosses and the biogeographic distribution of colour patterns that its pattern has arisen through hybridization. Crucially, the colour pattern is also involved in mate choice and leads to reproductive isolation between H. heurippa and its close relatives. We predict that the new era in which the genetic basis of adaptive traits can be understood at the DNA sequence level will increasingly uncover examples of the role of hybridization in the generation of novel adaptive diversity and species.
Acknowledgments
We would like to thank the Royal Society, BBSRC and Leverhulme Trust for funding to C.D.J.; M.L. and C.S. were funded by the Fondo Colombiano de Investigaciones Cientificas y Proyectos Especiales Francisco Jose de Caldas COLCIENCIAS grants 1204-05-11414 and 7155-CO, Banco de la Republica and private donations from Continautos, S. A., Proficol El Carmen, S. A., Didacol, S. A. and Arango, F. Colombia. We thank O. Delgado, Miza-UCV, for the photographs of Venezuelan specimens.
Footnotes
One contribution of 12 to a Theme Issue ‘Speciation in plants and animals: pattern and process’.
References
- Arnold M.L. Oxford series in ecology and evolution. Oxford University Press; New York, NY: 1997. Natural hybridization and evolution. [Google Scholar]
- Arnold M.L. Evolution through genetic exchange. Oxford University Press; Oxford, UK: 2006. [Google Scholar]
- Beltrán M, Jiggins C.D, Brower A.V.Z, Bermingham E, Mallet J. Do pollen feeding, pupal-mating and larval gregariousness have a single origin in Heliconius butterflies? Inferences from multilocus DNA sequence data. Biol. J. Linn. Soc. 2007;92:221–239. doi:10.1111/j.1095-8312.2007.00830.x [Google Scholar]
- Boughman J.W. Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature. 2001;411:944–948. doi: 10.1038/35082064. doi:10.1038/35082064 [DOI] [PubMed] [Google Scholar]
- Brown K.S, Benson W.W. Evolution in modern Amazonian non-forest islands: Heliconius hermathena. Biotropica. 1977;9:95–117. doi:10.2307/2387664 [Google Scholar]
- Bull V, Beltrán M, Jiggins C.D, McMillan W.O, Bermingham E, Mallet J. Polyphyly and gene flow between non-sibling Heliconius species. BMC Biol. 2006;4:11. doi: 10.1186/1741-7007-4-11. doi:10.1186/1741-7007-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P.F, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodyplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. doi:10.1126/science.1107239 [DOI] [PubMed] [Google Scholar]
- Coyne J.A, Orr H.A.Speciation2004Sinauer Associates; Sunderland, MA [Google Scholar]
- Estrada C, Jiggins C.D. Patterns of pollen feeding and habitat preference among Heliconius species. Ecol. Entomol. 2002;27:448–456. doi:10.1046/j.1365-2311.2002.00434.x [Google Scholar]
- Feder J.L, et al. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl Acad. Sci. USA. 2003;100:10 314–10 319. doi: 10.1073/pnas.1730757100. doi:10.1073/pnas.1730757100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S.Fitness landscapes and the origin of species2004Princeton University Press; Princeton, NJ [Google Scholar]
- Gilbert L.E. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic ‘tool box’ from synthetic hybrid zones and a theory of diversification. In: Boggs C.L, Watt W.B, Ehrlich P.R, editors. Ecology and evolution taking flight: butterflies as model systems. University of Chicago Press; Chicago, IL: 2003. pp. 281–318. [Google Scholar]
- Gompert Z, Fordyce J.A, Forister M.L, Shapiro A.M, Nice C.C. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. doi:10.1126/science.1135875 [DOI] [PubMed] [Google Scholar]
- Hegarty M.J, Barker G.L, Brennan A.C, Edwards K.J, Abbott R.J, Hiscock S.J. Changes to gene expression associated with hybrid speciation in plants: further insights from transcriptomic studies in Senecio. Phil. Trans. R. Soc. B. 2008;363:3055–3069. doi: 10.1098/rstb.2008.0080. doi:10.1098/rstb.2008.0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins C.D, Naisbit R.E, Coe R.L, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. doi:10.1038/35077075 [DOI] [PubMed] [Google Scholar]
- Jiggins C.D, Emelianov I, Mallet J. Pleiotropy promotes speciation: examples from phytophagous moths and mimetic butterflies. In: Fellowes M, Holloway G.J, Rolff J, editors. Insect evolutionary ecology. CABI; London, UK: 2004. pp. 451–473. [Google Scholar]
- Joron M, et al. A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 2006;4:e303. doi: 10.1371/journal.pbio.0040303. doi:10.1371/journal.pbio.0040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M. Gene flow persists millions of years after speciation in Heliconius butterflies. BMC Evol. Biol. 2008;8:98. doi: 10.1186/1471-2148-8-98. doi:10.1186/1471-2148-8-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M, Young L.G, Kapan D.D, McNeely C, O'Neill R.J, Gilbert L.E. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc. Natl Acad. Sci. USA. 2006a;103:6575–6580. doi: 10.1073/pnas.0509685103. doi:10.1073/pnas.0509685103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M.R, Young L.G, Blume L.M, Gilbert L.E. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006b;60:1254–1268. doi:10.1554/06-005.1 [PubMed] [Google Scholar]
- Kronforst M.R, Salazar C, Linares M, Gilbert L.E. No genomic mosaicism in a putative hybrid butterfly species. Proc. R. Soc. B. 2007;274:1255–1264. doi: 10.1098/rspb.2006.0207. doi:10.1098/rspb.2006.0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R.C, Birch L.C. Hybridisation as a source of variation for adaptation to new environments. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. doi:10.2307/2406633 [DOI] [PubMed] [Google Scholar]
- Linares, M. 1989 Adaptive microevolution through hybridization and biotic destruction in the neotropics. PhD thesis, University of Texas, Austin, TX.
- Linares M. The genetics of mimetic coloration in the butterfly Heliconius cydno weymeri. J. Hered. 1996;87:142–149. [Google Scholar]
- Mallet J. Causes and consequences of a lack of coevolution in Mullerian mimicry. Evol. Ecol. 1999;13:777–806. doi:10.1023/A:1011060330515 [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. doi:10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. doi:10.1038/nature05706 [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Phil. Trans. R. Soc. B. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. doi:10.1098/rstb.2008.0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Beltran M, Neukirchen W, Linares M. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol. Biol. 2007;7:28. doi: 10.1186/1471-2148-7-28. doi:10.1186/1471-2148-7-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavárez J, Salazar C, Bermingham E, Salcedo C, Jiggins C.D, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;411:868–871. doi: 10.1038/nature04738. doi:10.1038/nature04738 [DOI] [PubMed] [Google Scholar]
- Meyer A, Salzburger W, Schartl M. Hybrid origin of a swordtail species (Teleostei: Xiphophorus clemenciae) driven by sexual selection. Mol. Ecol. 2006;15:721–730. doi: 10.1111/j.1365-294X.2006.02810.x. doi:10.1111/j.1365-294X.2006.02810.x [DOI] [PubMed] [Google Scholar]
- Naisbit R.E, Jiggins C.D, Mallet J. Disruptive sexual selection against hybrids contributes to speciation between Heliconius cydno and Heliconius melpomene. Proc. R. Soc. B. 2001;268:1849–1854. doi: 10.1098/rspb.2001.1753. doi:10.1098/rspb.2001.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit R, Jiggins C.D, Linares M, Salazar C, Mallet J. Hybrid sterility, Haldane's rule and speciation in Heliconius cydno and H. melpomene. Genetics. 2002;161:1517–1526. doi: 10.1093/genetics/161.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit R.E, Jiggins C.D, Mallet J. Mimicry: developmental genes that contribute to speciation. Evol. Dev. 2003;5:269–280. doi: 10.1046/j.1525-142x.2003.03034.x. doi:10.1046/j.1525-142X.2003.03034.x [DOI] [PubMed] [Google Scholar]
- Nolte A.W, Freyhof J, Stemshorn K.C, Tautz D. An invasive lineage of sculpins, Cottus sp. (Pisces, Teleostei) in the Rhine with new habitat adaptations has originated from hybridization between old phylogeographic groups. Proc. R. Soc. B. 2005;272:2379–2387. doi: 10.1038/rspb.2005.3231. doi:10.1098/rspb.2005.3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, R. et al Submitted. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. [DOI] [PMC free article] [PubMed]
- Podos J. Correlated evolution of morphology and vocal signal structure in Darwin's finches. Nature. 2001;409:185–188. doi: 10.1038/35051570. doi:10.1038/35051570 [DOI] [PubMed] [Google Scholar]
- Puebla O, Bermingham E, Guichard F, Whiteman E. Colour pattern as a single trait driving speciation in Hypoplectrus coral reef fishes? Proc. R. Soc. B. 2007;274:1265–1271. doi: 10.1098/rspb.2006.0435. doi:10.1098/rspb.2006.0435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H, Archer M.A, Wayne R.K. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. doi:10.1038/sj.hdy.6886170 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. doi:10.1126/science.1086949 [DOI] [PubMed] [Google Scholar]
- Salazar C, Jiggins C.D, Arias C.F, Tobler A, Bermingham E, Linares M. Hybrid incompatibility is consistent with a hybrid origin of Heliconius heurippa Hewitson from its close relatives, Heliconius cydno Doubleday and Heliconius melpomene Linnaeus. J. Evol. Biol. 2005;18:247–256. doi: 10.1111/j.1420-9101.2004.00839.x. doi:10.1111/j.1420-9101.2004.00839.x [DOI] [PubMed] [Google Scholar]
- Salazar C, Jiggins C.D, Taylor J.E, Kronforst M.R, Linares M. Gene flow and the genealogical history of Heliconius heurippa. BMC Evol. Biol. 2008;8:132. doi: 10.1186/1471-2148-8-132. doi:10.1186/1471-2148-8-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Matta B.M, Shakir-Botteri N.L, McPheron B.A. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436:546–549. doi: 10.1038/nature03800. doi:10.1038/nature03800 [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. doi:10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Seehausen O, van Alphen J.J.M, Witte F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science. 1997;227:1808–1811. doi:10.1126/science.277.5333.1808 [Google Scholar]
- Smiley J.T. Plant chemistry and the evolution of host specificity: new evidence from Heliconius and Passiflora. Science. 1978;201:745–747. doi: 10.1126/science.201.4357.745. doi:10.1126/science.201.4357.745 [DOI] [PubMed] [Google Scholar]
- Stebbins G.L.Variation and evolution in plants1950Columbia University Press; New York, NY [Google Scholar]
- Stebbins G.L. The role of hybridisation in evolution. Proc. Am. Philos. Soc. 1959;103:231–251. [Google Scholar]
- Templeton A.R. Mechanisms of speciation—a population genetic approach. Annu. Rev. Ecol. Syst. 1981;12:23–48. doi:10.1146/annurev.es.12.110181.000323 [Google Scholar]
- Ungerer M.C, Baird S.J.E, Pan J, Rieseberg L.H. Rapid hybrid speciation in wild sunflowers. Proc. Natl Acad. Sci. USA. 1998;95:11 757–11 762. doi: 10.1073/pnas.95.20.11757. doi:10.1073/pnas.95.20.11757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. The effects of subdivision on the genetic divergence of populations and species. Evolution. 2000;54:1092–1101. doi: 10.1111/j.0014-3820.2000.tb00545.x. doi:10.1554/0014-3820(2000)054[1092:TEOSOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]