Abstract
The biogeographic patterns in sexually reproducing animals in island archipelagos may be interpreted as reflecting the importance of allopatric speciation. However, as the forms are allopatric, their reproductive isolation is largely untestable. A historical perspective integrating geology and molecular phylogeny reveals specific cases where ancient precursor islands coalesce, which allows the application of population genetics to critically test genetic isolation. The Anolis populations on Martinique in the Lesser Antilles are one such case where species-level populations on ancient precursor islands (ca 6–8 Myr BP) have met relatively recently. The distribution of the mtDNA lineages is tightly linked to the precursor island, but the population genetic analysis of microsatellite variation in large samples shows no evidence of restricted genetic exchange between these forms in secondary contact. This tests, and rejects, the hypothesis of simple allopatric speciation in these forms. By contrast, Martinique has pronounced environmental zonation, to which anoles are known to adapt. The population genetic analysis shows restricted genetic exchange across the ecotone between xeric coastal habitat and montane rainforest. This does not indicate full ecological speciation in these forms, but it does suggest the relative importance of the role of ecology in speciation in general.
Keywords: allopatric speciation, ecological speciation, Martinique anoles, Bayesian assignment
1. Introduction
The relative importance of the various factors that may potentially influence speciation in sexually reproducing animals (e.g. drift in spatial isolation, natural selection and sexual selection) is one of the central issues in the active debate on speciation (Claridge et al. 1997; Howard & Berlocher 1997; Smith et al. 1997; Magurran & May 1998; Orr & Smith 1998; Dieckmann & Doebeli 1999; Schneider et al. 1999; Schluter 2000; Barton 2001; Thorpe & Richard 2001; Ogden & Thorpe 2002; Doebeli & Dieckmann 2003; Gavrilets 2003; Tautz 2003; Dieckmann et al. 2004; Ortiz-Barrientos & Kane 2007; Ritchie 2007; Abbott et al. 2008). The traditional view of speciation in sexually reproducing animals is that it generally involves the divergence of populations over time while they are geographically apart (geographical or allopatric speciation; Mayr 1970). This approach is typified in genera on many island archipelagos where each island or island bank tends to have its own nominal species (Stenson et al. 2004; Ricklefs & Bermingham 2007), and generally speciation is assumed to occur when an island is colonized by dispersal from another; the subsequent geographical isolation allowing the founding population to evolve into a new species (see also Comes et al. 2008).
There are two conceptually different empirical approaches when seeking an insight into causative factors in evolutionary ecology, which are pertinent to the question of speciation in these island archipelagos. One approach is to interpret the overall biogeographic or phylogenetic pattern (Losos & Schluter 2000; Borsa et al. 2007; Hyde & Vetter 2007) in order to deduce the population-level process that caused it. The other is to take a more reductionist stance and try and look in detail directly at the population-level processes that may have led to such a biogeographic or phylogenetic pattern. Examples of both approaches can be found using island anoles where both biogeographic patterns (Losos & Schluter 2000) and population-level approaches (Ogden & Thorpe 2002) have been employed in studies of speciation and ecology (reviewed in Thorpe 2005). Both approaches have their merits and difficulties when applied to island speciation studies. One difficulty with a biogeographic approach is that it is possible to regard the islands that are observed today as the entity to study, when, in fact, general sea-level changes, sea-floor uplifts in specific areas and volcanic activity may substantially alter the number and size of islands and their connectivity. A further difficulty is that without sound fossil evidence, we may have an inadequate or misleading view of the past distribution of species and their environmental conditions. Another critically important limitation is that with allopatric species, such as are found in island archipelagos, one may be dealing with just arbitrary nominal species making some biological interpretations tenuous or inappropriate. What we mean by speciation depends on what we mean by species (Wiens 2004). The term species, of course, encompasses the Linnaean name, the taxonomic category and potentially a biological entity, however defined by the species concept of choice. When we talk about speciation we mean the origin of the biological entity and not the former two uses. Geographical variation within species is ubiquitous and population differentiation is not speciation (Magurran 1998). Hence, if allopatric populations are recognized as arbitrary nominal species on the basis of trivial population differentiation, then the process of ‘speciation’ can mean something different from the process that leads to, say, reproductive isolation between sympatric forms (see Mallet 2008). Consequently, patterns of speciation in arbitrarily recognized (nominal) allopatric species may tell us something about population differentiation, but they may have limited usefulness when trying to generalize to speciation involving reproductive isolation.
2. The area and the organism
Anoles are small insectivorous, generally arboreal, iguanine lizards. Males have a dewlap (an extendable throat flap) that acts as a sexual signal and (in multi-species sympatric communities) species recognition (Losos 1985). There are approximately 400 described species of Anolis, making it the largest genus of amniote and perhaps the largest monophyletic tetrapod genus (Losos & Thorpe 2004). In some organisms, interspecific hybrids are common (Mallet 2008), but sympatric anoles generally appear to behave like good biological species. Anoles are well studied, and these studies have influenced much of evolutionary ecology, including niche theory, adaptive radiation, behaviour, natural selection, biogeography and phylogeny (extensively reviewed by Losos in press). Their diversity and their focal role in many studies of evolutionary ecology make them good candidates for the study of speciation.
Approximately 150 Caribbean species are thought to have arisen from just two colonizations from the mainland (Jackman et al. 1997), so the number of species is due to diversification within the Caribbean rather than multiple colonization from the mainland. In the Greater Antilles islands of Jamaica, Cuba, Hispaniola and Puerto Rico, anoles live in communities with up to 11 species in sympatry. The molecular phylogenetic, morphological and ecological evidence provides an outstanding case of predictable adaptive radiation, where species with a specific morphology occupy specific niches, i.e. ecomorphs. This occurs independently, in parallel, on each Greater Antillean island (Losos in press).
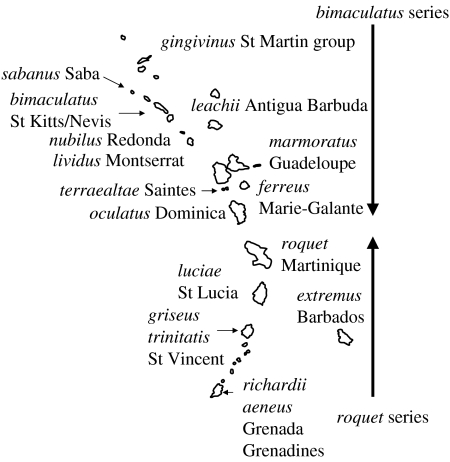
By contrast, the Lesser Antillean anoles are either solitary or live in pairs. There are two main radiations (figure 1): the roquet series extend up to Martinique from the south and the bimaculatus/wattsi series extend down from the north to Dominica. Two species of the roquet series (one large and another small) are sympatric on St Vincent and Grenada (and Grenadines), and all other southern islands have solitary anoles. In the north, no bimaculatus (medium or large) series live sympatrically with other members of the series, but they can live in sympatry (species pairs) with anoles from the wattsi group. This results in one large and one small species on the Antigua/Barbuda bank, the St Martin bank and the St Kitts/Nevis bank, and solitary bimaculatus anoles on the remaining northern islands.
Figure 1.
Lesser Antillean anoles. Distribution of the A. roquet series species in the south and the A. bimaculatus series in the north (A. wattsi group not shown).
This presents some clear biogeographic patterns. Each island or island bank has its endemic anole species. There are no empty islands as every single island and habitable islet has anoles. There is limited sympatry, but the anoles from the roquet and bimaculatus series are never sympatric with one another, their distributions do not even interdigitate.
Estimates of island-wide population size are going to be error prone; the extrapolation from biomass estimates on Dominica (Malhotra & Thorpe 1991) suggests a standing (breeding) population of 200 million anole adults (or more) and as females may produce an egg every two weeks, there may be a turnover of approximately a billion (or more) per year. Hence, while populations on small islands may often be thought of as fragile, it appears that anole populations may be very large and robust on small islands. This is compatible with the observations that currently no island or habitable islet in the Lesser Antilles is without anoles, and they may withstand even cataclysmic volcanic activity (Malhotra & Thorpe 2000). Consequently, this suggests that anole populations may increase rapidly in size after the founding colonization, and that, while bottlenecks may occur, they are not particularly vulnerable to prolonged bottlenecks after establishment.
The mountainous Lesser Antillean island possesses pronounced environmental zonation (Beard 1948). The Caribbean coastal strip (and the tip of southern and eastern peninsulas on some islands) may be hot and dry with seasonal rain and have xeric woodland. Higher elevation areas are cooler, with substantial, less seasonal, rain, with montane rainforest (giving way to cloud forest). The mid-Atlantic coast, which collects the warm moist trade winds, may have dense, windswept littoral woodland. The quantitative traits of anoles, such as scalation, body dimensions, hue and pattern, appear to be primarily genetically controlled when tested (Thorpe et al. 2005a; Calsbeek et al. 2006; Calsbeek & Smith 2007), and show pronounced intraspecific geographical variation within islands in concert with this environmental zonation irrespective of phylogeographic lineage (Malhotra & Thorpe 2000; Thorpe et al. 2004). The overwhelming conclusion from studying field experiments on natural selection, correlations between the geographical variation in a trait and its putative cause, and parallels between patterns of geographical variation on independent islands, is that these patterns of ecotypic geographical variation are the result of rapid, pronounced and, in some instances, predictable, natural selection. Sexual selection may also play a role as visual sexual signals, such as male dewlap hue and sexually dimorphic elements of the hue and colour pattern, which may vary substantially with habitat types that have different ambient light conditions (Endler 1992; Leal & Fleishman 2004).
The biogeographic pattern of species distribution (figure 1) and molecular phylogeny suggests progressive overseas colonization from one island to another as in Thorpe et al. (2004), with endemic species evolving on each island or island bank. The Anolis bimaculatus and wattsi series come from the Greater Antilles in the north, and the roquet series from the mainland in the south. This overseas colonization by founders and subsequent speciation is a form of allopatric or geographical speciation, so the biogeographic pattern strongly reinforces the traditional view of the primary role of allopatric speciation in this type of organism. Some of the limitations of interpreting the pattern to deduce the cause, which are listed above, apply to this case. Allopatric speciation can only be said to have occurred if the island forms are more than just nominal species. As the current islands are spatially isolated, this is untestable. Here, another facet of the ‘pattern’ approach is pertinent, i.e. current islands do not represent the past islands. The case in point is Martinique, which is thought to be composed of precursor islands that have only recently coalesced into a single island. However, this may give us a rare opportunity to test, which is often not directly testable under natural conditions, the reproductive isolation of allopatric island populations and the impact of allopatry on speciation.
3. Testing the roles of allopatry and ecology in speciation
(a) Martinique
(i) Island-wide geology and environment
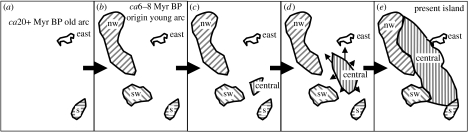
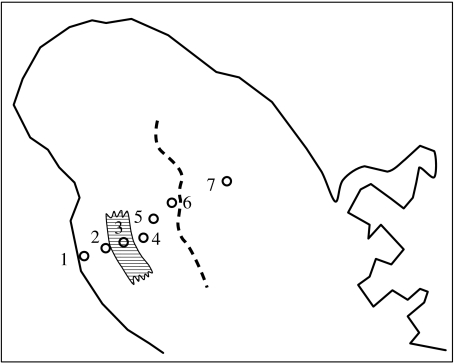
The island of Martinique in the mid-latitudes of the Lesser Antilles has been the single entity we recognize today for only a brief period of its geological existence (Maury et al. 1990; and references in Thorpe & Stenson 2003). For an extensive time during the period of the older arc, two islands existed: a southern island (St Anne) and an eastern island (Caravelle; figure 2a). The origin of the younger arc would have seen the eruption of two further islands, in the southwest (Trois Ilet) and northwest (figure 2b). The subsequent emergence of a central island (figure 2c) and the gradual uplifting of this region (figure 2d) eventually joined the four peripheral islands to form the single entity we know today as Martinique (figure 2e). Independent of this is the distinct environmental zonation typically found in mid-latitude, high-elevation, Lesser Antillean islands. There is a xeric strip on the Caribbean coast in the rain shadow of the major central mountains (figure 3), a xeric southern peninsula and a xeric eastern tip to the eastern Caravelle peninsula. The central mountains are largely covered with montane rainforest, with a sharp transition to xeric woodland on the Caribbean coast (figure 3). The middle section of the Atlantic coast has littoral conditions, but these change towards the north (more seasonal) and south (drier). The anoles appear to adapt by natural selection to these regimes, as tested in Thorpe & Stenson (2003) and figured in Thorpe (2005). Among other colour patterns, the Martinique anoles have zebra striping in these xeric regions, while those from montane rainforest have white (without UV) and black markings on a deeply saturated green background. Other quantitative traits, such as scalation and body proportion, also vary with habitat and these ecotypic patterns occur irrespective of lineage.
Figure 2.
Geological history of the components of Martinique. Two islands in (a) the older arc, Caravelle (east) and St Anne (south (s)), were subsequently joined by a further two islands at the origin of (b) the younger arc, northwest (nw) and southwest (sw). General uplifting of (c) the central area, (d) continued until the central area joined the peripheral precursor islands into (e) the present single island of Martinique.
Figure 3.
Secondary contact zone and ecotone in northwest Martinique. The dashed line marks the geological boundary between the northwest and central regions where populations on these precursor islands may make secondary contact. The cross-hatched area represents the ecotone where the xeric coastal habitat (transect localities 1 and 2) is in transition to the montane rainforest (transect localities 4–7).
This system can be used to critically test the impact of allopatry on the potential for speciation, and relate this to the degree of genetic isolation associated with habitat differences (ecological speciation). This rests on two assumptions. First, the precursor islands need to be occupied while they were separate to give allopatric forms. Geologically recent colonization, after the islands had coalesced into the single island of Martinique, would not allow such a test. Second, if there were allopatric populations on these precursors, the timing and level of divergence needs to be compatible with that found in other species in the series and other series of Lesser Antillean anoles.
(ii) Island-wide phylogeography
Many of these questions can be answered by molecular phylogenetic and phylogeographic study. Tail tips were taken from 39 individuals from 15 localities for A. extremus and 68 individuals from 64 localities for Anolis roquet. Anolis luciae and Anolis jacare were the outgroup. Genomic DNA was extracted using the chelex method described in Surget-Groba et al. (2001) and a 1063 bp fragment of the cytochrome b gene was amplified with primers MTA-S (ATCTCAGCATGATGAAACTTCG) and MTF-S (TTTGGTTTACAAGACCAATG). The sequencing of polymerase chain reaction (PCR) products was performed using the PCR primers and two internal primers (CB2F: ACAACGCAACCTTAACACGATT and CB3R: GGTGGAATGGGATTTTATCTG) by Macrogen (www.macrogen.com). All sequences used in this study are deposited in GenBank (accession nos. EU557098–EU557193). The Bayesian method (MrBayes v. 3.1) was used to reconstruct the tree (Huelsenbeck & Ronquist 2001) based on an optimized model of sequence evolution (HYK+I+gamma), determined for the specific dataset by the Bayesian information criterion in MrAIC v. 1.4.3 (Nylander 2004). Two independent runs of five simultaneous Markov chains (one cold and four heated) were run for 10 million generations, sampling the chains every 1000 generations. The first 5 million generations were discarded as the burn-in. Convergence was checked by plotting the parameters against generations, and using the diagnostic tools available in MrBayes v. 3.1. A 50% majority-rule consensus tree (‘Bayesian’ tree) was then constructed and node support was considered significant when more than 95% of the sampled trees recovered a particular clade (Huelsenbeck & Ronquist 2001).
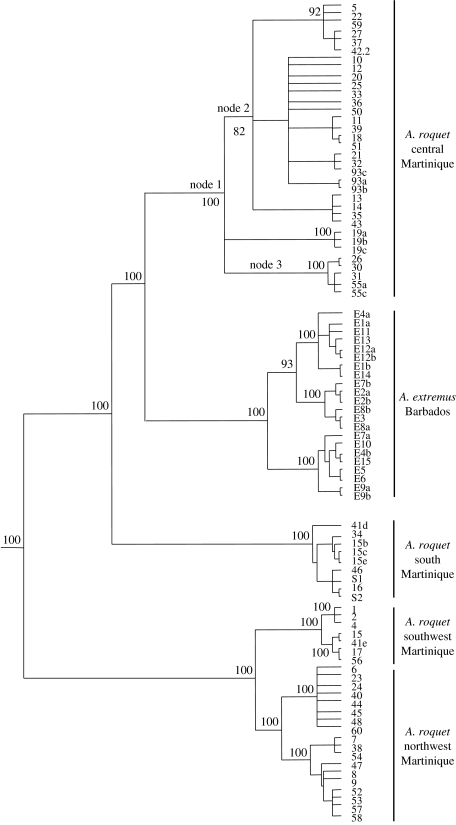
This phylogeographic analysis of the A. roquet/extremus complex gives a well-supported tree with four primary clades in Martinique, as shown in figure 4. The spatial distribution of these clades can be tested against these geological regions using matrix correspondence, which shows an extremely close association between the four clades and four out of the five precursor islands (Thorpe & Stenson 2003). This suggests that all the precursors, except Caravelle in the east, have populations surviving from a much earlier time when they were separate islands. Once again, although interpretations of times of island origin may vary, the molecular clock interpretations are compatible with island times, indicating that these separate Martinique precursor islands were occupied from around the time of origin of the young arc in this area, ca 6–8 Myr BP (figure 2b; Thorpe & Stenson 2003; Thorpe et al. 2005b). The chronology of the uplift of the central area is less easy to time geologically, but molecular clock estimates of divergence can be made using penalized likelihood (Sanderson 2002, 2003) using the four internal geological calibration points in Thorpe et al. (2005b). This suggests a time of 1.5 Myr BP for the central lineage node (figure 4, node 1) and ca 1.1 Myr BP for subsequent nodes encompassing the widest geographically distributed sublineages within the central (figure 4, nodes 2 and 3). Hence, it appears that the uplift required to join all the precursors into the single island of Martinique may have been as recent as 1.5 Myr BP or less. The combination of phylogeography and geology therefore suggests that these island populations have diverged in allopatry for as long as most allopatric island species in this series and other Lesser Antillean anole series (Thorpe et al. 2008), and have made secondary contact relatively recently. Indeed, the Barbadian species (Anolis extremis) is in fact paraphyletic to A. roquet, being a sister lineage to the central Martinique lineage and nested within the remaining Martinique lineages (figure 4). Hence, the Martinique lineages are de facto phylogenetically deeper than this nominal species.
Figure 4.
Phylogeny of Anolis roquet and associated species. This Bayesian tree shows the Barbadian species, A. extremus, is the sister lineage to central Martinique and is nested within the phylogenetically deeper divisions of A. roquet from the precursor islands, which are now in secondary contact. The terminal node numbers are the locality codes for Martinique in Thorpe & Stenson (2003) and Barbados in Thorpe et al. (2005b), which are followed by alphabetic codes for individual replicates, with E denoting A. extremus, and additional localities 93, S1 and S2 (located at UTM zone 20 coordinates E-720425/N-1630450, E-733300/N-1599225 and E-732157/N-1594585, respectively).
Taken overall, the Martinique phylogeography, timing and interspecific phylogeny indicate that the Martinique precursor lineages are the equivalent of the current nominal allopatric island species, and their interaction on secondary contact is a sound test of the contribution of allopatry to speciation in this group. Since Martinique has strong environmental zonation, to which the anoles are known to adapt, a perspective can be gained by comparing the extent of genetic interaction associated with these ecotones with that associated with the secondary contact of the previously allopatric populations.
(b) Transects
The relative impact of allopatry and ecotones in this complex is tested by taking transects across the ecotones and secondary contact zones and investigating the genetic interaction across these zones. Here, we exemplify the approach using just one transect that goes across both ecotone and secondary contact zone. The transect goes from the xeric woodland of the north Caribbean coast to the central montane rainforest. In doing so, it also crosses from the northwest geological region to the central geological region, hence crossing both ecotones and secondary contact zones. Figure 3 illustrates the seven localities on this transect in relation to the ecotone and the position of secondary contact suggested by geological evidence. At each locality, 48 tail-tip biopsies were sampled for molecular analysis, while quantitative traits and dewlap hue were recorded from 10 adult males.
(i) Lineages
The lineages were first investigated using the complete cytochrome b sequence from the mtDNA. PCR-restriction fragment length polymorphism analyses were then designed to efficiently assign numerous individuals to either the northwest or central clade. The same fragment was digested after amplification using the restriction enzyme SspI (New England Biolabs) during 3 hours at 37°C. The digested products were run on a 2% agarose gel containing ethidium bromide. PCR products from the central clade are uncut by this enzyme while those from the northwestern clade are cut at position 128, allowing an unambiguous assignment of the individuals to these two clades.
(ii) Selection regimes
Both physical and biotic conditions vary dramatically along the transect. The xeric woodland gives way abruptly to montane rainforest through a belt of transitional woodland (Lassere 1977; figure 3). In the xeric woodland of the Caribbean coast, there may be as little as just over 1000 mm per annum of seasonal rain, while in the higher altitude rainforest there may be in excess of 5000 mm per annum (or even more than 8000 mm) of less seasonal rain (Meteo France; Lassere 1977). The temperature (lower in the rainforest) also changes along the transect in concert with these changes in the extent and timing of precipitation and vegetation. Normalized principal component analysis was used to generalize these environmental variables (vegetation type, altitude, seasonality of precipitation, annual precipitation and temperature), with the latter three variables obtained from Worldclim (http://www.worldclim.org/bioclim.htm).
Here, as elsewhere in the Lesser Antilles, the quantitative traits of these anoles track these selection regimes. Parallel variation within Martinique shows that xeric forms have zebra striping whereas montane rainforest forms have intense green hue marbled with black and non-UV white spots, and a variety of scalation, body dimension and other characters show a similar pattern of variation (Thorpe 2005). A multivariate suite of quantitative traits is selected to represent the phenotypic change along the transect in response to the changing selection regime. There are seven body dimensions adjusted against snout–vent length by analysis of covariance (jaw length, head length, head depth, head width, upper leg length, lower leg length and dewlap length), the percentage red, green and blue hue on the posterior trunk assessed in Adobe Photoshop, together with seven scale and pattern characters (post-mental scales, scales between supra-orbitals, number of dorsal chevrons, chevron intensity, occipital ‘A’ mark, black dorsal reticulation and white spots) that could result in a heteroscedastic within-group covariance matrix. These heteroscedastic characters were normalized and then subjected to a principal component analysis. The subsequent component scores, together with the body dimensions and hues, were subjected to a canonical variance analysis, where each transect locality is a group. The individual scores (normalized so that the pooled within-group standard deviation is unity) are plotted against the transect.
A large dewlap is present in males. In solitary anoles it is largely used for conspecific (male–male and male–female) signalling. The hue of both the anterior and posterior dewlap, from near UV to the red spectrum, was recorded as the diffuse reflectance from the surface as a percentage of a WS-2 white standard, using an AvaSpec-2048 spectrometer, with an AvaLight-XE xenon pulsed lights source (Avantes, The Netherlands), with a 200 μ receptor fibre, held at 45° to the surface by a purpose-made matt-black attachment. On each individual, at least three recordings were taken per dewlap region.
Rather than simply subjecting the very large number of autocorrelated reflectances at a given specific wavelength to a principal component analysis, the matrix-algebraic procedure in Thorpe (2002) is followed. This gives several independent wavelength segments (colours) as unit characters that can be compared across large samples of individuals at numerous localities, and is consequently suitable for this comparative population-level study. The application of this process to the Martinique (Thorpe & Stenson 2003) and related (Thorpe 2002) anoles, using both large samples and multiple body regions, yields robust results. For this study, we follow the results of Thorpe & Stenson (2003), with the exception that the near UV is divided into two segments: UV 330–380 nm; UV/violet 380–430 nm; blue 430–490 nm; green 520–590 nm; yellow/orange 590–640 nm; and red 640–710 nm. These ‘colour’ characters from the anterior and posterior dewlap (12 in all) were subjected to a canonical variate analysis with each transect locality as a group. As with the quantitative traits, the individual scores (with the pooled within-group standard deviation as unity) are plotted against the transect.
(iii) Genetic structure: nDNA
Samples were genotyped at nine microsatellite loci (AAE-P2F9, ABO-P4A9, AEX-P1H11, ALU-MS06, ARO-035, ARO-062, ARO-065, ARO-120, ARO-HJ2; Ogden et al. 2002; Gow et al. 2006) in a single multiplex using a Qiagen Multiplex PCR kit following the manufacturer's recommendations except that the annealing temperature was 55°C. PCR products were then analysed on an ABI 3130xl genetic analyser and the genotypes scored using Genemapper v. 4.0 (Applied Biosystems). There were no instances of linkage disequilibrium (Gow et al. 2006). The genetic structure along the transect was studied using Bayesian clustering as in the program Structure v. 2.1 (Pritchard et al. 2000). We defined the number of populations (K) from 1 to 9, and 10 independent runs were performed for each value of K using the admixture model, a burn-in of 100 000 steps followed by 400 000 post-burn-in iterations. We used the method described by Evanno et al. (2005) to determine the optimal number of populations. Finally, we studied the partitioning of the genetic variance within and among predefined groups with an analysis of molecular variance (AMOVA) using the program Arlequin v. 3.11 (Excoffier et al. 2005). We carried out two different AMOVAs: first, grouping populations into the two geological regions and, second, grouping populations by habitat type, i.e. xeric woodland (localities 1 and 2), transitional forest (locality 3) and montane rainforest (localities 4–7). This classification is supported by both the vegetation type and the generalized multivariate environmental analysis.
4. Results and conclusions
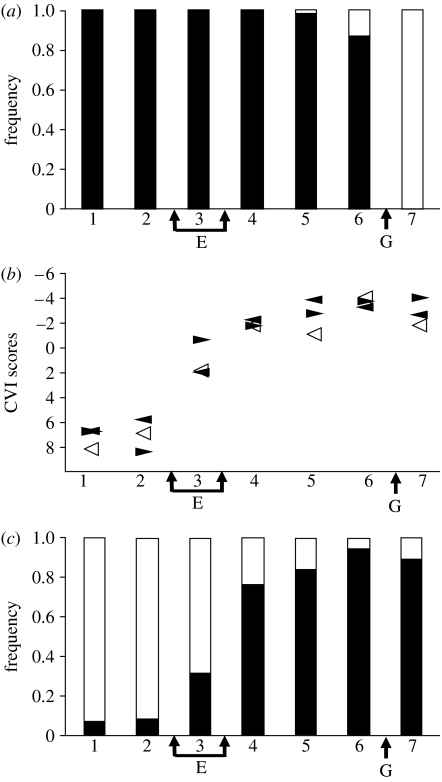
The island-wide study of lineages on Martinique, although based on a single, or a few, specimens per locality, showed a close association between the lineages and precursor islands. The transect study, which is based on much greater sample sizes (48 specimens per locality), confirms this. The transect shows that there is remarkably little introgression of the mtDNA lineages into the area occupied by the adjacent lineage (figure 5a). At locality 7, there are 100% central clade haplotypes, while at the adjacent locality there are 88% northwestern clade haplotypes.
Figure 5.
Transects showing (a) lineages, (b) selection regimes and (c) genotype structure. The transect localities (mapped in figure 3) are arranged in geographical order on the horizontal axis, with E representing the position of the ecotone (the switch between xeric coastal woodland and montane rainforest) and G the position of secondary contact between the northwest (localities 1–6) and central (7) geological regions. (a) The frequency (48 individuals per locality) of the northwest (black) and central (white) mtDNA lineages. (b) Mean first canonical variate (CVI) scores for adult males based on their quantitative traits (right-pointing filled triangles) and dewlap hue (left-pointing filled triangles), and the first principal coordinate score of environmental variables (open triangles). The CV scores are in units of 2 within group standard deviations. (c) The frequency of individuals (48 individuals per locality) assigned to the two Bayesian clusters, ‘xeric’ (white) and ‘montane rainforest’ (black), based on variation in nine unlinked microsatellites.
All three character sets providing insights into the selection regime (environmental variables, quantitative traits and dewlap hue) change in concert along the zone (figure 5b), with both the quantitative traits and dewlap hue being strongly correlated with the generalized environmental component (r=0.96 and 0.97, p<0.001, respectively). All three change, as expected, at the ecotone between xeric woodland and montane rainforest. There is a very pronounced change in the quantitative traits across the transect, there being approximately 15 within-locality standard deviations between the extreme montane and xeric forms (figure 5b). The dewlap hue also changes dramatically across the ecotone, there being 10 within-locality standard deviations between the xeric and montane extremes (figure 5b). The anterior dewlap is light grey, including near-UV reflectance, in xeric forms, but bright focal green without notable UV reflectance in montane forms, while the posterior dewlap is brighter, more focal yellow, in the rainforest forms compared with xeric forms (figure 6).
Figure 6.
Dewlap hue. Examples of spectrophotometrically measured percentage reflectance across the near UV to red spectrum (in nm). (a) The anterior dewlap of a xeric woodland male is a dull greyish (including near UV) hue, which contrasts to the bright focal green hue of the montane rainforest male. (b) The posterior dewlap of the rainforest male is brighter, more focal yellow, than that of the xeric woodland male.
If isolation in allopatry results in reproductively isolated species, then there should be little or no gene exchange between the genomes of the lineages from the precursor islands. If this is the case, then the nuclear DNA, as reflected in the microsatellite variation, should be distinct, and the Bayesian assignment should assign individuals into two groups reflecting their precursor island origin. This clearly does not occur (figure 5c). The distribution of the mtDNA lineages is distinct and remarkably faithfully segregated between the precursor island regions, yet the Bayesian assignment based on microsatellite variation shows no indication at all that the individuals either side of the secondary contact zone are assigned to different groups. The Bayesian analysis recognized two clusters, which we can refer to as the xeric and montane nuclear genotypes. Localities 6 and 7, either side of the secondary contact zone, both have predominantly (more than 90%) the same (montane) nuclear genotype group, suggesting that there is no discernable barrier to genetic exchange across this region of the zone. There is no significant correlation (p≥0.05) between the frequency of nuclear genotype group and the frequency of the mtDNA haplotype group along the transect. The AMOVA results confirm this, there being no significant variance associated with grouping by lineage (p=0.56). By notable contrast, the nuclear genotype changes at the ecotone from a predominantly xeric nuclear genotype at localities 1 and 2 (more than 92%) to the predominantly montane nuclear genotype at localities 4–7 (76–95%), with a strong correlation with the selection regimes (generalized environmental variables r=0.96, p<0.001; quantitative traits r=0.93, p<0.005; dewlap hue r=0.98, p<0.001). Once again, the AMOVA results confirm this with a significant variance associated with ecological grouping (3.58%, p=0.007 between ecological groups compared with 1.07% between localities within ecological groups).
The broad biogeographic pattern for Lesser Antillean anoles suggests the importance of allopatry in speciation, but this population-level study clearly questions this hypothesis. The lineages on the Martinique precursor island diverged in allopatry a long time ago, and are certainly phylogenetically deeper than some named species in the series and the bimaculatus series to the north (Thorpe et al. 2008), but there is no indication of any reduced genetic exchange between them in nDNA genes, let alone full reproductive isolation. This suggests that some of the nominal species in this series are not reproductively isolated and, of more general importance, allopatry, even for long periods, may not be sufficient to result in speciation (sensu even partial reproductive isolation).
By contrast, there is notable evidence of reduced nuclear genetic exchange between the anoles from the adjacent habitat types (xeric versus montane rainforest), without any physical barrier to gene flow. This may involve both natural and sexual selection. There is overwhelming evidence of the link between quantitative traits and natural selection regimes in the anoles from mountainous Lesser Antillean islands, and the quantitative traits and pattern of restricted gene exchange are strongly linked. The dewlap hue, possibly influenced by the interaction between natural and sexual selection, is also very closely linked with the pattern of nuclear gene exchange. The mechanism for this may well involve the different light regimes in these habitat types and its impact on visual communication, i.e. the role of sensory drive in speciation (Endler 1992; Boughman 2002). This may be the case, even though the generalizations about dewlap hue and habitat type (which are not derived from Lesser Antillean anoles; Leal & Fleishman 2004; Losos in press) do not necessarily appear to pertain to these Lesser Antillean anoles. The easiest mechanism to envisage is one of assortative mating where females choose males with the ‘correct’ dewlap hue. However, demonstrating female choice in anoles, and other lizards, is not straightforward and male–male competition, rather than female choice, may dominate sexual selection.
The lack of introgression in the mtDNA between precursor island regions is notable compared with the situation with the nDNA microsatellite markers, and incongruence between mtDNA and nDNA patterns can be seen in other organisms (Moritz 1994; Avise 2004), including Scandinavian brown bears where Waits et al. (2000) remarked that nDNA genetic differentiation, as measured by microsatellite loci, is not consistent with mtDNA phylogeographic groupings. Given the difference in mutation rates, variability, recombination and effective population size, there is no expectation that the patterns of variation in space should be the same in the mtDNA and the microsatellites. These differences are why mtDNA is widely used to reveal phylogeographic relationships (Avise 2000), while hypervariable microsatellites are widely used to reveal more recently formed population genetic structure (Freeland 2005). As males generally transmit only nDNA and not mtDNA, sex-biased dispersal may be important (Waits et al. 2000). Anoles, like many lizards, are polygynous (Andrews & Nichols 1990; Jenssen et al. 2001), which predicts greater male dispersal (Dobson 1982; Pusey 1987; Perrin & Mazalov 2000) that in turn would lead to proportionally greater nDNA gene flow. There is little direct observational data available on sex-biased dispersal in anoles, although Doughty & Sinervo (1994), Doughty et al. (1994) and Massot et al. (2003) gave some evidence of male-biased dispersal in other iguanids. However, there is genetic evidence of sex-biased dispersal in Anolis oculatus on Dominica (Stenson et al. 2002) and A. roquet on Martinique (R. S. Thorpe, Y. Surget-Groba & H. Johansson 2008, unpublished data) from microsatellite studies. The situation is further complicated by sperm retention and multiple insemination in Anolis (Calsbeek et al. 2007). Recent common garden experiments on anoles (Eales et al. 2008) suggest that a female may carry up to six viable eggs (excluding those lost in transit) that are laid at two-week intervals. Hence, a dispersing gravid female may carry up to seven different diploid nuclear genomes (one from the female and six from the males), but only the one type of mtDNA haplotype.
Taken together, these factors suggest a greater capacity for nDNA gene flow, and may explain why the lack of reproductive isolation has allowed nDNA introgression while retaining a strong mtDNA phylogeographic signature. An alternative explanation is that the nDNA markers are under selection, or are linked to loci under selection. However, on balance, we consider this as improbable, because all the nDNA markers are unlinked. So they would not only have to be linked to different loci that are under selection, but the direction of the selection on these independent loci would have to be congruent with one another and also with the habitat type.
The anoles from different habitat types have reduced genetic exchange rather than no (or almost no) genetic exchange, so we are not suggesting that these are full species generated by ecological speciation. However, this evidence of reduced genetic exchange among habitat types may well have general implications for the role of ecology in speciation in other sexually reproducing animals. In the Greater Antillean anoles, most speciation has occurred within islands (Losos in press) and, in the light of the evidence presented here, models emphasizing the role of ecology over allopatry should be given serious consideration for these and other sexually reproducing animals. This is compatible with the growing literature highlighting the role of ecology in speciation across a range of organisms (Smith et al. 1997; Orr & Smith 1998; Schluter 2000; Rundle & Nosil 2005; Gross 2006; Hendry et al. 2007; Butlin et al. in press; Lowry et al. 2008).
Acknowledgments
This research was primarily supported by the BBSRC (BB/C500544/1) with additional fellowship (Y.S.-G.) support from the EC (MEIF-CT-2005-009981) and studentship support (H.J.) from NERC. We thank DIREN Martinique for permission to take non-invasive samples from anoles, and Mike Ritchie, Godfrey Hewitt and an anonymous referee for their helpful comments on the manuscript.
Footnotes
One contribution of 12 to a Theme Issue ‘Speciation in plants and animals: pattern and process’.
References
- Abbott R.J, Ritchie M.G, Hollingsworth P.M. Speciation in plants and animals: pattern and process. Phil. Trans. R. Soc. B. 2008;363:2965–2969. doi: 10.1098/rstb.2008.0096. doi:10.1098/rstb.2008.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R.M, Nichols J.D. Temporal and spatial variation in survival rates of the tropical lizard Anolis limifrons. Oikos. 1990;2:215–221. doi:10.2307/3565942 [Google Scholar]
- Avise J.C. Harvard University Press; Cambridge, MA: 2000. Phylogeography: the history and formation of species. [Google Scholar]
- Avise J.C. 2nd edn. Sinauer Associates; Sunderland, MA: 2004. Molecular markers, natural history, and evolution. [Google Scholar]
- Barton, N. H. (ed.) 2001 Special issue on speciation. Trends Ecol. Evol 16, 325–413. [DOI] [PubMed]
- Beard J.S. Clarendon Press; Oxford, UK: 1948. The natural vegetation of the Windward and Leeward Islands. [Google Scholar]
- Borsa P, Lemer S, Aurelle D. Patterns of lineage diversification in rabbitfishes. Mol. Phylogenet. Evol. 2007;44:427–435. doi: 10.1016/j.ympev.2007.01.015. doi:10.1016/j.ympev.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Boughman J.W. How sensory drive can promote speciation. Trends Ecol. Evol. 2002;17:571–577. doi:10.1016/S0169-5347(02)02595-8 [Google Scholar]
- Butlin R.K, Galindo J, Grahame J.W. Sympatric, parapatric or allopatric: the most important way to classify speciation? Phil. Trans. R. Soc. B. 2008;363:2997–3007. doi: 10.1098/rstb.2008.0076. doi:10.1098/rstb.2008.0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calsbeek R, Smith T.B. Probing the adaptive landscape using experimental islands: density-dependent natural selection on lizard body size. Evolution. 2007;61:1052–1061. doi: 10.1111/j.1558-5646.2007.00093.x. doi:10.1111/j.1558-5646.2007.00093.x [DOI] [PubMed] [Google Scholar]
- Calsbeek R, Knouft J.H, Smith T.B. Variation in scale numbers is consistent with ecologically based natural selection acting within and between lizard species. Evol. Ecol. 2006;20:377–394. [Google Scholar]
- Calsbeek R, Bonneaud C, Prabhu S, Manoukis N, Smith T.B. Multiple paternity and sperm storage lead to increased genetic diversity in Anolis lizards. Evol. Ecol. Res. 2007;9:495–503. [Google Scholar]
- Claridge M.F, Dawah H.A, Wilson M.R. Chapman Hall; London, UK: 1997. Species: the units of biodiversity. [Google Scholar]
- Comes H.P, Tribsch A, Bittkau C. Plant speciaton in continental island floras as exemplified by Nigella in the Aegean archipelago. Phil. Trans. R. Soc. B. 2008;363:3083–3096. doi: 10.1098/rstb.2008.0063. doi:10.1098/rstb.2008.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann U, Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400:354–357. doi: 10.1038/22521. doi:10.1038/22521 [DOI] [PubMed] [Google Scholar]
- Dieckmann U, Metz J.A.J, Doebeli M, Tautz D. Cambridge University Press; Cambridge, UK: 2004. Adaptive speciation. [Google Scholar]
- Dobson S. Competition for mates and predominant juvenile dispersal in mammals. Anim. Behav. 1982;30:1183–1192. doi:10.1016/S0003-3472(82)80209-1 [Google Scholar]
- Doebeli M, Dieckmann U. Speciation along environmental gradients. Nature. 2003;421:259–264. doi: 10.1038/nature01274. doi:10.1038/nature01274 [DOI] [PubMed] [Google Scholar]
- Doughty P, Sinervo B. The effects of habitat, time of hatchling and body size on the dispersal of hatchling Uta stansburiana. J. Herpetol. 1994;28:485–490. doi:10.2307/1564962 [Google Scholar]
- Doughty P, Sinervo B, Burghardt G. Sex-biased dispersal in a polygynous lizard, Uta stansburiana. Anim. Behav. 1994;47:227–229. doi:10.1006/anbe.1994.1029 [Google Scholar]
- Eales J, Thorpe R.S, Malhotra A. Weak founder effect signal in a recent introduction of Caribbean Anolis. Mol. Ecol. 2008;17:1416–1426. doi: 10.1111/j.1365-294X.2007.03684.x. doi:10.1111/j.1365-294X.2007.03684.x [DOI] [PubMed] [Google Scholar]
- Endler J.A. Signals, signal conditions, and the direction of evolution. Am. Nat. 1992;139:S125–S153. doi:10.1086/285308 [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. doi:10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin. v. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Freeland J.R. Wiley; Chichester, UK: 2005. Molecular ecology. [Google Scholar]
- Gavrilets S. Perspective: models of speciation: what have we learned in 40 years. Evolution. 2003;57:2197–2215. doi: 10.1111/j.0014-3820.2003.tb00233.x. doi:10.1111/j.0014-3820.2003.tb00233.x [DOI] [PubMed] [Google Scholar]
- Gow J.L, Johansson H, Surget-Groba Y, Thorpe R.S. Ten polymorphic tetranucleotide microsatellite markers isolated from the Anolis roquet series of Caribbean lizards. Mol. Ecol. Notes. 2006;6:873–876. doi:10.1111/j.1471-8286.2006.01382.x [Google Scholar]
- Gross L. Demonstrating the theory of ecological speciation in Cichlids. PLoS Biol. 2006;4:e449. doi: 10.1371/journal.pbio.0040449. doi:10.1371/journal.pbio.0040449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry A.P, Nosil P, Rieseberg L.H. The speed of ecological speciation. Funct. Ecol. 2007;21:455–464. doi: 10.1111/j.1365-2435.2006.01240.x. doi:10.1111/j.1365-2435.2007.01240.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D.J, Berlocher S.H. Oxford University Press; Oxford, UK: 1997. Endless forms: species and speciation. [Google Scholar]
- Huelsenbeck J.P, Ronquist F.R. MrBayes: Bayesian inference in phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde J.R, Vetter R.D. The origin, evolution, and diversification of rockfishes of the genus Sebastes (Cuvier) Mol. Phylogenet. Evol. 2007;44:790–811. doi: 10.1016/j.ympev.2006.12.026. doi:10.1016/j.ympev.2006.12.026 [DOI] [PubMed] [Google Scholar]
- Jackman T.R, Larson A, De Queiroz K, Losos J.B. Phylogenetic relationships and tempo of early diversification in Anolis lizards. Syst. Biol. 1997;48:254–285. doi:10.1080/106351599260283 [Google Scholar]
- Jenssen T.A, Lovern M.B, Congdon J.D. Field-testing the protandry-based mating system for the lizard, Anolis carolinensis: does the model organism have the right model? Behav. Ecol. Sociobiol. 2001;50:162–172. doi:10.1007/s002650100349 [Google Scholar]
- Lassere G. CNRS-IGN; Paris, France: 1977. Atlas des Department Francais d'Outre Mer. II La Martinique. [Google Scholar]
- Leal M, Fleishman L.J. Differences in visual signal design and detectability between allopatric populations of Anolis lizards. Am. Nat. 2004;163:26–39. doi: 10.1086/379794. doi:10.1086/379794 [DOI] [PubMed] [Google Scholar]
- Losos J.B. An experimental demonstration of the species-recognition role of Anolis dewlap color. Copeia. 1985;1985:905–910. doi:10.2307/1445240 [Google Scholar]
- Losos J.B, Schluter D. Analysis of an evolutionary species–area relationship. Nature. 2000;408:847–850. doi: 10.1038/35048558. doi:10.1038/35048558 [DOI] [PubMed] [Google Scholar]
- Losos J.B, Thorpe R.S. Evolutionary diversification of Anolis lizards: introduction. In: Dieckmann U, Metz H.A.J, Doebeli M, Tautz D, editors. Adaptive speciation. Cambridge University Press; Cambridge, UK: 2004. pp. 322–324. [Google Scholar]
- Losos, J. B. In press. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles California, CA: University of California Press.
- Lowry D.B, Modliszewski J.L, Wright K.M, Wu C.A, Willis J.H. The strength and genetic basis of reproductive isolating barriers in flowering plants. Phil. Trans. R. Soc. B. 2008;363:3009–3021. doi: 10.1098/rstb.2008.0064. doi:10.1098/rstb.2008.0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran A.E. Population differentiation without speciation. Phil. Trans. R. Soc. B. 1998;353:275–286. doi:10.1098/rstb.1998.0209 [Google Scholar]
- Magurran, A. E. & May, R. M. (eds) 1998 Evolution of biological diversity: from population differentiation to speciation. Phil. Trans. R. Soc. B353, 173–345.
- Malhotra A, Thorpe R.S. Experimental detection of rapid evolutionary response in natural lizard populations. Nature. 1991;353:347–348. doi:10.1038/353347a0 [Google Scholar]
- Malhotra A, Thorpe R.S. The dynamics of natural selection and vicariance in the Dominican anole: patterns of within island molecular and morphological divergence. Evolution. 2000;54:245–258. doi: 10.1111/j.0014-3820.2000.tb00025.x. doi:10.1111/j.0014-3820.2000.tb00025.x [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization, ecological races, and the nature of species: empirical evidence for the ease of speciation. Phil. Trans. R. Soc. B. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. doi:10.1098/rstb.2008.0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massot M, Huey R.B, Tsuji J, van Berkum F.H. Genetic, prenatal, and postnatal correlates of dispersal in hatchling fence lizards (Scleroporus occidentalis) Behav. Ecol. 2003;14:650–655. doi:10.1093/beheco/arg056 [Google Scholar]
- Maury R.C, Westbrook G.K, Baker P.E, Bouysse P, Westercamp D. Geology of the Lesser Antilles. In: Dengo G, Case J.E, editors. The Caribbean region. Geological Society of America; Boulder CO: 1990. pp. 141–166. [Google Scholar]
- Mayr E. Belknap Press of Harvard University Press; Cambridge, MA: 1970. Populations, species, and evolution. [Google Scholar]
- Moritz C. Applications of mitochondrial DNA analysis in conservation. A critical review. Mol. Ecol. 1994;3:401–411. doi:10.1111/j.1365-294X.1994.tb00080.x [Google Scholar]
- Nylander, J. A. A. 2004 MrAIC.pl. Program distributed by the author. Uppsala University, Sweden: Evolutionary Biology Centre.
- Ogden R, Thorpe R.S. Molecular evidence for ecological speciation in tropical habitats. Proc. Natl Acad. Sci. USA. 2002;99:13 612–13 615. doi: 10.1073/pnas.212248499. doi:10.1073/pnas.212248499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R, Griffiths T.J, Thorpe R.S. Eight microsatellite loci in the Caribbean lizard, Anolis roquet. Conserv. Genet. 2002;3:345–346. doi:10.1023/A:1019928801124 [Google Scholar]
- Orr M.R, Smith T.B. Ecology and speciation. Trends Ecol. Evol. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. doi:10.1016/S0169-5347(98)01511-0 [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Kane N.C. Meeting review: American Genetics Association Symposium on the genetics of speciation. Mol. Ecol. 2007;16:2852–2854. doi: 10.1111/j.1365-294X.2007.03358.x. doi:10.1111/j.1365-294X.2007.03358.x [DOI] [PubMed] [Google Scholar]
- Perrin N, Mazalov L. Local competition, inbreeding, and the evolution of sex-biased dispersal. Am. Nat. 2000;135:116–127. doi: 10.1086/303296. doi:10.1086/303296 [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donnelly P. Inference of population structure from multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusey A.E. Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends Ecol. Evol. 1987;2:295–299. doi: 10.1016/0169-5347(87)90081-4. doi:10.1016/0169-5347(87)90081-4 [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E, Bermingham E. The causes of evolutionary radiations in archipelagoes: passerine birds in the Lesser Antilles. Am. Nat. 2007;169:285–297. doi: 10.1086/510730. doi:10.1086/510730 [DOI] [PubMed] [Google Scholar]
- Ritchie M. Feathers, females, and fathers. Science. 2007;318:54–55. doi: 10.1126/science.1149597. doi:10.1126/science.1149597 [DOI] [PubMed] [Google Scholar]
- Rundle H.D, Nosil P. Ecological speciation. Ecol. Lett. 2005;8:336–352. doi:10.1111/j.1461-0248.2004.00715.x [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. doi:10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Schluter D. Oxford University Press; Oxford, UK: 2000. The ecology of adaptive radiation. [Google Scholar]
- Schneider C.J, Smith T.B, Larison B, Moritz C. A test of alternative models of diversification in tropical rainforests: ecological gradients vs. rainforest refugia. Proc. Natl Acad. Sci. USA. 1999;96:13 869–13 873. doi: 10.1073/pnas.96.24.13869. doi:10.1073/pnas.96.24.13869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.B, Wayen R.K, Girman D.J, Bruford M.W. A role for ecotones in generating rainforest biodiversity. Science. 1997;276:1855–1857. doi:10.1126/science.276.5320.1855 [Google Scholar]
- Stenson A.G, Malhotra A, Thorpe R.S. Population differentiation and nuclear gene flow in the Dominican anole (Anolis oculatus) Mol. Ecol. 2002;11:1679–1688. doi: 10.1046/j.1365-294x.2002.01564.x. doi:10.1046/j.1365-294X.2002.01564.x [DOI] [PubMed] [Google Scholar]
- Stenson A.G, Thorpe R.S, Malhotra A. Mitochondrial and nuclear evolution: a phylogeny of bimaculatus group anoles. Mol. Phylogenet. Evol. 2004;32:1–10. doi: 10.1016/j.ympev.2003.12.008. doi:10.1016/j.ympev.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Surget-Groba Y, et al. Intraspecific phylogeography of Lacerta vivipara and the evolution of viviparity. Mol. Phylogenet. Evol. 2001;18:449–459. doi: 10.1006/mpev.2000.0896. doi:10.1006/mpev.2000.0896 [DOI] [PubMed] [Google Scholar]
- Tautz D. Splitting in space. Nature. 2003;421:225–226. doi: 10.1038/421225b. doi:10.1038/421225b [DOI] [PubMed] [Google Scholar]
- Thorpe R.S. Analysis of color spectra in comparative evolutionary studies: molecular phylogeny and habitat adaptation in the St. Vincent Anole, Anolis trinitatis. Syst. Biol. 2002;51:554–569. doi: 10.1080/10635150290069986. doi:10.1080/10635150290069986 [DOI] [PubMed] [Google Scholar]
- Thorpe R.S. Population evolution and island biogeography. Science. 2005;310:1778–1779. doi: 10.1126/science.1122457. doi:10.1126/science.1122457 [DOI] [PubMed] [Google Scholar]
- Thorpe R.S, Richard M. Evidence that ultraviolet markings are associated with patterns of molecular gene flow. Proc. Natl Acad. Sci. USA. 2001;98:3929–3934. doi: 10.1073/pnas.071576798. doi:10.1073/pnas.071576798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe R.S, Stenson A.G. Phylogeny, paraphyly and ecological adaptation of the colour and pattern in the Anolis roquet complex on Martinique. Mol. Ecol. 2003;12:117–132. doi: 10.1046/j.1365-294x.2003.01704.x. doi:10.1046/j.1365-294X.2003.01704.x [DOI] [PubMed] [Google Scholar]
- Thorpe R.S, Malhotra A, Stenson A.G, Reardon J.T. Adaptation and speciation in Lesser Antillean anoles. In: Dieckmann U, Metz H.A.J, Doebeli M, Tautz D, editors. Adaptive speciation. Cambridge University Press; Cambridge, UK: 2004. pp. 324–335. [Google Scholar]
- Thorpe R.S, Reardon J.T, Malhotra A. Common garden and natural selection experiments support ecotypic differentiation in the Dominican anole (Anolis oculatus) Am. Nat. 2005a;165:495–504. doi: 10.1086/428408. doi:10.1086/428408 [DOI] [PubMed] [Google Scholar]
- Thorpe R.S, Leadbeater D.L, Pook C.E. Molecular clocks and geological dates: cytochrome b of Anolis extremus substantially contradicts dating of Barbados emergence. Mol. Ecol. 2005b;14:2087–2096. doi: 10.1111/j.1365-294X.2005.02574.x. doi:10.1111/j.1365-294X.2005.02574.x [DOI] [PubMed] [Google Scholar]
- Thorpe R.S, Jones A.G, Malhotra A, Surget-Groba Y. Adaptive radiation in Lesser Antillean lizards: molecular phylogenetics and species recognition in the Lesser Antillean dwarf gecko complex, Sphaerodactylus fantasticus. Mol. Ecol. 2008;17:1489–1504. doi: 10.1111/j.1365-294X.2007.03686.x. doi:10.1111/j.1365-294X.2007.03686.x [DOI] [PubMed] [Google Scholar]
- Waits L, Taberlet P, Swenson J.E, Sandergren F, Nuclear D.N.A. microsatellite analysis of genetic diversity and gene flow in the Scandinavian brown bear (Ursus arctos) Mol. Ecol. 2000;9:421–431. doi: 10.1046/j.1365-294x.2000.00892.x. doi:10.1046/j.1365-294x.2000.00892.x [DOI] [PubMed] [Google Scholar]
- Wiens J.J. What is speciation and how should we study it? Am. Nat. 2004;163:914–923. doi: 10.1086/386552. doi:10.1086/386552 [DOI] [PubMed] [Google Scholar]