Abstract
The genic view of the process of speciation is based on the notion that species isolation may be achieved by a modest number of genes. Although great strides have been made to characterize ‘speciation genes’ in some groups of animals, little is known about the nature of genic barriers to gene flow in plants. We review recent progress in the characterization of genic species barriers in plants with a focus on five ‘model’ genera: Mimulus (monkey flowers); Iris (irises); Helianthus (sunflowers); Silene (campions); and Populus (poplars, aspens, cottonwoods). The study species in all five genera are diploid in terms of meiotic behaviour, and chromosomal rearrangements are assumed to play a minor role in species isolation, with the exception of Helianthus for which data on the relative roles of chromosomal and genic isolation factors are available. Our review identifies the following key topics as being of special interest for future research: the role of intraspecific variation in speciation; the detection of balancing versus directional selection in speciation genetic studies; the timing of fixation of alleles of major versus minor effects during plant speciation; the likelihood of adaptive trait introgression; and the identification and characterization of speciation genes and speciation gene networks.
Keywords: speciation, Mimulus, Iris, Helianthus, Silene, Populus
1. Introduction
Recent years have seen great changes in our understanding of the nature of barriers to gene flow between diverging populations and species (Coyne & Orr 2004). Perhaps the most consequential of these is an extension of Ernst Mayr's biological species concept (BSC; Mayr 1942) to accommodate a ‘genic view’ of the process of speciation (Wu 2001a). Mayr's classic BSC sees species as ‘…groups of actually or potentially interbreeding natural populations which are reproductively isolated from other such groups…’ (Mayr 1942). This view of species contains the notion of ‘whole-genome isolation’ or ‘whole-genome reproductive isolation’ (Wu 2001a,b); that is, diverging populations must be protected from gene flow throughout their genomes to deserve the biologist's label of good species. By contrast, the genic view of speciation is based on the notion that species isolation may be achieved by a modest number of genes (Wu 2001a). Alleles at such loci will not move freely between species because they are selected against in a ‘foreign’ genetic background (Wu 2001b). This genic view of speciation has been motivated mainly by the discovery of ‘speciation genes’ in Drosophila (Wu 2001a; Orr et al. 2004) and holds great potential to improve our understanding of species boundaries and divergence in many other groups (Noor & Feder 2006). Theoretical and empirical work predicts an important role for genic factors—as opposed to chromosomal or genomic factors—during speciation (Felsenstein 1981; Via 2001). ‘Porous genomes’ are also an expected feature of adaptive species radiations (Gavrilets & Vose 2005). Here, we review recent studies that address the genetic architecture of genic barriers to gene flow in plants. We do not intend to suggest a new species concept, as plenty of these are already available in the speciation literature (Coyne & Orr 2004). The genic view of plant speciation represents an update to Mayr's (1942) BSC. By recognizing that genealogies may vary among loci in the genome, it facilitates the study of genetic factors such as drift, gene flow, recombination, selection and their interactions during divergence.
Plant evolutionary genetics has long been an important contributor to evolutionary theory and thinking; for example, the modern evolutionary synthesis would not have been possible without Gregor Mendel's experimental work on plants. In the twenty-first century, plants have lost nothing of their attractiveness for evolutionary biologists interested in testing theoretical predictions: plants can often be crossed rather easily, and their sessile nature facilitates the estimation of fitness effects of individual traits, chromosomal segments or even individual genes in the wild (Bradshaw et al. 1998; Bradshaw & Schemske 2003; Lexer et al. 2004; Martin et al. 2006; Noor & Feder 2006). Thus, plants offer attractive study objects for addressing fundamental questions in speciation genetics. The purpose of this review is to discuss recent progress of the field and the questions that are emerging from this work. We avoid discussing the nature of plant species as this topic has been covered recently (Rieseberg et al. 2006). Also, we refer to Rieseberg & Willis (2007) for an overview on current progress in plant speciation and to Ramsey & Schemske (1998), Soltis et al. (2004) and Comai (2005) for polyploid speciation in particular (see also Hegarty et al. (2008) in the present issue). Here, we focus on recent speciation genetic work in selected groups of diploid plants (diploid in terms of current meiotic behaviour rather than evolutionary history) in which rapid progress has been made in the last few years. A focus on diploids allows us to discuss questions of general interest to botanists and zoologists, which may contribute to much-needed discussion between these groups of researchers.
Only few potential plant speciation genes, genes involved in the evolution or maintenance of species barriers, have been characterized at the molecular level. These include: hybrid sterility genes conferring cytoplasmic male sterility (CMS), that is, the loss of coordination between nuclear and organellar genes inherited from different parents (Chase 2007); myb-type transcription factors as major determinants of flower colour-linked pollinator shifts (Hoballah et al. 2007); and genes involved in hybrid necrosis, i.e. autoimmune responses presumably mediated by rapidly evolving pathogen-resistant genes (Bomblies & Weigel 2007). Fortunately, studies of genic species barriers can also commence without knowledge of the actual genes involved. It is important to point this out, as this fact was not expressed clearly in Wu's (2001a) landmark paper: the relative roles of selection and drift in driving speciation may be assessed using the analytical toolkits of quantitative evolutionary genetics and population genomics (Lynch & Walsh 1998; Luikart et al. 2003; Beaumont 2005) even before speciation genes have been identified. A more important prerequisite is knowledge of the nature and type of barriers among the taxa studied. As long as there is some evidence that barriers are primarily genic rather than chromosomal (e.g. evidence from cytogenetics or comparative genetic mapping), plant evolutionary geneticists can begin to study the architecture of genic species isolation.
Studies of Arabidopsis thaliana and its relatives have contributed greatly to our understanding of plant evolutionary genetics, and much of this work has been reviewed recently (Mitchell-Olds 2001; Mitchell-Olds & Schmitt 2006; Lysak & Lexer 2006; Bomblies & Weigel 2007). Here, we review recent work of relevance in five other plant genera that have become or are currently emerging as model systems for plant evolutionary genetics: Mimulus (monkey flowers); Iris (irises); Helianthus (sunflowers); Silene (campions); and Populus (poplars, aspens, cottonwoods; table 1). Specifically, we look for answers to the following questions of current interest: (i) how often do genes of major effect contribute to plant speciation, (ii) do the architectures of genic species barriers differ between pairs of species of the same plant group, (iii) what are the relative roles of endogenous (intrinsic) versus exogenous (environmental) genic barriers in particular cases, (iv) are species differences in general, and trait differences involved in reproductive isolation in particular, more likely to arise via selection or drift, (v) under which conditions can the breakdown of reproductive barriers result in adaptive introgression, (vi) what are the roles of pleiotropy or linkage in facilitating or constraining speciation, (vii) how do interspecific barriers relate to and interact with barriers to gene flow within species, and (viii) which experimental approaches may be used to study species barriers in situ in their ecological context? Table 1 lists which of these eight questions were addressed by each reviewed study. We mine the literature for answers and close with an outlook on future research.
Table 1.
Essential studies reviewed in this contribution, including types of genic barriers studied, methodologies used and questions addressed.
| study group | genic barrier studied | methodologies used | level of analysis | main questions addresseda | references |
|---|---|---|---|---|---|
| Mimulus | Dobzhansky–Muller incompatibility; differential adaptation | experimental crosses | intraspecific | 1, 4, 6 | Macnair & Christie (1983) and Christie & Macnair (1984) |
| pollinator isolation | QTL mapping of floral traits and pollinator preference | interspecific | 1, 3, 4, 8; 1, 3, 4, 8; 1, 3, 4, 8 | Bradshaw et al. (1995); Bradshaw et al. (1998) and Schemske & Bradshaw (1999) | |
| pollinator isolation | field experiments with NILsb | interspecific | 1, 3, 4, 8 | Bradshaw & Schemske (2003) | |
| mating system divergence | QTL mapping | interspecific | 1, 4, 6; 1, 4, 6 | Lin & Ritland (1997) and Fishman et al. (2002) | |
| mating system divergence | QTL mapping | intraspecific | 2, 6, 7 | Hall et al. (2006) | |
| intrinsic/Dobzhansky–Muller incompatibility | genetic mapping of TRDLb | interspecific | 1, 3, 4, 6 | Fishman & Willis (2001) | |
| intrinsic/hybrid male sterility | genetic mapping/analysis of backcrosses and NILsb | interspecific | 1, 3, 6 | Sweigart et al. (2006) | |
| intrinsic/Dobzhansky–Muller incompatibility | genetic analysis of intraspecific variation using controlled crosses | interspecific/intraspecific | 1, 3, 7 | Sweigart et al. (2007) | |
| intrinsic/cytonuclear incompatibility | segregation analysis and genetic mapping | interspecific | 1, 3, 4, 6 | Fishman & Willis (2006) | |
| intrinsic/cytonuclear incompatibility | molecular analysis of mitochondrial genes and transcripts | interspecific | 1, 3, 4, 7 | Case & Willis (2008) | |
| Iris | intrinsic postzygotic isolation | genetic mapping of TRDLb | interspecific | 1, 3, 5, 6 | Bouck et al. (2005) |
| post-establishment mortality | QTL mapping of hybrid survivorship | interspecific | 1, 3, 5 | Martin et al. (2005) | |
| ecological divergence | QTL mapping under highly selective field conditions | interspecific | 1, 3, 5, 6, 8 | Martin et al. (2006) | |
| flowering phenology | QTL mapping of flowering time | interspecific | 1, 3, 4, 5, 6, 8 | Martin et al. (2007) | |
| pollinator isolation | QTL mapping of floral traits | interspecific | 1, 3, 4, 5, 6 | Bouck et al. (2007) | |
| pollinator isolation | QTL mapping of pollinator preference | interspecific | 1, 3, 4, 5, 6, 8 | Martin et al. (2008) | |
| Helianthus | intrinsic barriers to introgression | QTL mapping of phenotypic differences and pollen sterility | interspecific | 4, 5 | Kim & Rieseberg (1999) |
| intrinsic and exogenous barriers to introgression | population genomic analysis of hybrid zones | interspecific | 3, 5, 8; 3, 5, 8; 3, 5, 7, 8 | Rieseberg et al. (1999b); Gardner et al. (2000) and Buerkle & Rieseberg (2001) | |
| intrinsic and exogenous barriers to introgression | population genomic analysis of species differentiation | interspecific | 2, 3, 4, 5, 8 | Yatabe et al. (2007) | |
| ecological divergence (40 phenotypic traits) | QTL mapping | interspecific/intraspecific | 1, 4, 7 | Lexer et al. (2005a) | |
| differential adaptation | population genomic analysis/selective sweep mapping | intraspecific | 4, 7 | Kane & Rieseberg (2007) | |
| Silene | differential adaptation | QTL mapping of adaptive traits | intraspecific | 1, 3, 4, 6, 7 | Bratteler et al. (2006a–c) |
| intrinsic barriers and ecological divergence | population genetic analysis of hybrid zones | interspecific | 5, 8 | Minder et al. (2007) | |
| intrinsic barriers and ecological divergence | population genomic analysis of species differentiation | interspecific | 3, 5, 8 | Minder & Widmer (2008) | |
| Populus | ecological divergence | QTL mapping of bud set and bud flush | interspecific | 1, 4, 6 | Frewen et al. (2000) |
| intrinsic barriers and ecological divergence | genetic mapping of TRDLb | interspecific | 1, 3, 5, 6 | Yin et al. (2004) | |
| intrinsic barriers and ecological divergence | analysis of physical genome sequence and genetic maps | interspecific | 3, 4, 5, 6 | Yin et al. (2008) | |
| intrinsic barriers and ecological divergence | population genomic analysis of hybrid zones | interspecific | 5, 8 | Martinsen et al. (2001) | |
| intrinsic barriers and ecological divergence | population genomic/genetic analysis of hybrid zones | interspecific | 3, 5, 7, 8; 3, 5, 7, 8; 5, 7, 8 | Lexer et al. (2005b, 2007) and van Loo et al. (2008) |
The numbering refers to the eight questions of interest posed at the end of the introduction.
Abbreviations: NIL, near-isogenic line; TRDL, transmission ratio distortion locus.
2. Genetic architecture of reproductive barriers and habitat adaptation in Mimulus
The genus Mimulus is highly diverse both in terms of species numbers and in adaptations to ecological factors ranging from diverse pollinators and mating systems to extreme habitats. Members of the genus have been the subject of intensive ecological and evolutionary genetic research for over 50 years and have become a model system for studies on the genetics of reproductive isolation and speciation (Wu et al. 2008; see also Lowry et al. 2008).
A detailed study on the different components of reproductive isolation between two Mimulus species, Mimulus lewisii and Mimulus cardinalis, found that most reproductive isolation between these two taxa occurs prior to hybrid formation (Ramsey et al. 2003; Martin & Willis 2007). A central component of such premating reproductive isolation in animal-pollinated angiosperms are thought to be specific pollinators (see also Cozzolino & Scopece 2008; Lowry et al. 2008). Analyses of the genetic architecture of floral traits differentiating Mimulus species that attract different pollinators consistently found evidence for a role of major-effect genes in the control of phenotypic trait differences (Bradshaw et al. 1995, 1998). This implies that either individual genes of individually large effect or linked clusters of genes with a large cumulative effect can play a central role in the evolution of plant reproductive isolation and speciation (Bradshaw et al. 1998).
The evidence for a role of major-effect genes in reproductive isolation gained from quantitative trait locus (QTL) analyses was confirmed in experimental analyses of pollinator preference. For example, analysis of pollinator visitation in a highly polymorphic F2 population derived from a cross between M. lewisii and M. cardinalis found that an allele that increased petal carotenoid concentration reduced bee visitation by 80%, whereas an allele that increased nectar production doubled hummingbird visitation (Schemske & Bradshaw 1999). A follow-up analysis of the adaptive value of alternative alleles at the former locus, referred to as YELLOW UPPER5-7 (YUP) locus, using near-isogenic lines (NILs) in field experiments revealed that both YUP alleles in the foreign background increased visitation of the alternative pollinator approximately 70-fold. These experimental pollinator analyses provide strong support for the notion that major-effect genes are involved in premating reproductive isolation (Bradshaw & Schemske 2003).
In contrast to the genetic architecture of floral trait differences involved in pollinator attraction, QTL analyses of traits involved in interspecific mating system differences between inbreeding and outbreeding Mimulus species indicate that these trait differences (floral morphology and stigma–anther separation) are primarily controlled by loci with small effect (Lin & Ritland 1997; Fishman et al. 2002). An analysis of intraspecific divergence between annual and perennial Mimulus guttatus came to the same conclusion (Hall et al. 2006). A comparison of the genetic architecture controlling floral trait divergence, both within divergent populations of M. guttatus and between M. guttatus and Mimulus nasutus (Fishman et al. 2002), implied that there may be a shared genetic basis for floral divergence within and among species of Mimulus (Hall et al. 2006).
An important aspect is that these analyses of floral trait divergence repeatedly found strong linkage among floral traits (Lin & Ritland 1997; Fishman et al. 2002). Furthermore, inferred QTL often affected more than one trait, thus contributing to the genetic correlation between those traits (Lin & Ritland 1997; Fishman et al. 2002). An intraspecific analysis in M. guttatus revealed high degrees of pleiotropy as well (Hall et al. 2006), thus indicating a probable role for genetic correlations in both inter- and intraspecific barriers to gene flow between these Mimulus taxa.
Reproductive isolation among Mimulus species is not only brought about by floral trait divergence associated with different pollinators or shifts in mating system, but also by postzygotic intrinsic barriers that may have contributed to species isolation at a later stage. The first direct genetic analyses of hybrid incompatibilities in Mimulus were performed by Macnair & Christie (Macnair & Christie 1983; Christie & Macnair 1984; see recent review of this work by Wu et al. 2008). Notably, these studies not only provided evidence for the existence of Dobzhansky–Muller incompatibilities in this species, but also further established a clear link between reproductive isolation and habitat adaptation. F1 lethality was observed in crosses between copper tolerant and non-tolerant populations of M. guttatus, with a gene responsible for copper tolerance in adapted populations pleiotropically affecting reproductive isolation between tolerant and non-tolerant populations. This suggests that habitat adaptation may lead to reproductive isolation and ultimately to speciation through interactions with intrinsic isolating factors.
More recently, Fishman & Willis (2001) analysed patterns of sterility in F1 and F2 hybrids between M. guttatus and M. nasutus and found evidence for Dobzhansky–Muller incompatibilities contributing to the sterility between the two species. Analysis of marker segregation in the F2 generation further revealed that nearly half of the markers exhibited significant transmission ratio distortion, and allowed identification of 11 transmission ratio distorting loci (TRDL) throughout the genome. Negative interactions between the heterospecific genomes are the most likely cause of the observed pattern, which is in line with Dobzhansky–Muller incompatibilities existing between the two species (Fishman et al. 2001).
Further loci involved in heterospecific incompatibility between these two Mimulus species were identified upon examination of male-sterile F2 hybrids. Nearly complete hybrid male sterility was found to result from a simple genetic incompatibility between a single pair of heterospecific loci in the investigated Mimulus species pair (Sweigart et al. 2006). Interestingly, the alleles causing genetic incompatibility were found not to be fixed within species. Instead, one of the loci, hms1, was polymorphic in the outcrossing M. guttatus and only locally distributed, which limits its potential to consistently contribute to the barrier between sympatric populations of the two species (Sweigart et al. 2007).
In addition to negative interactions among heterospecific nuclear loci, Fishman & Willis (2006) found that cytonuclear incompatibility occurs in some M. guttatus×M. nasutus hybrids that carry a particular M. guttatus cytoplasm. Hybrids have a pollenless anther phenotype that is caused by a cryptic CMS factor. In hermaphroditic M. guttatus populations carrying the CMS cytoplasm, the necessary nuclear restorer is present, but interspecific hybrids lack the co-evolved restorer locus and express the male-sterile phenotype (Fishman & Willis 2006). Case & Willis (2008) found that hybrid male sterility in Mimulus is associated with a specific mitochondrial rearrangement and that the CMS cytotype is highly restricted geographically in M. guttatus and is absent in M. nasutus (Case & Willis 2008). These results support the idea that selfish cytoplasmic elements may play an important role in shaping patterns of plant hybrid incompatibilities (Levin 2003).
3. Genetic analysis of species boundaries and adaptive introgression in Louisiana irises
The Louisiana irises form a classical example of introgressive hybridization in plants (Anderson 1949; Arnold 1997). Early molecular marker studies supported the long-standing hypothesis that introgressive hybridization has played an important role in the evolution of the three ‘model’ irises Iris fulva, Iris hexagona and Iris brevicaulis (reviewed by Arnold et al. 2004). It has been argued that habitat disturbance may play a role in the ecology and evolution of the Louisiana arises. This may include both natural disturbance due to temporary flooding in the drainage areas where these species occur and man-made disturbance in more recent times (Arnold et al. 2004). Here, we focus on recent genetic mapping studies of species boundaries and adaptive introgression in I. fulva and I. brevicaulis.
Initial progress in genetic map-based studies of species boundaries in I. fulva and I. brevicaulis was hampered by the size and complexity of their genomes, but this obstacle was recently overcome by using retrotransposon display markers to map species boundaries (Bouck et al. 2005). A surprising result of this work was that only a relatively small proportion of markers (15%) showed transmission ratio distortions in the interspecific backcross populations compared with other interspecific mapping studies in plants (Bouck et al. 2005). Also, most of these distortions were due to an over- rather than an under-representation of introgressed alleles, which suggested that introgressed alleles may often have a positive fitness effect in their new genetic background (Bouck et al. 2005). Nevertheless, the authors cautioned that the over-representation of heterospecific alleles may also stem in part from inbreeding depression caused by the experimental design used for mapping. Also, considering the frequent occurrence of transposable elements in Iris spp. (Bouck et al. 2005), it is possible that ‘selfish’ transposable element activity contributed to the observed transmission ratio distortions as well, in analogy to the selfish element argument discussed for Mimulus above.
In a QTL study of post-establishment mortality involving reciprocal backcrosses between I. fulva and I. brevicaulis in a controlled greenhouse environment, only a small proportion of the genome contributed to hybrid survivorship (Martin et al. 2005). For eight out of nine candidate genomic regions associated with survivorship in the two backcrosses, introgressed alleles increased survivorship. This provided additional support for the idea that introgression in Louisiana irises may sometimes contribute to the evolutionary dynamics of naturally occurring hybrid zones (Martin et al. 2005). Nevertheless, the fitness of introgressants under natural conditions depends on several additional isolating mechanisms described below.
A transplantation study with reciprocal backcrosses of the same two species under highly selective field conditions provided a first glimpse of the genetic architecture of differences in survivorship associated with habitat divergence in the wild (wet versus dry conditions; Martin et al. 2006). While field survivorship in the backcross to I. fulva was influenced greatly by negative epistasis between two QTL, introgressed I. fulva alleles tended to increase survivorship in the backcross towards I. brevicaulis. The latter finding suggests that traits responsible for adaptation to wet I. fulva habitat can be transferred into I. brevicaulis via introgressive hybridization (Martin et al. 2006).
Genetic mapping in interspecific crosses of I. fulva and I. brevicaulis was also used to explore the genetic architecture of flowering phenology, floral traits and pollinator preference in this group (Bouck et al. 2007; Martin et al. 2007, 2008). Although in both cases the authors cautioned that sample sizes were limited, heritable variation detectable as QTL was identified for all three suites of traits (Bouck et al. 2007; Martin et al. 2007). In the case of flowering phenology, the results were consistent with selection rather than drift promoting divergence in flowering time in the evolutionary history of these two species. This was concluded since both additive and epistatic effects of QTL alleles generally were in the direction expected from their parental species origin. In the case of morphological floral traits, frequent co-localization of QTL on the genetic map provided a straightforward explanation for why natural late-generation Iris hybrids often exhibit parental-like phenotypes: QTL for five of the nine traits examined were co-localized on a single linkage group, which implies a high potential for joint transmission of these traits in hybrids (Bouck et al. 2007). Genetic mapping of QTL affecting pollinator (hummingbird, lepidopteran and bee) preferences indicated that pollinator behaviour poses a much weaker barrier to gene flow in these species compared with flowering phenology. Nevertheless, co-localization of floral and preference QTL also showed that pollinators do respond to heritable floral traits (Martin et al. 2008).
Clearly, ongoing work in Louisiana irises is beginning to reveal which genetic factors are most critical to reproductive isolation between I. fulva and I. brevicaulis. Flowering time loci and QTL for floral traits acting as prezygotic barriers (Bouck et al. 2007; Martin et al. 2007) and QTL associated with habitat divergence as a potential pre- or postzygotic barrier (Martin et al. 2006) appear to be more important in terms of barrier strength than intrinsic isolating factors (Bouck et al. 2005). It will be interesting to compare the genetic architectures and genomic distributions of all these different types of isolating factors once this series of mapping experiments is complete (see Martin et al. 2008), and to compare the sizes of selection coefficients associated with different types of barriers. If adaptive introgression is important in irises, then we may expect that the fitness benefits of introgressed alleles in certain environments outweigh the reduction in fitness due to postzygotic intrinsic factors. Similarly, we would expect that genetic correlations (linkage or pleiotropy) along the chromosomes are of a type that facilitates rather than constrains adaptive introgression.
4. The genetic architecture of barriers to inter- and intraspecific gene flow in Helianthus
Studies of Helianthus have contributed greatly to our understanding of the molecular genetic architecture of barriers to gene flow in outcrossing plants. Here, we focus on the studies that inform us about the genetic basis of barriers to introgression between diploid Helianthus species, and largely ignore the increasing body of data on hybrid or ‘recombinational’ speciation in this group (e.g. see Rieseberg et al. 2003).
Among early molecular marker studies of introgression in Helianthus (Arias & Rieseberg 1994; Kim & Rieseberg 1999; Rieseberg et al. 1999a), the last one is of particular relevance as it was among the first to use QTL mapping to address the potential of adaptive trait introgression in plants. Kim & Rieseberg (1999) used QTL mapping to study the genetic architecture of introgression from the wild sunflower species Helianthus debilis into subspecies texanus of Helianthus annuus. The results revealed a simple architecture of genetic isolation—only 11 out of 60 QTL controlling morphological species differences were linked to pollen sterility factors or otherwise negatively selected chromosomal blocks. For the remaining 45 QTL no barrier to introgression was detected, and introgression of just three small linkage blocks appeared sufficient to recover the phenotype of H. annuus ssp. texanus. This supported the hypothesis that subspecies texanus of H. annuus originated through introgression, although the authors cautioned that data on the adaptive value of critical QTL are required to ultimately test this hypothesis (Kim & Rieseberg 1999). We note that some of the phenotypic traits under selection in H. annuus ssp. texanus have been identified more recently (Whitney et al. 2006).
In addition to QTL studies of experimental crosses, genomic studies of natural interspecific hybrid zones proved highly informative for studying the architecture of barriers to gene flow in Helianthus (Rieseberg et al. 1999b; Gardner et al. 2000; Buerkle & Rieseberg 2001). Rieseberg et al. (1999a,b) used a genome-wide panel of dominant molecular markers to study patterns of introgression across three ‘replicate’ hybrid zones of H. annuus and Helianthus petiolaris. Out of 26 genomic segments with significantly reduced introgression in these hybrid zones, 16 were associated with pollen sterility, and only 50% of the barrier to introgression was due to chromosomal rearrangements segregating between the two species. This indicated that the species barrier had a complex basis that included both chromosomal and genic isolating factors (Rieseberg et al. 1999b). A characteristic feature of these early studies of Helianthus hybrid zones was that increased or reduced introgression often was observed over large centimorgan distances or even over entire linkage groups. This is expected based on the available knowledge of linkage disequilibrium (LD) in hybrid zones (Barton & Hewitt 1985), since LD close to the centre of a hybrid zone will effectively increase the size of chromosomal blocks affected by either positive or negative selection (Rieseberg & Buerkle 2002; Yatabe et al. 2007).
A recent study of the same two sunflower species based on more than 100 codominant microsatellite loci and 14 sequenced genes revealed a more refined picture of the genetics of species isolation in this group (Yatabe et al. 2007). In that study, genetic divergence for individual marker loci between populations of the two sympatric congeners H. annuus and H. petiolaris was much weaker than that in the previous studies of hybrid zones (Rieseberg et al. 1999b; Buerkle & Rieseberg 2001), even after correcting for differences in variability between loci. The only highly divergent ‘outlier’ loci were five microsatellites associated with expressed sequence tags (ESTs), and these five loci were put forward as candidate loci potentially under divergent selection (Beaumont 2005; Yatabe et al. 2007). For most loci, divergence between H. annuus and H. petiolaris was also much weaker than between either of these two species and the closely related allopatric Helianthus argophyllus (Yatabe et al. 2007). Furthermore, increased divergence near chromosome breakpoints extended only as far as 5 cM. These results suggest that even strong and genetically complex isolating barriers are unlikely to prevent widespread introgression.
It is important to note that the population genomic work by Yatabe et al. (2007) was based on contrasts of populations that lacked early generation hybrids, as opposed to the hybrid zone studies outlined earlier. LD in populations of these self-incompatible outcrossers will be much lower than that in hybrid zones (Rieseberg & Buerkle 2001; Flint-Garcia et al. 2003). Theory predicts that the effect of linkage on gene dispersal will decrease rapidly over generations, as neutral or advantageous alleles are recombined into a foreign genetic background (Barton & Hewitt 1985; Martinsen et al. 2001). At this stage, ‘hitch-hiking’ will be confined to short genomic distances and the fate of introgressed alleles will depend mainly on their own fitness effects (Martinsen et al. 2001; Yatabe et al. 2007), as also predicted by the genic view of the process of speciation (Wu 2001a).
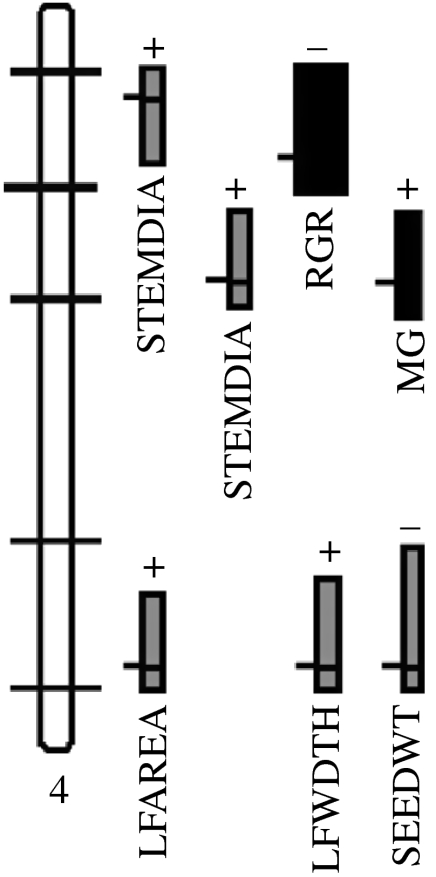
Studies of barriers to gene flow in Helianthus increasingly aim at addressing both barriers between and within species. A large QTL study on the genetics of species differences in sunflowers facilitated the simultaneous mapping of interspecific morphological, physiological and life-history QTL segregating between H. annuus and H. petiolaris, and intraspecific QTL segregating between the different H. petiolaris backcross parents used to form the mapping cross (figure 1; Lexer et al. 2005a). A comparison of QTL effect sizes indicated that intraspecific QTL generally had smaller effects than interspecific QTL, consistent with the prediction that larger QTL are more likely to spread to fixation across a subdivided species (Rieseberg & Burke 2001; Morjan & Rieseberg 2004). By contrast, smaller QTL may be trapped in local populations and are thus more likely to be visible as allelic substitutions at the within-species level (figure 1; Lexer et al. 2005a).
Figure 1.
QTL analysis in a complex interspecific pedigree facilitates the simultaneous mapping of inter- and intraspecific QTL potentially involved in ecological divergence in Helianthus. Linkage group 4 of Helianthus is shown as an example. Marker positions are indicated by horizontal bars along the linkage group. Shown as blocks to the right of the group are interspecific QTL positions and support limits with interspecific effect sizes indicated by the horizontal width of each block. All interspecific QTL positions were also tested for the presence of intraspecific QTL, and positions at which intraspecific QTL polymorphism was found are shown in black. The average effect size of intraspecific QTL (not shown) was less than one-third that of interspecific QTL. STEMDIA, stem diameter; RGR, relative growth rate; MG, magnesium uptake; LFAREA, leaf area; LFWDTH, leaf width; SEEDWT, seed weight. Modified from Lexer et al. (2005a).
Studies of selective sweeps and local adaptation within wild sunflower species are informative in this context as well (Kane & Rieseberg 2007). This study used 128 EST-derived microsatellite loci to reveal the geographical extent of selective sweeps in H. annuus sampled from typical habitat of this species in the plains of central/western North America and from distinct arid desert and brackish salt marsh habitats. Spatially separated populations often diverged at some loci while evolving in concert at other loci, and selective sweeps associated with local adaptation (e.g. salt tolerance) were detected at three different geographical scales (Kane & Rieseberg 2007). Studies such as this can contribute to our understanding of both cohesion of sets of populations within species and the origin of barriers between species.
5. Ecotype formation, hybridization and introgression in Silene
The genus Silene encompasses approximately 700 species that display great morphological diversity and have colonized a wide range of habitats, ranging from the sea coast to heavy metal-contaminated soils, and high alpine tundra vegetation. The genus includes species that are either hermaphroditic, gynodioecious (hermaphrodites and females coexist) or dioecious (males and females coexist; Desfeux et al. 1996).
A common and widespread European herb with a gynodioecious mating system is Silene vulgaris, the bladder campion. It is well known for its ability to colonize heavy metal-rich soils, such as mines or serpentine soils. Bratteler and co-workers have investigated the genetic architecture of morphological and physiological traits that differentiate two S. vulgaris ecotypes, one that grows on serpentine soil (naturally heavy metal-rich soil), and one on non-serpentine soil. A genetic linkage map based on amplified fragment length polymorphism markers (Bratteler et al. 2006b) provided the foundation for the identification of QTL for morphological and physiological traits potentially involved in serpentine adaptation (Bratteler et al. 2006a,c). The observed genetic architecture suggests that traits potentially involved in habitat adaptation are controlled by few genes of major effect and have evolved under divergent selection (Bratteler et al. 2006c).
The studies on S. vulgaris ecotypes also revealed strong phenotypic correlations between traits, and QTL underlying this variation displayed extensive linkage. Clustering of QTL for different traits can be due to either pleiotropy or linkage and both can either constrain or facilitate adaptation, depending on whether trait correlations have positive or negative fitness effects (Bratteler et al. 2006c). Based on the observed combination of QTL directions, it was suggested that further ecotype divergence may be genetically constrained and may thus slow down or prevent further divergence between ecotypes.
Silene vulgaris is related to a clade of dioecious Silene species, of which the white campion, Silene latifolia Poiret (syn. Melandrium album Garcke, syn. Melandrium pratense Roehl.), is arguably the best known. The sex of individual plants is genetically determined by sex chromosomes. Males are heterogametic (XY) and females are homogametic (XX; Vyskot & Hobza 2004). Silene latifolia and the closely related Silene dioica represent an interesting model to study the process of interspecific differentiation at the genome level because they are able to hybridize when they come into contact, yet maintain morphological integrity at the boundaries of such hybrid zones (Minder et al. 2007). The two species differ most prominently in floral traits and habitat preferences (Karrenberg & Favre 2008). Hybrid zones between the two species were found to be dominated by late-generation backcross hybrids, whereas intermediate, presumably early generation hybrids, are rare. This indicates that gene flow is likely to occur between the two species, with the hybrids acting as bridges to gene flow (Minder et al. 2007). A population genomic analysis of differentiation and identification of outlier loci between S. latifolia and S. dioica revealed that the species boundary has been shaped by a combination of introgression and selection (Minder & Widmer 2008). Introgression occurs as a consequence of hybridization upon secondary contact, but species differences appear to be maintained outside of hybrid zones, presumably as a consequence of selection acting on phenotypic traits involved in pollinator or habitat adaptation. These results challenge the idea of whole-genome isolation (Mayr 2001; Wu 2001a), and support the idea that genomes can be porous and that species differentiation has a genic basis (Minder & Widmer 2008).
6. Populus as a ‘model tree’ for studying the ecological and evolutionary genomics of species isolation
Two parallel developments call for the inclusion of the wind-pollinated, primarily dioecious tree genus Populus in this review: (i) it is among the few tree genera with three-generation pedigrees available, and these resources are increasingly being used to map species isolation factors and barriers to introgression (Frewen et al. 2000; Yin et al. 2004) and (ii) closely related species of Populus are currently being used to adopt the concepts of ‘admixture mapping’ (Chakraborty & Weiss 1988) from human medical genetics to plant speciation genetics, and the first results of this work are now becoming available (Lexer et al. 2007). A number of additional factors render Populus an attractive model for speciation genetics: ecologically divergent species often co-occur in sympatry or parapatry and are interfertile (Eckenwalder 1996); widespread synteny suggests that species barriers are genic rather than chromosomal (Cervera et al. 2001); and a complete genomic sequence, genetic maps and many thousands of ESTs are available (Cervera et al. 2001; Yin et al. 2004; Sjodin et al. 2006; Tuskan et al. 2006). As a result of these developments, the speciation genetic literature on Populus is growing rapidly, paralleled by only a few other tree genera (e.g. oaks; Muir et al. 2000; Scotti-Saintagne et al. 2004).
Genetic linkage and QTL mapping were used successfully to map traits involved in interspecific differentiation between the two North American species Populus deltoides and Populus trichocarpa, with a QTL analysis of bud set/bud flush being perhaps the most relevant example with respect to ecological differentiation (Frewen et al. 2000). Two candidate genes involved in photoperiod were found to co-localize with bud set and bud flush QTL, thus providing a rare example for the molecular characterization of ecological QTL in trees. The much larger linkage mapping study by Yin et al. (2004) between the same two species revealed large-scale heterospecific segregation distortion. Large contiguous blocks of donor genome on two linkage groups were distorted in a manner consistent with positive selection, thus suggesting a role for genetic correlations in the maintenance of species barriers in Populus in sympatry or parapatry (Yin et al. 2004). Some of the genetic correlations involved in barriers among Populus species may be the result of reduced recombination in specific regions of the genome carrying ecologically relevant genes, such as the peritelomeric region of chromosome XIX of Populus. This region has been shown to carry a large number of NBS–LRR (nucleotide-binding site/leucine-rich repeat)-type resistance genes and it also harbours several features expected for an incipient sex chromosome, such as a gender determination locus, distorted segregation and high levels of haplotype divergence (Yin et al. 2008).
The second approach, admixture mapping (association mapping in recombinant admixed populations), has been highly successful in human medical genetics (Reich et al. 2005; Zhu et al. 2005) but is only beginning to be applied to evolutionary biology. In Populus, first indications for the usefulness of the approach came from genetic studies of the North American cottonwoods Populus fremontii and P. trichocarpa (Keim et al. 1989; Martinsen et al. 2001). A molecular marker study of a natural hybrid zone between these species indicated great inter-locus variation in the geographical extent of introgression (Martinsen et al. 2001). Genomic analyses of hybrid zones are currently taken further in the ecologically divergent hybridizing European species Populus alba (white poplar) and Populus tremula (European aspen; figure 2; Lexer et al. 2005b, 2007). Hybrid zones of these two species contain a high proportion of recombinant backcrosses (Lexer et al. 2005b), and admixture LD decays quickly with successive backcross generations (Lexer et al. 2007). Although both clonality and assortative mating appear to contribute to fine-scale spatial patterns in hybrid zones of P. alba and P. tremula, these potential sources of LD can be accounted for by widening the spatial level of sampling (van Loo et al. 2008). Thus, interspecific Populus hybrid zones can serve as natural laboratories for the identification of candidate loci involved in species isolation (Lexer et al. 2007; Joseph & Lexer 2008). An important notion is that in Populus, candidate gene loci can be subjected to rigorous tests of gene function, because the necessary tools for this type of functional genomics work are available (Brunner et al. 2004; Sjodin et al. 2006; Tuskan et al. 2006).
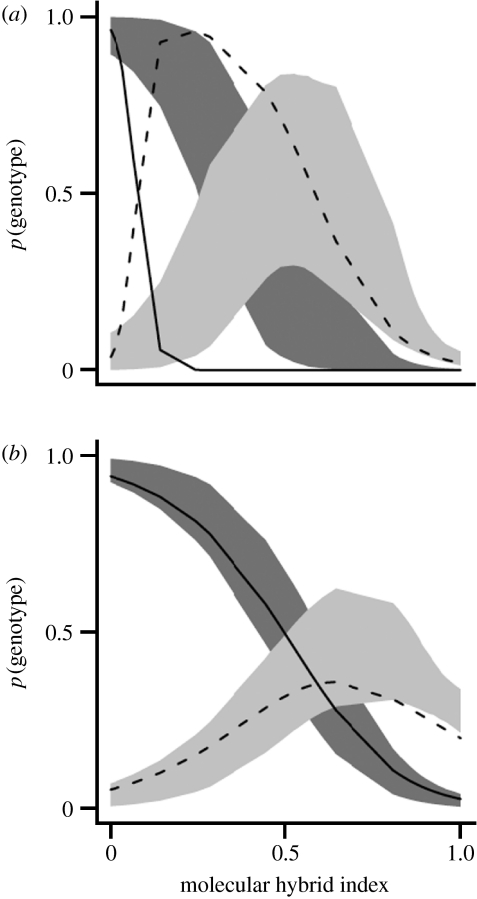
Figure 2.
Introgression of two codominant DNA-based markers (a, O220; b, O149) across a Central European hybrid zone of P. alba and P. tremula, relative to neutral expectations. X-axes: molecular hybrid index as an estimate of average genome-wide introgression. Y-axes: genotype frequencies for each locus. The introgression of the predominant P. tremula allelic class (black solid lines, homozygotes; black dashed lines, heterozygotes) from individuals with low hybrid index (P. tremula) to those with high hybrid index (P. alba) varies among loci. Grey regions indicate the 95% confidence envelope for logistic regressions fit to simulated data under the assumption of neutral introgression for each locus (dark grey region, homozygotes; light grey region, heterozygotes). The slope of the regression for homozygotes but not for heterozygotes departs significantly from neutrality for the marker in (a). Modified from Lexer et al. (2007).
7. Synthesis and emerging questions for future research
The genic view of the process of speciation can provide a framework for rigorous tests of the mechanistic underpinnings of and evolutionary forces operating during speciation (Rieseberg & Burke 2001; Coyne & Orr 2004; Noor & Feder 2006). Studies with a focus on speciation and differential adaptation were considered jointly in this review, since adaptation may result in speciation if there is pleiotropy or linkage between loci involved in reproductive isolation and loci under divergent selection (Felsenstein 1981; Via 2001). Note that, in principle, adaptation may be sufficient to achieve isolation if extrinsic postzygotic isolation is strong (e.g. late-generation hybrids are outperformed by F1 genotypes in hybrid zones among Rhododendron species in the absence of strong intrinsic postzygotic barriers, Milne et al. 2003).
In plants, most research programmes currently use genetic mapping of TRDL and QTL and population genomic scans to study the genetic architecture of porous genomes (table 1). The reviewed studies may not represent a random draw of species from nature, but they may inspire related work on different groups of taxa so that generalizations can be drawn with ever-increasing precision. We suspect that new developments in molecular systematics (e.g. DNA-barcoding; Chase et al. 2005) will increasingly facilitate both the identification of species pairs for in-depth genetic study and the interpretation of speciation genetic data in a comparative framework.
Our present review of recent progress with a focus on five plant genera suggests that the genetic architecture of species isolation differs greatly between taxa. Nevertheless, a direct involvement of selection in speciation (as opposed to divergence due to drift alone) can no longer be ignored, and future work can now aim at understanding functional aspects and ecological impacts of the selected traits and genes. A ‘genic view of speciation’ is particularly useful for this purpose, because the explicit consideration of recombination and selection (‘locus-specific effects’) potentially allows evolutionary biologists to disentangle the different mechanisms operating during divergence. The following topics emerge as being of special interest for future research on the nature of genic barriers to gene flow in plants.
(a) The role of intraspecific variation in speciation
Intraspecific variation is an often neglected aspect in plant speciation genetics. Only three of the reviewed studies addressed it explicitly, namely the genomic analysis of replicate hybrid zones in Helianthus by Buerkle & Rieseberg (2001), the simultaneous inter- and intraspecific QTL analyses of ecological species differences in Helianthus by Lexer et al. (2005a,b; figure 1), and the genetic analysis of natural variation for hybrid incompatibility in Mimulus by Sweigart et al. (2007). More speciation genetic studies are needed that explicitly address this topic. This is important to (i) understand the fate of incompatibility alleles within species and ask whether such alleles evolve in an adaptive or neutral fashion at the within-species level (Sweigart et al. 2007), (ii) learn whether intraspecific variation for species isolation is due to polymorphism of isolation factors or rather due to differences in exogenous (ecological) selection pressures in different parts of a species' range (Buerkle & Rieseberg 2001), (iii) address the potential of evolution from standing variation versus evolution from new mutations (Hermission & Pennings 2005) and (iv) improve our understanding of the interactions between species differentiation and species cohesion (Ehrlich & Raven 1969; Morjan & Rieseberg 2004). Species cohesion and species differentiation cannot be viewed in isolation, as both will depend on interactions of the same evolutionary forces, such as gene flow, selection and drift (Ehrlich & Raven 1996; Morjan & Rieseberg 2004). Thus, speciation genetic studies should increasingly integrate analysis of inter- and intraspecific components of variation.
(b) Balancing versus directional selection in population genomic scans
An increasing number of speciation genetic studies use genome scans for species differentiation to characterize porous genomes (Scotti-Saintagne et al. 2004; Savolainen et al. 2006; Yatabe et al. 2007; Minder & Widmer 2008). A shortcoming of most existing genome scans is that they often fail to identify loci under balancing selection, i.e. outlier loci in the lower tail of the distribution of divergence estimates (Beaumont & Balding 2004). This is the case in part because the species pairs tested are typically sympatric and have a history of frequent gene exchange, thus the lower 95% confidence limit of divergence (FST or GST) will be biased towards zero (Scotti-Saintagne et al. 2004; Savolainen et al. 2006; Yatabe et al. 2007; Butlin et al. 2008), which makes it difficult to distinguish between neutral loci and loci under balancing selection (Beaumont & Balding 2004). Consequently, neutral expectations used to identify outliers in the upper tail may be biased. A rare exception is the study of the well-differentiated hybridizing Silene species, S. latifolia and S. dioica (Minder & Widmer 2008); that study was able to identify loci under divergent and loci under balancing selection.
One question of great interest to students of porous genomes is whether most of the genome is permeable to gene flow with occasional low-recombination ‘hotspots of differentiation’, or whether most of the genome is protected from gene flow except for occasional ‘pores’ (Mayr 2001; Wu 2001a,b). Answering this question with population genomic scans will require reliable neutral expectations for outlier detection. As a start, more studies are needed that scan pairs of populations and species with different degrees of relatedness and geographical overlap to ‘calibrate’ the outlier tests currently in use.
(c) The timing of fixation of major- versus minor-effect alleles
The order of fixation of ‘major’- and ‘minor’-effect alleles that differ in their effect on the phenotype during adaptation has sometimes been considered to be clear. Theory predicts an ‘adaptive walk’ that starts with large steps that become smaller as a population nears a fitness optimum (Orr 1998). Recent theoretical work, however, indicates that minor-effect alleles may be fixed before major ones when the selection pressure increases gradually over time or, in other words, if environmental change is slow (Collins et al. 2007; Kopp & Hermisson 2007). In this context, the meaningful estimation of effect sizes (=magnitudes) of QTL involved in adaptation and speciation is of major interest.
As shown by Lexer et al. (2005a,b) in the case of Helianthus and Bouck et al. (2007) for Iris, the magnitudes of QTL effects sometimes differ greatly when estimated as ‘per cent variance explained’ (PVE) in the mapping population, relative to the interspecific phenotypic gap or relative to levels of standing variation. To effectively address the size distribution and timing of factors fixed during adaptation and speciation, QTL studies should try to estimate and interpret the magnitudes of QTL effects in all three ways in parallel whenever possible; for example, the estimates of standing variation and interspecific gaps could be obtained from population samples used to characterize quantitative traits prior to genetic mapping. Consistently small interspecific QTL, as observed for floral traits involved in mating system divergence in Mimulus (Fishman et al. 2002), may then indicate an important role for the preferential fixation of minor-effect QTL. This may be the case if the adaptive walk is limited by the rate of environmental change (Kopp & Hermisson 2007); for example, the selective environment in which mating system divergence occurred in Mimulus may have changed gradually. This would make sense—mating system divergence (the transition from outcrossing to selfing or vice versa) is known to be affected by complex fitness trade-offs (Fishman et al. 2002) and mixed mating is common in plants (Lande & Schemske 1985). On the contrary, some of the floral traits differentiating Iris species with different pollination syndromes were of large effect regardless whether they were measured in terms of PVE, standing variation or the interspecific gap (Bouck et al. 2007). This lends additional support to saltational changes associated with abrupt shifts in pollination syndromes, as also observed in previous landmark studies of pollinator isolation (Bradshaw et al. 1995; Schemske & Bradshaw 1999; Bradshaw & Schemske 2003). We note that common criteria for the definition of major- and minor-effect QTL are available only for effect size in terms of PVE, but not in terms of standing variation or species difference. Clearly, more empirical and theoretical work is needed to assess the likelihood of fixation of QTL measured on these scales.
(d) The likelihood of adaptive introgression
Several of the reviewed studies touched on the likelihood of adaptive introgression in plants, the QTL analyses of species boundaries in Iris (Martin et al. 2005, 2006) and population genomic studies in Helianthus (Kane & Rieseberg 2007; Yatabe et al. 2007) being excellent examples. The Helianthus studies clearly show that even complex barriers to introgression are permeable to gene flow (Yatabe et al. 2007) and that beneficial alleles at some loci will spread across large distances in outcrossing plants (Kane & Rieseberg 2007), thus contributing to species cohesion (Morjan & Rieseberg 2004). An open question is whether such ‘cohesion genes’ often introgress across species boundaries due to their adaptive value, or whether they are more likely to be contained within species.
Owing to the paucity of genetic studies addressing adaptive introgression, it is too early to say for sure which plant traits are likely to favour this process. Nevertheless, judging from molecular marker data in Iris (reviewed by Arnold et al. 2004) and Populus (van Loo et al. 2008), mixed sexual/asexual reproductive systems appear to favour introgression, possibly by facilitating the persistence of ecologically successful genotypes in the face of meiotic difficulties. Population genomic scans and admixture mapping in natural hybrid zones hold great promise for studying adaptive introgression in situ. This is facilitated by new approaches of detecting locus-specific effects in hybrid zones (figure 2), which allow the detection of both negative and positive departures from neutral introgression (Lexer et al. 2007).
(e) Speciation genes
Identifying the molecular identity and function of genes involved in species barriers in plants will remain an important task for evolutionary geneticists, and the methodologies required to achieve this goal are available (reviews by Feder & Mitchell-Olds (2003) and Noor & Feder (2006)). The QTL and population genomic studies reviewed here represent a starting point for the molecular characterization of loci involved in species isolation. Experimental transplants or natural hybrid zones should allow the estimation of selection coefficients of these genes under realistic conditions. Studies of incipient species or divergent ecotypes will be relevant as they allow the detection of genes directly involved in reproductive isolation, as opposed to isolation factors that evolved post-speciation. It will be interesting to see which functional classes of genes most often contribute to genic barriers in plants, which types of regulatory networks are involved and how these genes interact with other types of isolation factors, e.g. chromosomal rearrangements (Navarro & Barton 2003). These questions can be addressed using the tools of ecological and evolutionary functional genomics (Feder & Mitchell-Olds 2003; Noor & Feder 2006), which are becoming available for several of the plant groups reviewed here. We look forward to seeing the results of this research over the coming years.
Acknowledgments
We thank Vincent Savolainen, Salvatore Cozzolino, Noland Martin, Douglas Schemske, Jay Sobel and two anonymous referees for their helpful comments on the manuscript. C.L.'s work on within-species variation for genomic isolation in European Populus is supported by grant NE/E016731/1 of the British NERC (Natural Environment Research Council) and A.W.'s work on Silene is funded by SNF grants 3100AO-104114 and 3100A0-116455.
Footnotes
One contribution of 12 to a Theme Issue ‘Speciation in plants and animals: pattern and process’.
References
- Anderson E. Wiley; New York, NY: 1949. Introgressive hybridization. [Google Scholar]
- Arias D.M, Rieseberg L.H. Gene flow between cultivated and wild sunflowers. Theor. Appl. Genet. 1994;89:655–660. doi: 10.1007/BF00223700. doi:10.1007/BF00223700 [DOI] [PubMed] [Google Scholar]
- Arnold M.L. Oxford University Press; New York, NY: 1997. Natural hybridisation and evolution. [Google Scholar]
- Arnold M.L, Bouck A.C, Cornman R.S. Verne Grant and Louisiana irises: is there anything new under the sun? New Phytol. 2004;161:143–149. doi:10.1046/j.1469-8137.2003.00856.x [Google Scholar]
- Barton N.H, Hewitt G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. doi:10.1146/annurev.es.16.110185.000553 [Google Scholar]
- Beaumont M.A. Adaptation and speciation: what can FST tell us? Trends Ecol. Evol. 2005;20:435–440. doi: 10.1016/j.tree.2005.05.017. doi:10.1016/j.tree.2005.05.017 [DOI] [PubMed] [Google Scholar]
- Beaumont M.A, Balding D.J. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. doi:10.1111/j.1365-294X.2004.02125.x [DOI] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 2007;8:382–393. doi: 10.1038/nrg2082. doi:10.1038/nrg2082 [DOI] [PubMed] [Google Scholar]
- Bouck A.C, Peeler R, Arnold M.L, Wessler S.R. Genetic mapping of species boundaries in Louisiana irises using IRRE retrotransposon display markers. Genetics. 2005;171:1289–1303. doi: 10.1534/genetics.105.044552. doi:10.1534/genetics.105.044552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck A.C, Wessler S.R, Arnold M.L. QTL analysis of floral traits in Louisiana iris hybrids. Evolution. 2007;61:2308–2319. doi: 10.1111/j.1558-5646.2007.00214.x. doi:10.1111/j.1558-5646.2007.00214.x [DOI] [PubMed] [Google Scholar]
- Bradshaw H.D, Schemske D.W. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. doi:10.1038/nature02106 [DOI] [PubMed] [Google Scholar]
- Bradshaw H.D, Wilbert S.M, Otto K.G, Schemske D.W. Genetic mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus) Nature. 1995;376:762–765. doi:10.1038/376762a0 [Google Scholar]
- Bradshaw H.D, Otto K.G, Frewen B.E, McKay J.K, Schemske D.W. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus) Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratteler M, Baltisberger M, Widmer A. QTL analysis of intraspecific differences between two Silene vulgatis ecotypes. Ann. Bot. 2006a;98:411–419. doi: 10.1093/aob/mcl113. doi:10.1093/aob/mcl113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratteler M, Lexer C, Widmer A. A genetic linkage map of Silene vulgaris based on AFLP markers. Genome. 2006b;49:320–327. doi: 10.1139/g05-114. doi:10.1139/G05-114 [DOI] [PubMed] [Google Scholar]
- Bratteler M, Lexer C, Widmer A. Genetic architecture of traits associated with serpentine adaptation of Silene vulgaris. J. Evol. Biol. 2006c;19:1149–1156. doi: 10.1111/j.1420-9101.2006.01090.x. doi:10.1111/j.1420-9101.2006.01090.x [DOI] [PubMed] [Google Scholar]
- Brunner A.M, Busov V.B, Strauss S.H. Poplar genome sequence: functional genomics in an ecologically dominant species. Trends Plant Sci. 2004;9:49–56. doi: 10.1016/j.tplants.2003.11.006. doi:10.1016/j.tplants.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Buerkle C.A, Rieseberg L.H. Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution. 2001;55:684–691. doi: 10.1554/0014-3820(2001)055[0684:livfgi]2.0.co;2. doi:10.1554/0014-3820(2001)055[0684:LIVFGI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Butlin R.K, Galindo J, Grahame J.W. Sympatric, parapatric or allopatric: the most important way to classify speciation? Phil. Trans. R. Soc. B. 2008;363:2997–3007. doi: 10.1098/rstb.2008.0076. doi:10.1098/rstb.2008.0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case A.L, Willis J.H. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically-restricted mitochondrial rearrangement. Evolution. 2008;62:1026–1039. doi: 10.1111/j.1558-5646.2008.00360.x. doi:10.1111/j.1558-5646.2008.00360.x [DOI] [PubMed] [Google Scholar]
- Cervera M.T, Storme V, Ivens B, Gusmao J, Liu B.H, Hostyn V, Slycken J.V, Montagu M.V, Boerjan W. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra, and P. trichocarpa) based on AFLP and microsatellite markers. Genetics. 2001;158:787–809. doi: 10.1093/genetics/158.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Weiss K.M. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl Acad. Sci. USA. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. doi:10.1073/pnas.85.23.9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase C.D. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. doi:10.1016/j.tig.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Chase M.W, Salamin N, Wilkinson M, Dunwell J.M, Kesanakurthi R.P, Haidar N, Savolainen V. Land plants and DNA barcodes: short-term and long-term goals. Phil. Trans. R. Soc. B. 2005;360:1889–1895. doi: 10.1098/rstb.2005.1720. doi:10.1098/rstb.2005.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P, Macnair M.R. Complementary lethal factors in two North-American populations of the yellow monkey flower. J. Hered. 1984;75:510–511. [Google Scholar]
- Collins S, de Meaux J, Acquisti C. Adaptive walks toward a moving optimum. Genetics. 2007;176:1089–1099. doi: 10.1534/genetics.107.072926. doi:10.1534/genetics.107.072926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. doi: 10.1038/nrg1711. doi:10.1038/nrg1711 [DOI] [PubMed] [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer Associates; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Cozzolino S, Scopece G. Specificity in pollination and consequences for postmating reproductive isolation in deceptive Mediterranean orchids. Phil. Trans. R. Soc. B. 2008;363:3037–3046. doi: 10.1098/rstb.2008.0079. doi:10.1098/rstb.2008.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfeux C, Maurice S, Henry J.-P, Lejeune B, Gouyon P.-H. Evolution of reproductive systems in the genus Silene. Proc. R. Soc. B. 1996;263:409–414. doi: 10.1098/rspb.1996.0062. doi:10.1098/rspb.1996.0062 [DOI] [PubMed] [Google Scholar]
- Eckenwalder J.E. Systematics and evolution of Populus. In: Stettler R, Bradshaw H, Heilman P, Hinckley T, editors. Biology of Populus, and its implications for management and conservation. NRC Research Press; Ottawa, Canada: 1996. pp. 7–30. [Google Scholar]
- Ehrlich P.R, Raven P.H. Differentiation of populations. Science. 1969;165:1228–1232. doi: 10.1126/science.165.3899.1228. doi:10.1126/science.165.3899.1228 [DOI] [PubMed] [Google Scholar]
- Feder M.E, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nat. Rev. Genet. 2003;4:651–657. doi: 10.1038/nrg1128. doi:10.1038/nrg1128 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution. 1981;35:124–138. doi: 10.1111/j.1558-5646.1981.tb04864.x. doi:10.2307/2407946 [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis J.H. Evidence for Dobzhansky–Muller incompatibilites contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution. 2001;55:1932–1942. doi: 10.1111/j.0014-3820.2001.tb01311.x. doi:10.1111/j.0014-3820.2001.tb01311.x [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis J.H. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution. 2006;60:1372–1381. doi: 10.1554/05-708.1. doi:10.1111/j.0014-3820.2006.tb01216.x [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly A.J, Morgan E, Willis J.H. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics. 2001;159:1701–1716. doi: 10.1093/genetics/159.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Kelly A.J, Willis J.H. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution. 2002;56:2138–2155. doi: 10.1111/j.0014-3820.2002.tb00139.x. doi:10.1111/j.0014-3820.2002.tb00139.x [DOI] [PubMed] [Google Scholar]
- Flint-Garcia S.A, Thornsberry J.M, Buckler E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. doi:10.1146/annurev.arplant.54.031902.134907 [DOI] [PubMed] [Google Scholar]
- Frewen B.E, Chen T.H.H, Howe G.T, Davis J, Rohde A, Boerjan W, Bradshaw H.D. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics. 2000;154:837–845. doi: 10.1093/genetics/154.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K, Buerkle C.A, Whitton J, Rieseberg L.H. Epistasis in wild sunflower hybrid zones. In: Wolf J, Brodie E III, Wade M, editors. Epistasis and the evolutionary process. Oxford University Press; New York, NY: 2000. pp. 264–279. [Google Scholar]
- Gavrilets S, Vose A. Dynamic patterns of adaptive radiation. Proc. Natl Acad. Sci. USA. 2005;102:18 040–18 045. doi: 10.1073/pnas.0506330102. doi:10.1073/pnas.0506330102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.C, Basten C.J, Willis J.H. Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics. 2006;172:1829–1844. doi: 10.1534/genetics.105.051227. doi:10.1534/genetics.105.051227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty M.J, Barker G.L, Brennan A.C, Edwards K.J, Abbott R.J, Hiscock S.J. Changes to gene expression associated with hybrid speciation in plants: further insights from transcriptomic studies in Senecio. Phil. Trans. R. Soc. B. 2008;363:3055–3069. doi: 10.1098/rstb.2008.0080. doi:10.1098/rstb.2008.0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings P.S. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. doi:10.1534/genetics.104.036947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah M.E, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell'Olivo A, Arnold M.L, Kuhlemeier C. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell. 2007;19:779–790. doi: 10.1105/tpc.106.048694. doi:10.1105/tpc.106.048694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J. A. & Lexer, C. 2008 A set of novel DNA polymorphisms within candidate genes potentially involved in ecological divergence between Populus alba and P. tremula, two hybridising European forest trees. Mol. Ecol. Res.8, 188–192. (doi:10.1111/j.1471-8286.2007.01919.x) [DOI] [PubMed]
- Kane N.C, Rieseberg L.H. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics. 2007;175:1832–1834. doi: 10.1534/genetics.106.067728. doi:10.1534/genetics.106.067728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrenberg, S. & Favre, A. 2008 Habitat overlap in the hybrizing species pair Silene dioica and S. latifolia Evolution62, 763–773. [DOI] [PubMed]
- Keim P, Paige K.N, Whitham T.G, Lark K.G. Genetic analysis of interspecific hybrid swarms of Populus: occurrence of unidirectional introgression. Genetics. 1989;123:557–565. doi: 10.1093/genetics/123.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-C, Rieseberg L.H. Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression. Genetics. 1999;153:965–977. doi: 10.1093/genetics/153.2.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M, Hermisson J. Adaptation of a quantitative trait to a moving optimum. Genetics. 2007;176:715–719. doi: 10.1534/genetics.106.067215. doi:10.1534/genetics.106.067215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Schemske D.W. The evolution of self-fertilization and inbreeding depression in plants. 1. Genetic models. Evolution. 1985;39:24–40. doi: 10.1111/j.1558-5646.1985.tb04077.x. doi:10.2307/2408514 [DOI] [PubMed] [Google Scholar]
- Levin D.A. The cytoplasmic factor in plant speciation. Syst. Bot. 2003;28:5–11. [Google Scholar]
- Lexer C, Lai Z, Rieseberg L.H. Candidate gene polymorphisms associated with salt tolerance in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. New Phytol. 2004;161:225–233. doi: 10.1046/j.1469-8137.2003.00925.x. doi:10.1046/j.1469-8137.2003.00925.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Rosenthal D.M, Raymond O, Donovan L.A, Rieseberg L.H. Genetics of species differences in the wild annual sunflowers, Helianthus annus and H. petiolaris. Genetics. 2005a;169:2225–2239. doi: 10.1534/genetics.104.031195. doi:10.1534/genetics.104.031195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Fay M.F, Joseph J.A, Nica M.-S, Heinze B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen): the role of ecology and life history in gene introgression. Mol. Ecol. 2005b;14:1045–1057. doi: 10.1111/j.1365-294X.2005.02469.x. doi:10.1111/j.1365-294X.2005.02469.x [DOI] [PubMed] [Google Scholar]
- Lexer C, Buerkle A, Joseph J.A, Heinze B, Fay M.F. Admixture in European Populus hybrid zones makes feasible the mapping of loci that contribute to reproductive isolation and trait differences. Heredity. 2007;98:74–84. doi: 10.1038/sj.hdy.6800898. doi:10.1038/sj.hdy.6800898 [DOI] [PubMed] [Google Scholar]
- Lin J.Z, Ritland K. Quantitative trait loci differentiating the outbreeding Mimulus guttatus from the inbreeding M. platycalyx. Genetics. 1997;146:1115–1121. doi: 10.1093/genetics/146.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D.B, Modliszewski J.L, Wright K.M, Wu C.A, Willis J.H. The strength and genetic basis of reproductive isolating barriers in flowering plants. Phil. Trans. R. Soc. B. 2008;363:3009–3021. doi: 10.1098/rstb.2008.0064. doi:10.1098/rstb.2008.0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England P.R, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 2003;4:981–994. doi: 10.1038/nrg1226. doi:10.1038/nrg1226 [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Sinauer Associates; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- Lysak M.A, Lexer C. Towards the era of comparative evolutionary genomics in Brassicaceae. Plant Syst. Evol. 2006;259:175–198. doi:10.1007/s00606-006-0418-9 [Google Scholar]
- Macnair M.R, Christie P. Reproductive isolation as a pleiotropic effect of copper tolerance in Mimulus guttatus. Heredity. 1983;50:295–302. doi:10.1038/hdy.1983.31 [Google Scholar]
- Martin N.H, Willis J.H. Ecological divergence associated with mating system causes nearly complete reproductive isolation between sympatric Mimulus species. Evolution. 2007;61:68–82. doi: 10.1111/j.1558-5646.2007.00006.x. doi:10.1111/j.1558-5646.2007.00006.x [DOI] [PubMed] [Google Scholar]
- Martin N.H, Bouck A.C, Arnold M.L. Loci affecting long-term hybrid survivorship in Louisiana irises: implications for reproductive isolation and introgression. Evolution. 2005;59:2116–2124. doi:10.1111/j.0014-3820.2005.tb00922.x [PubMed] [Google Scholar]
- Martin N.H, Bouck A.C, Arnold M.L. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics. 2006;172:2481–2489. doi: 10.1534/genetics.105.053538. doi:10.1534/genetics.105.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N.H, Bouck A.C, Arnold M.L. The genetic architecture of reproductive isolation in Louisiana irises: flowering phenology. Genetics. 2007;175:1803–1812. doi: 10.1534/genetics.106.068338. doi:10.1534/genetics.106.068338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, N. H., Sapir, Y. & Arnold, M. L. 2008 The genetic architecture of reproductive isolation in Louisiana irises: pollination syndromes and pollinator preferences. Evolution62, 740–752. (doi:10.1111/j.1558-5646.2008.00342.x) [DOI] [PubMed]
- Martinsen G.D, Whitham T.G, Turek R.J, Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x. doi:10.1111/j.0014-3820.2001.tb00655.x [DOI] [PubMed] [Google Scholar]
- Mayr E. Columbia University Press; New York, NY: 1942. Systematics and the origin of species. [Google Scholar]
- Mayr E. Wu's genic view of speciation. J. Evol. Biol. 2001;14:866–867. doi:10.1046/j.1420-9101.2001.00336.x [Google Scholar]
- Milne R.I, Terzioglu S, Abbott R.J. A hybrid zone dominated by fertile F1s: maintenance of species barriers in Rhododendron. Mol. Ecol. 2003;12:2719–2729. doi: 10.1046/j.1365-294x.2003.01942.x. doi:10.1046/j.1365-294X.2003.01942.x [DOI] [PubMed] [Google Scholar]
- Minder A.M, Widmer A. A population genomic analysis of species boundaries: neutral processes, adaptive divergence and introgression between two hybridizing plant species. Mol. Ecol. 2008;17:1552–1563. doi: 10.1111/j.1365-294X.2008.03709.x. doi:10.1111/j.1365-294X.2008.03709.x [DOI] [PubMed] [Google Scholar]
- Minder A.M, Rothenbuehler C, Widmer A. Genetic structure of hybrid zones between Silene latifolia and Silene dioica (Caryophyllaceae): evidence for introgressive hybridization. Mol. Ecol. 2007;16:2504–2516. doi: 10.1111/j.1365-294X.2007.03292.x. doi:10.1111/j.1365-294X.2007.03292.x [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol. Evol. 2001;16:693–700. doi:10.1016/S0169-5347(01)02291-1 [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. doi:10.1038/nature04878 [DOI] [PubMed] [Google Scholar]
- Morjan C.L, Rieseberg L.H. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol. Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. doi:10.1111/j.1365-294X.2004.02164.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir G, Fleming C.C, Schlotterer C. Taxonomy—species status of hybridizing oaks. Nature. 2000;405:1016. doi: 10.1038/35016640. doi:10.1038/35016640 [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton N.H. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution. 2003;57:447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x. doi:10.1111/j.0014-3820.2003.tb01537.x [DOI] [PubMed] [Google Scholar]
- Noor M.A.F, Feder J.L. Speciation genetics: evolving approaches. Nat. Rev. Genet. 2006;7:851–861. doi: 10.1038/nrg1968. doi:10.1038/nrg1968 [DOI] [PubMed] [Google Scholar]
- Orr H.A. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. doi:10.2307/2411226 [DOI] [PubMed] [Google Scholar]
- Orr H.A, Masly J.P, Presgraves D.C. Speciation genes. Curr. Opin. Genet. Dev. 2004;14:675–679. doi: 10.1016/j.gde.2004.08.009. doi:10.1016/j.gde.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998;29:467–501. doi:10.1146/annurev.ecolsys.29.1.467 [Google Scholar]
- Ramsey J, Bradshaw H.D, Jr, Schemske H.D. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae) Evolution. 2003;57:1520–1534. doi: 10.1111/j.0014-3820.2003.tb00360.x. doi:10.1111/j.0014-3820.2003.tb00360.x [DOI] [PubMed] [Google Scholar]
- Reich D, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat. Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. doi:10.1038/ng1646 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, Burke J.M. The biological reality of species: gene flow, selection, and collective evolution. Taxon. 2001;50:47–67. doi:10.2307/1224511 [Google Scholar]
- Rieseberg L.H, Buerkle C.A. Genetic mapping in hybrid zones. Am. Nat. 2002;159:S37–S49. doi: 10.1086/338371. doi:10.1086/338371 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, Willis J.H. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. doi:10.1126/science.1137729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H, Kim M.J, Seiler G.J. Introgression between the cultivated sunflower and a sympatric wild relative, Helianthus petiolaris (Asteraceae) Int. J. Plant Sci. 1999a;160:102–108. doi:10.1086/314107 [Google Scholar]
- Rieseberg L.H, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999b;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. doi:10.1126/science.1086949 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, Wood T.E, Baack E.J. The nature of plant species. Nature. 2006;440:524–527. doi: 10.1038/nature04402. doi:10.1038/nature04402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen V, et al. Sympatric speciation in palms on an oceanic island. Nature. 2006;441:210–213. doi: 10.1038/nature04566. doi:10.1038/nature04566 [DOI] [PubMed] [Google Scholar]
- Schemske D.W, Bradshaw H.D. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus) Proc. Natl Acad. Sci. USA. 1999;96:11 910–11 915. doi: 10.1073/pnas.96.21.11910. doi:10.1073/pnas.96.21.11910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, Goicoechea P.G, Barreneche T, Bodenes K, Burg K, Kremer A. Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. and Q. petraea (Matt.) Liebl.] Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. doi:10.1534/genetics.104.026849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodin A, Bylesjo M, Skogstrom O, Eriksson D, Nilsson P, Ryden P, Jansson S, Karlsson J. UPSC-BASE—Populus transcriptomics online. Plant J. 2006;48:806–817. doi: 10.1111/j.1365-313X.2006.02920.x. doi:10.1111/j.1365-313X.2006.02920.x [DOI] [PubMed] [Google Scholar]
- Soltis D.E, Soltis P.S, Tate J.A. Advances in the study of polyploidy since plant speciation. New Phytol. 2004;161:173–191. doi:10.1046/j.1469-8137.2003.00948.x [Google Scholar]
- Sweigart A.L, Fishman L, Willis J.H. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics. 2006;172:2465–2479. doi: 10.1534/genetics.105.053686. doi:10.1534/genetics.105.053686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A.L, Mason A.R, Willis J.H. Natural variation for a hybrid incompatibility between two species of Mimulus. Evolution. 2007;61:141–151. doi: 10.1111/j.1558-5646.2007.00011.x. doi:10.1111/j.1558-5646.2007.00011.x [DOI] [PubMed] [Google Scholar]
- Tuskan G.A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. doi:10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- van Loo M, Joseph J.A, Heinze B, Fay M.F, Lexer C. Clonality and spatial genetic structure in Populus×canescens and its sympatric backcross parent P. alba in a Central European hybrid zone. New Phytol. 2008;177:506–516. doi: 10.1111/j.1469-8137.2007.02266.x. [DOI] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. doi:10.1016/S0169-5347(01)02188-7 [DOI] [PubMed] [Google Scholar]
- Vyskot B, Hobza R. Gender in plants: sex chromosomes are emerging from the fog. Trends Genet. 2004;20:432–438. doi: 10.1016/j.tig.2004.06.006. doi:10.1016/j.tig.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Whitney K.D, Randell R.A, Rieseberg L.H. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am. Nat. 2006;167:794–807. doi: 10.1086/504606. doi:10.1086/504606 [DOI] [PubMed] [Google Scholar]
- Wu C.-I. The genic view of the process of speciation. J. Evol. Biol. 2001a;14:851–865. doi:10.1046/j.1420-9101.2001.00335.x [Google Scholar]
- Wu C.-I. Genes and speciation. J. Evol. Biol. 2001b;14:889–891. doi:10.1046/j.1420-9101.2001.00351.x [Google Scholar]
- Wu C.A, Lowry D.B, Cooley A.M, Wright K.M, Lee Y.W, Willis J.H. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity. 2008;100:220–230. doi: 10.1038/sj.hdy.6801018. doi:10.1038/sj.hdy.6801018 [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Kane N.C, Scotti-Saintagne C, Rieseberg L.H. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. doi:10.1534/genetics.106.064469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T.M, DiFazio S.P, Gunter L.E, Riemenschneider D, Tuskan G.A. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor. Appl. Genet. 2004;109:451–463. doi: 10.1007/s00122-004-1653-5. doi:10.1007/s00122-004-1653-5 [DOI] [PubMed] [Google Scholar]
- Yin T, et al. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 2008;18:422–430. doi: 10.1101/gr.7076308. doi:10.1101/gr.7076308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat. Genet. 2005;37:177–181. doi: 10.1038/ng1510. doi:10.1038/ng1510 [DOI] [PubMed] [Google Scholar]