Abstract
Interspecific hybridization is an important mechanism of speciation in higher plants. In flowering plants, hybrid speciation is usually associated with polyploidy (allopolyploidy), but hybrid speciation without genome duplication (homoploid hybrid speciation) is also possible, although it is more difficult to detect. The combination of divergent genomes within a hybrid can result in profound changes to both genome and transcriptome. Recent transcriptomic studies of wild and resynthesized homoploid and allopolyploid hybrids have revealed widespread changes to gene expression in hybrids relative to expression levels in their parents. Many of these changes to gene expression are ‘non-additive’, i.e. not simply the sum of the combined expression levels of parental genes. Some gene expression changes are far outside the range of gene expression in either parent, and can therefore be viewed as ‘transgressive’. Such profound changes to gene expression may enable new hybrids to survive in novel habitats not accessible to their parent species. Here, we give a brief overview of hybrid speciation in plants, with an emphasis on genomic change, before focusing discussion on findings from recent transcriptomic studies. We then discuss our current work on gene expression change associated with hybrid speciation in the genus Senecio (ragworts and groundsels) focusing on the findings from a reanalysis of gene expression data obtained from recent microarray studies of wild and resynthesized allopolyploid Senecio cambrensis. These data, showing extensive non-additive and transgressive gene expression changes in Senecio hybrids, are discussed in the light of findings from other model systems, and in the context of the potential importance of gene expression change to hybrid speciation in plants.
Keywords: hybrid speciation, homoploid, allopolyploid, genome, transcriptome, gene expression
1. Introduction
Natural hybridization is widespread among flowering plants and, when combined with genome duplication (allopolyploidy), is recognized as one of the most important mechanisms of plant speciation (Grant 1981). The union of two distinct genomes within a new hybrid individual can provide a source of phenotypic novelty upon which natural selection can act to facilitate the establishment of a new species (Rieseberg 1997; Rieseberg & Willis 2007). In plants, hybrid speciation is most frequently associated with polyploidy (allopolyploidy), but hybrid speciation where new hybrid species share the same ploidy as their parents (homoploid hybrid speciation) has also been described. While homoploid hybrid speciation is generally regarded as rare, allopolyploidy is common and may be responsible for the origins of most plant species believed to be recent or ancient polyploids (Tate et al. 2005). Well-known examples of recent allopolyploid speciation (within the last 100 years) include Spartina anglica and several species in the genera Tragopogon and Senecio. Other allopolyploid species of interest are of older origin (ca 100 000 years ago), such as Nicotiana and Arabidopsis as well as numerous important commercial crop plants, such as wheat, maize, cotton and canola (reviewed in Rieseberg 1997; Wendel 2000; Hegarty & Hiscock 2004, 2008).
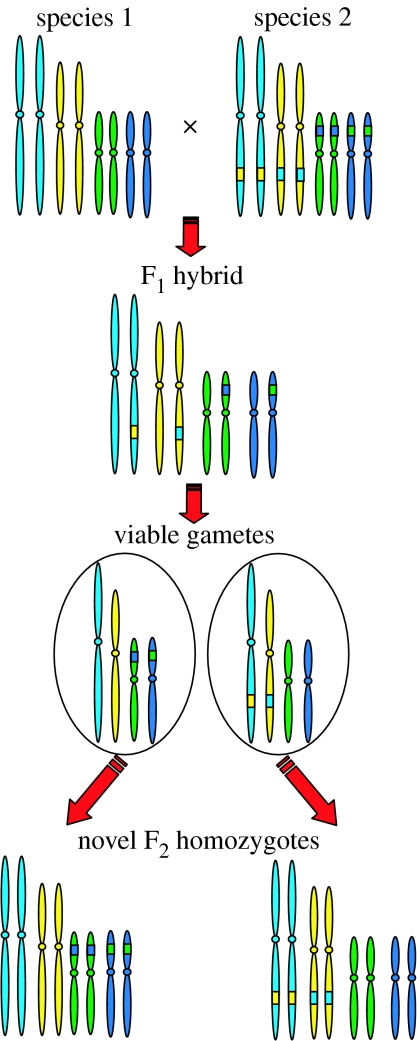
The apparent rarity of hybrid speciation in the absence of polyploidy may be partly due to the difficulty in identifying homoploid hybrids, particularly those of ancient origin (Ferguson & Sang 2001; see also Jiggins et al. 2008), but is also probably due to the difficulties of establishment faced by hybrids, and homoploid hybrids in particular. Even if the two parental species are similar enough to permit hybridization, genomic similarity may not be sufficient to ensure correct pairing of their chromosomes during meiosis. If the parental species differ by two chromosomal rearrangements, the resulting hybrid will be heterozygous for these rearrangements and thus partially sterile, because 75% of its gametes will be unbalanced and inviable due to deletion/insertion events. Half of the remaining viable gametes will recover parental chromosome structures, while the other half will possess recombinant karyotypes (Rieseberg 1997; figure 1). Should inbreeding occur in the hybrid, a small number of F2 individuals will be produced, which possess novel karyotypes. These offspring will be fertile among themselves and at least partially resistant to introgression with the parental species, giving those individuals and their progeny a chance to establish without parental introgression. This was proposed by Grant (1981) as a possible means by which hybrids could establish as reproductively isolated species in their own right—‘recombinational speciation’ (figure 1). It is clear that chromosomal incompatibilities represent a major barrier to hybrid fertility—a barrier that will pose less of an obstacle in allopolyploids but must still be taken into account.
Figure 1.
Recombinational speciation as a consequence of hybridization. A simple model for recombinational speciation sensu Grant (1981) is presented, in which two parental species with the same diploid chromosome number differ by two reciprocal translocations. The F1 hybrid is heterozygous for these rearrangements, and thus 75% of the possible gametic combinations will be inviable due to deletion/insertion events (not shown). Half of the remaining gametes (not shown) will recover parental chromosome combinations, while the remaining half (shown) will possess recombinant karyotypes. If selfed, a small number of F2 individuals will possess novel karyotypes. These will be fertile, but only partly interfertile with either or both of the progenitor species (adapted with permission from Hegarty & Hiscock 2004).
Polyploidization, leading to a duplication of each parental genome immediately ensures that each chromosome has an identical partner with which to pair with at meiosis. Many allopolyploids are therefore expected to have arisen from initially sterile hybrids with unbalanced chromosome numbers or from sterile or partially fertile homoploid hybrids. Indeed, a recent study of the parents of allopolyploid and homoploid hybrid taxa found more than twice the level of genetic divergence between the parents of allopolyploid taxa than that was observed in the progenitors of homoploid hybrids (Chapman & Burke 2007), suggesting that the incidence of allopolyploidy is linked to the level of structural difference between the parental chromosomes. Given this finding, the timing of the subsequent genome duplication event that restores a balanced karyotype is critical. Initially sterile hybrids, such as Spartina x townsendii, which gave rise to S. anglica by polyploidization, may be able to survive by asexual vegetative reproduction until a chance genome duplication event, but other hybrids will be lost unless genome duplication follows rapidly after hybrid formation or is coupled to it.
Genome duplication is most frequently brought about by the union of unreduced gametes that are estimated to arise at a frequency of approximately 0.1–2% in natural populations (Ramsey 2007). However, interspecific F1 hybrids have been found to possess substantially higher rates of unreduced gamete formation (Grant 1952; Ramsey & Schemske 2002). Indeed, our own observations of the largely sterile triploid hybrid Senecio x baxteri, which forms as a hybrid of diploid Senecio squalidus and tetraploid Senecio vulgaris, reveal that these hybrids do regularly produce a few viable seeds (less than one seed per capitulum, compared with 50–100 seeds per capitulum in a fully fertile individual; M. J. Hegarty & S. J. Hiscock 2004, unpublished data). These findings are consistent with those of Ingram (1978), who showed that offspring arising from the occasional production of seed by S. x baxteri displayed a range of ploidy levels from diploid to hexaploid. This suggests that production of fertile allopolyploids may be relatively common in wild populations once initial hybrids have been formed. Unreduced gametes can also result in the coupling of hybridization and polyploidization if, for example, an unreduced pollen grain from a diploid species fertilizes a reduced tetraploid egg to create a novel allotetraploid hybrid. This form of allopolyploidization was most probably responsible for the formation of tetraploid Arabidopsis suecica via fertilization of a normally reduced egg from tetraploid Arabidopsis arenosa by an unreduced pollen grain from diploid Arabidopsis thaliana (Comai et al. 2003).
2. Genomic consequences of hybridization
In hybrids, homoploid as well as allopolyploid, numerous different genomic changes have been described as a result of the union of divergent parental genomes (reviewed in Comai et al. 2003; Hegarty & Hiscock 2004, 2008). Rapid chromosomal rearrangements have been observed in both homoploid and allopolyploid hybrid systems (Rieseberg et al. 1996; Feldman et al. 1997; Ungerer et al. 1998; Shaked et al. 2001; Salmon et al. 2005). In homoploid hybrids, rearrangements that restore fertility by removing incompatibilities between the two parental genomes are selected for at the embryonic level. An ongoing process of recombination between parental linkage blocks then serves to reinforce reproductive isolation from the progenitor taxa and leads to the establishment of the hybrid as a new species (Rieseberg & Willis 2007). An analysis of recombination in the homoploid hybrid sunflower (Helianthus anomalus) showed that newly synthesized hybrids converged on a linkage pattern similar to that of wild hybrids within five generations (Rieseberg et al. 1996), and statistical modelling predicted that the genome would stabilize fully within 10–60 generations (Ungerer et al. 1998). Somewhat surprisingly, similar chromosomal rearrangements occur in allopolyploid hybrids, possibly at even higher rates than those observed in homoploid hybrids (Gale & Devos 1998a). However, closely related parental species may share stretches of collinear gene order in some parts of their respective genomes (Gale & Devos 1998b), making the mispairing of highly homeologous chromosomes during meiosis a distinct possibility (Moore 2002). Rearrangements within one or both parental genomes would reduce this possibility by making the two genomes non-homeologous. A related process, involving loss of non-coding, repetitive DNA regions from one or both parental genomes, has also been observed in newly synthesized allopolyploid lines of wheat (Feldman et al. 1997; Shaked et al. 2001). While the mechanism by which this sequence removal occurs is unknown, the process represents a further mechanism for the differentiation of homeologous chromosomes and insurance of regular meiotic pairing in the hybrid.
Chromosomal rearrangements in allopolyploids could also potentially occur in response to changes in nuclear–cytoplasmic interactions (Leitch & Bennett 1997; Wendel 2000). Because the cytoplasmic genome of a hybrid is derived solely from the female parent, the presence of a foreign nuclear genome can result in nuclear–cytoplasmic incompatibilities. Chromosomal rearrangements may therefore be necessary to restore nuclear–cytoplasmic compatibility (Soltis & Soltis 1999). Indeed, studies by Song et al. (1995) using restriction fragment length polymorphism marker assays in synthetic Brassica polyploids showed that rearrangement events tended to occur primarily in the paternally contributed genome, although this was later contradicted by Gaeta et al. (2007), and so may not be a general rule. One possible cause of the rapid recombination observed in hybrid systems is the activation of transposable elements. McClintock (1984) first predicted that the instability of a new hybrid genome, which she described as ‘genome shock’, might lead to widespread activation of transposons (McClintock 1984). Transposon activity within the hybrid genome can increase the likelihood of chromosome breakage (Weil & Wessler 1993), and may result in other changes to the genome such as sequence amplification/gene duplication (Jin & Bennetzen 1994). This has been proposed as a more likely mechanism for rapid genomic reorganization in allopolyploids than homoploid hybrids (Matzke & Matzke 1998) because transposon insertions into genic regions are less deleterious in polyploids that possess duplicate copies of every gene. Recombination between parental genomes is also a possibility in allopolyploids if these genomes are similar enough to enable bivalent formation, as demonstrated by a study of multiple independent synthetic lines of Brassica napus allotetraploids (Gaeta et al. 2007). This analysis showed few genetic changes in the initial allotetraploids, but subsequent selfed progeny displayed segregation for a number of non-reciprocal translocations between parental homeologues. These translocations occurred at each successive generation, being detected on nearly every chromosome by the fifth selfed generation. Since mispairing of the parental homeologues must occur in order for these recombinations to take place, this process may serve to distinguish the two genomes in a similar manner to that seen in homoploid hybrids and also the loss of non-coding repetitive DNA in wheat allopolyploids (indeed, such translocations may be one mechanism by which this occurs).
3. Changes to gene expression in hybrids
(a) Effects of changes to genomic DNA on gene expression
Given the widespread changes to the genome of a new hybrid, it would be surprising if these genomic alterations did not impact on the levels of gene expression. The combination of two divergent genomes may result in novel assortments of regulatory factors (Riddle & Birchler 2003), a process that has been demonstrated to result in alterations to gene expression in hybrids of species of Drosophila (Hammerle & Ferrus 2003). In addition, chromosomal rearrangements could lead to different parental regulatory elements being brought from cis to trans relative to the genes they control (Hegarty & Hiscock 2007), thus altering the expression of those genes.
Transgressive segregation (Rieseberg et al. 1999, 2003) in newly formed hybrids could also lead to altered patterns of gene expression. In this model, adaptively important alleles present at different loci in the parent species are recombined in the hybrids to produce transgressive genotypes/phenotypes through complementary gene action (Rieseberg et al. 1999). These ‘transgressive’ phenotypes may enable the hybrid to survive environmental conditions outside the range of either of its parents. Such hybrids are therefore potentially pre-adapted for survival in novel, often extreme, habitats, increasing the potential for ecological speciation (Rieseberg et al. 1999, 2003). Indeed, the research of Rieseberg and co-workers into homoploid hybridization in sunflowers demonstrated transgressive levels of gene expression in potentially adaptive genes in the wild hybrid Helianthus deserticola compared with its parental taxa Helianthus annuus and Helianthus petiolaris (Lai et al. 2006), with some evidence that these changes were associated with particular chromosomal rearrangements. Similarly, changes to flowering time observed to be associated with translocation events in synthetic Brassica allopolyploids (Pires et al. 2004) could be the result of altered expression of flowering time genes such as FRI and FLC, as has been shown in allopolyploid A. suecica (Wang et al. 2006a). The cause of these changed levels of FRI and FLC in A. suecica has yet to be determined.
Transgressive phenotypes could also result from other changes to gene expression resulting from hybridization and, in allopolyploids particularly, there is the potential for altered gene expression as a result of changes to gene dosage. Indeed, many regulatory networks are known to be highly dosage dependent (Riddle & Birchler 2003). The widespread transposon activation proposed in McClintock's genome shock theory could also result in alterations to gene expression, either by knocking out gene activity or creating novel promoter function. Indeed, it has been shown that transposable elements can sometimes drive transcription if inserted upstream of a coding region (Barkan & Martiennssen 1991; Raizada et al. 2001).
(b) Epigenetic modification of gene expression
Changes to the genome of a hybrid individual are not the only potential way in which gene expression can be perturbed: epigenetic changes can also play a role (Hegarty & Hiscock 2008). Research in this area has focused largely on allopolyploid hybrids, especially in the area of gene silencing. The classical model of genome evolution set out by Ohno (1970) predicts that duplicate genes will be subject to silencing and eventually lost due to mutational events over time. However, if all duplicated genes in polyploids were silenced and eventually lost, the effect of polyploidy on the evolution of new species would be minimal (Otto & Whitton 2000), whereas in fact many plant species appear to have retained high numbers of duplicated genes over long periods of evolutionary time. Recent theories (see Kellogg 2003; Otto 2003 for review) have suggested that many of these duplicated genes are retained due to sub-functionalization, whereby the duplicate copies of a gene suffer deleterious but complementary mutations such that both copies are required for phenotypic normality (Lynch & Force 2000). Such sub-functionalization can also arise as a result of altered expression between homeologous genes. The work of Adams and co-workers on allotetraploid cotton (Gossypium hirsutum) has revealed that duplicated genes can display organ-specific reciprocal silencing (Adams et al. 2003). Using cDNA-SSCP (single-strand conformation polymorphism), Adams et al. analysed the expression of homeologous gene copies in the allotetraploid hybrid and showed that 10 out of the 40 gene pairs analysed displayed biased expression from one parental genome or the other. Further analysis of 16 gene pairs in 10 different tissues showed organ-specific silencing effects in 11 genes in at least one tissue type. This showed that a relatively high percentage of genes display silencing effects or biased expression in a developmentally regulated manner, although there did not appear to be preferential transcription of genes from one particular genome. Recent research has shown that similar alterations to the expression of parental homeologues (though not involving total silencing of one copy) can occur in response to abiotic stress (Liu & Adams 2007), potentially explaining the high degree of phenotypic flexibility exhibited by allopolyploids.
Developmental regulation of genes through silencing had previously been observed in the phenomenon of genomic imprinting (Alleman & Doctor 2000), where control of gene expression depends upon the parent of origin. As with most examples of DNA-based gene silencing, genomic imprinting involves the methylation of cytosine residues (Adams et al. 2000). Changes in DNA methylation frequently occur in the genes of newly formed allopolyploids, as shown by amplified fragment length polymorphism (AFLP) and cDNA-AFLP analyses (Comai et al. 2000; Shaked et al. 2001; Salmon et al. 2005). One prime example is nucleolar dominance, in which rRNA genes from one parental genome are silenced independently of paternal or maternal origin (Pikaard 2000). Concerted evolution of rRNA genes has been observed in a number of allopolyploid systems, but most notably Tragopogon (Matya´šek et al. 2007) and Nicotiana (Kovarik et al. 2008), the latter providing strong evidence that these patterns of rRNA expression are established as early as the F1 hybrid. Other recent findings indicate that some redundant protein-coding genes and putative transcription factors can also be silenced in this manner (Comai et al. 2000; Shaked et al. 2001). These studies, predominantly in Arabidopsis, suggest that the levels of gene silencing in polyploids may be relatively low, ranging from approximately 0.4% in synthetic allopolyploid hybrids (Comai et al. 2000) to approximately 2.5% in natural allopolyploids (Lee & Chen 2001). However, several of the silenced genes identified in these studies were transcription factors; so a knock-on effect of reduced transcription for genes under the control of these transcription factors might be predicted, suggesting an indirect route for gene silencing in allopolyploids.
(c) Genome-wide changes to gene expression
With the increasing availability of microarray technologies enabling genome-wide gene expression assays, many recent studies have focused on genome-wide alterations to the transcriptome of hybrid plants. Microarray comparisons of gene expression in synthetic allotetraploid A. suecica and its progenitors A. thaliana and A. arenosa revealed widespread changes to the transcriptome of the hybrid, involving approximately 5.4% of genes analysed (Wang et al. 2006b). The majority of these genes were transcriptionally downregulated, apparently in accordance with silencing of A. thaliana rRNA genes caused by nucleolar dominance. Wang et al. (2006b) noted that many of the affected genes were involved in cell defence, ageing and hormonal regulation, suggesting that these classes of genes may be particularly susceptible to change resulting from hybridization. Since similar changes were not observed in autotetraploid A. thaliana, the authors concluded that hybridization, rather than polyploidy, was the cause of the altered patterns of gene expression. Studies in other systems have confirmed that hybridization can have a dramatic effect on gene expression. In cotton (Gossypium hirsutum), studies of newly synthesized allotetraploids using cDNA-AFLP showed silencing or downregulation of approximately 5% of duplicate gene copies for more than 2000 transcripts assessed (Adams et al. 2004). Analysis of diploid hybrid maize using microarrays to compare hybrids of two different cultivars with their parents revealed comparable levels of gene expression change (approx. 0.9% of the genes on the array) in both vegetative and endosperm tissues (Stupar et al. 2007). While this figure is lower than those seen in Arabidopsis and cotton, this may be due to the fact that interspecific hybridization is not involved (rather intervarietal hybridization). Our own studies of gene expression change associated with hybrid speciation in Senecio (Hegarty et al. 2005, 2006; see below) revealed similar findings to those in Arabidopsis and maize and, additionally, showed that while hybridization is principally responsible for the greatest effect on gene expression, polyploidization also impacts on gene expression in a separate and distinct way to hybridization (Hegarty et al. 2006).
The studies of Wang et al. (2006b) and Stupar et al. (2007) into altered gene expression in homoploid and allopolyploid hybrids, respectively, showed that many of the observed gene expression changes in hybrids were non-additive. That is to say, the differences in expression level observed in the hybrids were not simply the result of a blending of parental gene expression levels (the null hypothesis in both studies). Similar results were also reported by Lai et al. (2006) in a microarray analysis of ‘candidate genes’ selected as potentially involved in the physiological adaptation of the homoploid hybrid sunflower (H. deserticola) to extreme environmental conditions outside the range of its parent species. This study revealed non-additive gene expression in many stress–response genes, suggesting that gene expression change could be responsible for immediate physiological pre-adaptation of some hybrid individuals to extreme novel environments not accessible to their parent species. Such transgressive patterns of gene expression in hybrids offer a transcriptional link with the transgressive segregation and phenotypes seen as central to the process of hybrid speciation (Rieseberg et al. 1999; Lai et al. 2006; Hegarty & Hiscock 2007).
4. Changes to gene expression associated with hybrid speciation in Senecio
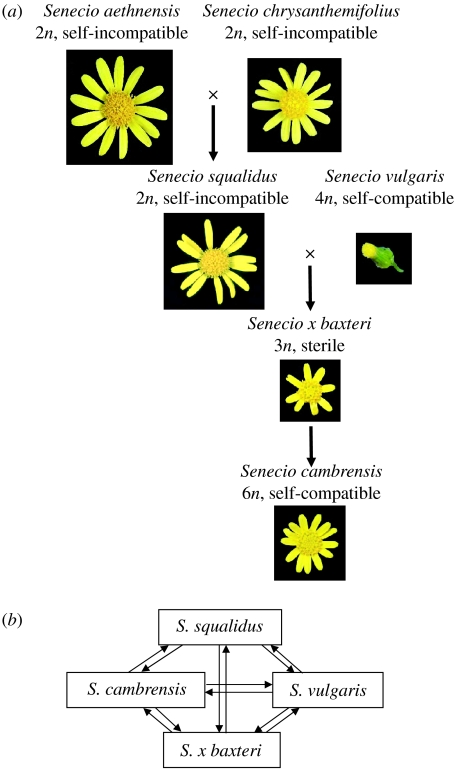
We are using the genus Senecio (Asteraceae) as a model system to study changes to gene expression associated with homoploid hybrid and allopolyploid speciation. Senecio squalidus (2n=2x=20) is a recent homoploid hybrid introduced to the UK from Mt Etna, Sicily via the Oxford Botanic Garden ca 300 years ago. Senecio squalidus is the product of hybridization between two native Sicilian species, Senecio aethnensis (2n=2x=20) and Senecio chrysanthemifolius (2n=2x=20), which form a large hybrid zone between the edges of their ranges on Mt Etna. Hybrid material on Mt Etna is interfertile with both S. chrysanthemifolius and S. aethnensis, but its removal to Britain has allowed it to diverge sufficiently in allopatry to give rise to the homoploid species S. squalidus (James & Abbott 2005). Since its escape from the Oxford Botanic Garden in the eighteenth-century, S. squalidus has established itself as a highly successful invasive species in the British Isles, being found as far north as Inverness in Scotland and as far west as Donegal in Ireland. During its ‘invasion’ of the UK, S. squalidus has hybridized (always as pollen parent) with the native groundsel S. vulgaris (2n=4x=40) to give rise to the sterile F1 hybrid S. x baxteri (3x=30), two new self-fertile allopolyploid species, Senecio cambrensis (2n=6x=60) and Senecio eboracensis (2n=4x=40), and an introgressant Senecio vulgaris var. hibernicus (2n=4x=40; Abbott et al. in press).
We have been using an anonymous cDNA microarray approach to study changes to gene expression associated with the allopolyploid origin of S. cambrensis using both wild and resynthesized hybrid material (see Hegarty et al. 2005, 2006). Natural S. cambrensis is assumed to have arisen via chromosome doubling in the sterile triploid hybrid S. x baxteri, and our ability to generate S. cambrensis-like individuals by colchicine treatment of S. vulgaris×S. squalidus F1 hybrids suggests that this was indeed its natural origin (Hegarty et al. 2006). Our first analyses focused on the allopolyploid origin of S. cambrensis because both hybridization and genome duplication were involved and their relative effects on gene expression could be decoupled when resynthesizing S. cambrensis (Hegarty et al. 2005, 2006). More recently, we have been investigating gene expression change in wild and resynthesized S. squalidus (M. J. Hegarty et al. 2005, unpublished data). We decided to investigate gene expression changes to the floral transcriptome because flower and inflorescence phenotypes differ significantly between hybrids and their parents. During the allopolyploid speciation ‘process’, the large radiate capitulum of S. squalidus with self-incompatible flowers combines with the small non-radiate self-compatible (SC) capitulum of S. vulgaris to produce intermediate short-rayed capitula that are sterile in S. x baxteri and fertile (SC) in S. cambrensis (figure 2). The inflorescence and flower structure of S. squalidus is intermediate between that of its parents—S. aethnensis has large capitula with long ray flowers, whereas S. chrysanthemifolius has smaller capitula with shorter ray florets (figure 2a).
Figure 2.
Hybrid speciation in Senecio and experimental design of microarray analysis. (a) Hybridization events involved in the origins of the diploid hybrid, S. squalidus, and the allohexaploid hybrid, S. cambrensis. (b) Experimental design employed in microarray comparisons of gene expression between the allopolyploid S. cambrensis and its progenitor taxa, S. vulgaris and S. squalidus, and their sterile triploid F1 hybrid, S. x baxteri. Experimental details can be found in Hegarty et al. (2005), but, briefly, mature flower bud tissue was harvested from a mixed population of approximately 30 plants and pooled prior to RNA extraction to create an ‘average’ for each taxon. Labelled cDNA for each taxon was hybridized to a custom floral cDNA microarray. Two taxa were differentially labelled and compared per array hybridization (with 10 replicate hybridizations performed per comparison) using dye swaps to account for any bias in labelling efficiency. Each taxon was compared with the other three, for a total of 30 array hybridizations per taxon. Raw expression data for each taxon were extracted from these 30 replicates and imported separately into the GeneSpring microarray analysis software (Silicon Genetics) to enable comparison between all four taxa.
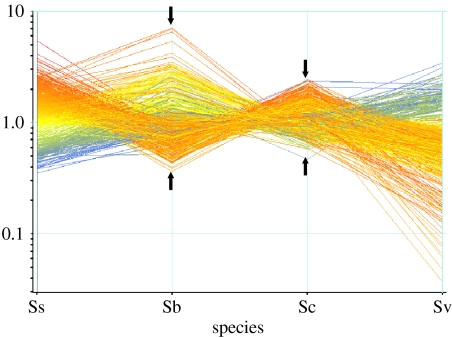
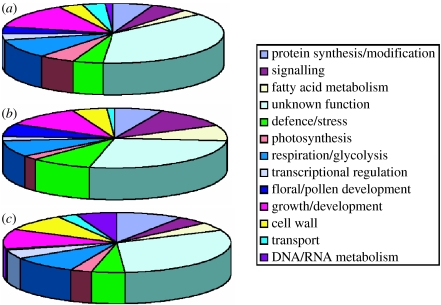
Microarray analysis of the allopolyploid origin of S. cambrensis (figure 2b) revealed an initial large change in floral gene expression in S. x baxteri, with approximately 475 cDNA clones showing up- or downregulation relative to its parental taxa or, also importantly, relative to natural S. cambrensis, from which it differs primarily by a change in ploidy level (Hegarty et al. 2005). Thus, the greatest changes in gene expression relative to the parents appeared to be associated with the hybridization step to form S. x baxteri (figure 3). This initial ‘transcriptome’ shock in S. x baxteri was confirmed in our analysis of resynthesized S. cambrensis, which further showed that the polyploidization event (here induced by colchicine) had an immediate calming (ameliorating) effect on altered patterns of gene expression detected in S. x baxteri (Hegarty et al. 2006). Importantly, this altered pattern of gene expression, apparent in first-generation allopolyploids, was preserved in four successive generations of the synthetic allopolyploids and in wild S. cambrensis (Hegarty et al. 2006). While previous research in resynthesized wheat (Feldman & Levy 2005) had identified separate effects of hybridization and polyploidization on the genome and transcriptome, our data represent the first indication that these changes to gene expression are genome wide. Interestingly, the putative functional classes of genes affected by hybridization and allopolyploidization were remarkably similar, with no functional class of genes being overly affected by hybridization or allopolyploidization (figure 4). However, perhaps not surprisingly, when compared with functional classes of genes not affected by either process, there was a greater representation of genes potentially involved in flower/inflorescence and pollen developments, which may reflect the transitions in floral phenotypes observed after hybridization and allopolyploidization.
Figure 3.
‘Transcriptome shock’ in Senecio x baxteri is ameliorated in the allohexaploid derivative S. cambrensis. Normalized microarray expression data for 475 cDNA clones previously identified as displaying significant differences in expression between either S. x baxteri (Sb) or S. cambrensis (Sc) compared with one or both of the parental taxa S. squalidus (Ss) and S. vulgaris (Sv). The arrows highlight the reduction in the severity (variance) of altered gene expression in S. cambrensis relative to the initial triploid hybrid S. x baxteri.
Figure 4.
Functional classes of genes affected by allopolyploidization and hybridization. Basic gene ontologies for cDNA clones displaying (a) conserved expression changes in both wild and synthetic Senecio cambrensis relative to S. x baxteri (genes affected by allopolyploidy, 540 clones), (b) expression changes relative to the parental taxa S. squalidus and S. vulgaris in both hybrid taxa (genes affected by hybridization, 99 clones) and (c) genes showing no expression difference between the parental and hybrid taxa (unaffected by hybridization or polyploidy, 289 clones). With the exception of a higher proportion of floral/pollen-related genes in (a,b) compared with (c), there are no substantial differences between the classes of affected genes (adapted with permission from Hegarty et al. 2006).
A recent study of the allopolyploid origin of A. suecica by Wang et al. (2006a) focused on the identification of genes whose expression in hybrids differed from the additive expression midpoint of the two different parental gene copies. A similar approach was also used recently to analyse gene expression change in maize hybrids (Stupar et al. 2007). Such an approach provides a consistent and unified methodology for identifying genes affected by hybridization and/or polyploidization in different model study systems. Using this approach, it will be possible to compare genes identified as affected by hybridization/polyploidization in different systems, in order to determine whether particular genes, or groups of genes, are consistently affected. Here we describe the use of this approach to reanalyse the microarray data from our original analysis of the allopolyploid origin of S. cambrensis to identify specific genes and classes of genes affected by hybridization and polyploidization.
(a) Are changes to gene expression in allopolyploid Senecio hybrids additive or non-additive?
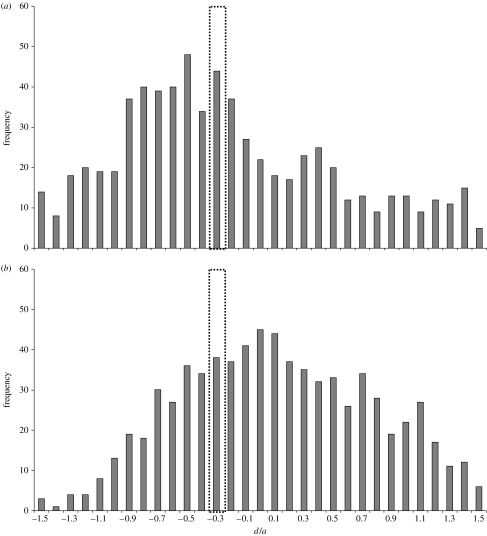
Using methods similar to Stupar et al. (2007), we tested whether changes in gene expression observed in synthetic S. x baxteri and wild allohexaploid S. cambrensis were additive or non-additive. By averaging the normalized parental expression values (as derived by the GeneSpring array analysis software) for each feature on the array showing differential expression in the hybrids, a parental midpoint expression value was obtained. This was slightly different from the approach of Wang et al. (2006b), who hybridized an equal mixture of parental cDNA to their arrays to directly measure midpoint expression levels. The derived midpoint values were then used to calculate a d/a value for each array feature, where d represents the difference between the expression level in the hybrid and the estimated parental midpoint, and a is the difference between the highest parental expression and the midpoint. The d/a ratio therefore gives a mathematical indicator of the expression differences in the hybrid compared with both parents. A ratio of 0 would normally represent complete additivity of expression in the hybrid, but because in S. x baxteri and S. cambrensis the parental genome contribution is unequal (1 S. squalidus : 2 S. vulgaris), the additive value will be −0.33 (relative to the higher expressing parent). Ratios of −1 or 1 indicate that expression in the hybrid is more likely to that of the lower or higher expressing parent, respectively. Ratios below −1 or above 1 represent expression in the hybrid outside the range of either parent. While it is possible to adjust the calculated midpoint value to reflect the respective genome contributions of S. squalidus and S. vulgaris, thereby returning the additive d/a value to 0, this also alters the expected d/a ratio for expression equal to the lower expressing parent to −0.5, while keeping the ratio for expression equal to the higher expressing parent as 1. To avoid confusion, therefore, we maintain the midpoint as a simple mean of the parental expression values and accept that additive expression would equal −0.33. While there is a caveat with this approach, in that highly similar parental expression levels lead to small a values, and therefore to potentially disproportionately high d/a ratios, the selection of clones displaying more than 1.5-fold differences between the hybrids and either parent ensures that this scenario will not result in false identification of clones as non-additive.
Using our previous microarray expression data (Hegarty et al. 2005), we filtered the initial results of the one-way analysis of variance (1208 cDNA clones) to identify clones showing more than 1.5-fold changes in expression in each hybrid compared with either parent. This identified 1001 cDNA clones for S. x baxteri and 829 for S. cambrensis. Note that, due to the anonymous design of the microarrays, there is some redundancy and thus the number of clones does not equal the number of actual genes affected. An Anderson–Darling test on these data then indicated that the frequency distribution of both sets of d/a values was not normal. A one-sample sign test was then performed to determine whether the data conformed to the expected median of −0.33. In both cases, the median was significantly different (p<0.0001) to the expected additive value; so the null hypothesis of largely additive gene expression changes can be rejected for both hybrids. Instead, for both hybrids, the majority of the data were skewed towards one of the parents; in the case of S. x baxteri, it was towards that of the lower expressing parent (figure 5a), whereas in S. cambrensis it was towards that of the higher expressing parent (figure 5b). Further analysis of the data showed that for approximately 70% of clones affected in either hybrid, S. vulgaris was the lower expressing parent. Expression outside the parental range (d/a value <−1 or >1) was observed in a substantial proportion of cDNA clones in both hybrids: 470 (46.95%) in S. x baxteri and 192 clones (23.16%) in S. cambrensis. As a percentage of the total number of features on the microarray, these numbers correspond to 7.42 and 3.03% in S. x baxteri and S. cambrensis, respectively. This is in accordance with the approximately 5% value observed in synthetic A. suecica allopolyploids (Wang et al. 2006b).
Figure 5.
Frequency distributions of d/a ratios for (a) S. x baxteri and (b) S. cambrensis, between the values of −1.5 and +1.5 (data outside this range not shown). The dotted line in both cases highlights the expected ratio if gene expression is additive. The graphs demonstrate that expression in S. x baxteri is largely skewed towards a level similar to that in the lower expressing parent, while the opposite trend is observed in S. cambrensis.
(b) Is there evidence of transgressive gene expression in allopolyploid Senecio hybrids?
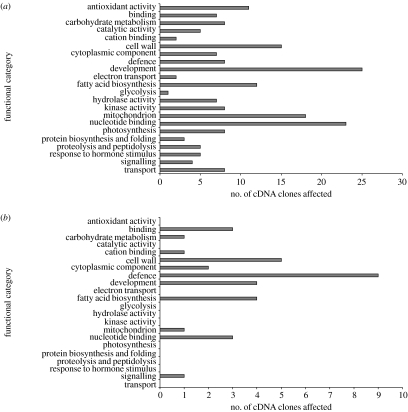
Having identified a pool of cDNA clones displaying non-additive changes to gene expression in both hybrid taxa, we then tested these clones for evidence of expression beyond the parental ranges, i.e. transgressive gene expression. Of the 470 cDNA clones identified for S. x baxteri, 378 displayed expression differing from the parental midpoint by more than 1.5-fold, with 81 out of the 192 cDNA clones for S. cambrensis showing the same effect. Within both these groups, the majority of cDNA clones showed upregulation compared with the parental midpoint (66.9 and 70.4% in S. x baxteri and S. cambrensis, respectively). Aside from the genes for which no functional class could be ascribed (49.2%), the major functional groups affected in S. x baxteri (figure 6a) were genes involved in development (6.6%), nucleotide binding (6.1%), mitochondrial activity (4.76%) and cell wall function (3.97%). Within the development category, a high proportion of clones (32%) were found to encode tubulins, profilins or senescence-associated proteins. Of the clones involved in nucleotide binding, 34% were transcription factors. In S. cambrensis (figure 6b), the majority of clones (58%) could not be assigned to a functional category. Of the remainder, the largest categories were defence (11.1%) and cell wall-related genes (6.17%). The majority of clones (five out of nine) in the defence category encode a pore-forming toxic-like protein similar to Hfr2, a protein involved in insect herbivory resistance (Williams et al. 2002). This cDNA clone was also identified as affected in S. x baxteri, its expression being downregulated in both cases. Three of the remaining clones in the defence category encode a germin-like protein. Such proteins have been shown to play a potential role in resistance to herbivory (Lou & Baldwin 2006) and pathogen attack (Godfrey et al. 2007), with studies suggesting that germin-like proteins possess superoxide dismutase activity (Godfrey et al. 2007; Gucciardo et al. 2007). All three of these clones were upregulated compared with the parental midpoint and displayed expression similar to that observed in the S. vulgaris parent.
Figure 6.
Functional classes of genes showing non-additive expression in triploid hybrid S. x baxteri and allohexaploid S. cambrensis. Non-additive gene expression change in (a) S. x baxteri and (b) S. cambrensis: numbers are taken from the 378 and 81 clones differing by more than 1.5-fold from the parental midpoint value for S. x baxteri and S. cambrensis, respectively. Genes of unknown function (which represent the majority of genes identified, see text) are not included.
To look for evidence of transgressive expression outside the range of either parent, we then further filtered the data to identify clones with expression values differing by more than 1.5-fold from either parent. This identified 165 cDNA clones in S. x baxteri, but only eight clones in S. cambrensis. This is consistent with our previous observation (Hegarty et al. 2006) that genome duplication has an ameliorating effect on altered levels of gene expression observed in S. x baxteri. Genes involved in mitochondrial function and development again formed a high proportion of the clones identified in S. x baxteri; in particular, those encoding subunits of ATP synthase, α-tubulin and a senescence-related protein were all upregulated compared with both parents. Only three of the transcription factors differing from the parental midpoint were present after filtering with two of these encoding a LIM domain protein known to be pollen specific in sunflower (Baltz et al. 1992). Four of the eight clones identified in S. cambrensis were of unknown function, the remainder encoding the α-subunit of ATP synthase (upregulated), a purple acid phosphatase (upregulated) and two copies of the pore-forming toxic-like Hfr2 gene (both downregulated). Purple acid phosphatase genes are part of a 28-member family in Arabidopsis, all of which are expressed in floral tissue (though not exclusively) and are localized to the petals and anther filaments (Zhu et al. 2005). While RNAi in Arabidopsis failed to suggest a phenotype for knockout mutants (Zhu et al. 2005), other studies (Sánchez-Calderón et al. 2006; Veljanovski et al. 2006) have shown that such phosphatases can be upregulated as a response to phosphate deficiency. This does not appear to be related to the upregulation of ATP synthase, however, because the purple phosphatase gene was not upregulated in S. x baxteri. Interestingly, other acid phosphatases were also upregulated in S. cambrensis and again these showed the opposite expression pattern in S. x baxteri.
(c) Genes and gene classes showing non-additive expression change
Reanalysis of our microarray comparison of gene expression in the allopolyploid hybrids S. x baxteri and S. cambrensis and their progenitors revealed a relatively high proportion of non-additive gene expression change in the hybrids relative to their parental expression levels. In addition, the degree of non-additive gene expression was lower in allohexaploid S. cambrensis compared with its triploid intermediate S. x baxteri. This is consistent with our previous observation that the ‘transcriptome shock’ resulting from allopolyploidization is largely due to hybridization, with polyploidization resulting in a distinct secondary shift (amelioration) in gene expression (Hegarty et al. 2006). The fairly diverse nature of the genes affected is consistent with other findings in Arabidopsis (Wang et al. 2006b), cotton (Adams et al. 2004) and maize (Stupar et al. 2007) that non-additive changes to gene expression are genome wide. The next important step is to determine how these changes in gene expression impact upon the phenotypes of the new hybrids at a structural/morphological level, as well as at a physiological level, to determine the extent to which they contribute to phenomena such as hybrid vigour (heterosis) and outbreeding depression.
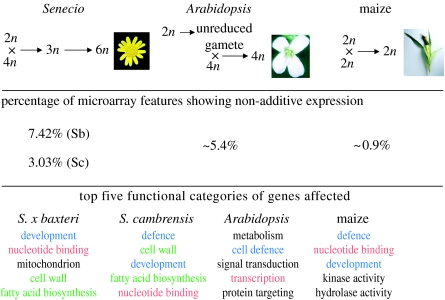
It was interesting to see that similar functional classes of genes were affected by hybridization in Senecio, Arabidopsis and maize (figure 7), suggesting that certain gene networks may be particularly susceptible to perturbation by hybridization; the functional categories of nucleotide binding, defence and mitochondria being good examples. In terms of the classes of genes affected in Senecio, it is also interesting to note that one of the major affected groups in S. x baxteri was nucleotide binding. In addition to a number of (primarily downregulated) transcription factors that have shown similar alterations in expression pattern in Arabidopsis suecica (Wang et al. 2006b), this category also contained clones encoding cytidine deaminase (CDA) and 8-oxoG-DNA glycosylase (OGG1). OGG1 has been implicated in DNA base excision repair (García-Ortiz et al. 2001), while CDA has popularly been suspected to be involved in RNA editing, although it now appears that pentatricopeptide repeat proteins that contain CDA-like domains are the more likely candidates (Salone et al. 2007). These genes were of interest, given that we also observed a relatively high number of clones encoding proteins involved in either DNA modification or cell division. In addition to cytidine deaminase and OGG1 (both of which were upregulated compared with the parental midpoint in S. x baxteri), there was also upregulation of adenosylhomocysteinase and adenosyl kinase, the genes involved in S-adenosylmethionine (SAM) dependent methylation (Moffatt et al. 2002; Mull et al. 2006). The expression of SAM synthetase was also increased relative to both parents. SAM-dependent methylation is used in gene silencing, and also in pectin methylation (Pereira et al. 2006), and, indeed, we observed an increase in the expression of pectin methylesterase as well as another SAM-dependent enzyme, caffeic acid 3-O-methyltransferase. While we were unable to demonstrate complete silencing of any clones on the array, it is important to note that cDNA-based microarrays cannot distinguish between different parental homeologues. Therefore, in the cases where the hybrid shows downregulation of a gene relative to the parental taxa, it may well be that this is due to silencing of one parental gene copy. Further investigations are underway to study the expression of specific parental homeologues and genome-wide changes to DNA methylation in the hybrids using cDNA-SSCP and methylation-sensitive AFLP, respectively.
Figure 7.
Comparison of non-additive expression changes resulting from hybridization in maize and hybridization/polyploidy in Senecio and Arabidopsis. The formation of the hybrids is shown in each case, together with the level of non-additive gene expression in the hybrids expressed as a percentage of the features on the microarray platform used. Finally, the top five functional gene classes affected (ignoring unknowns) for each hybrid are displayed for comparison. Red indicates a functional gene class affected in all four hybrid systems, blue indicates a functional class affected in at least one of the Senecio hybrids and one of the other two hybrid taxa, and green indicates a functional class affected in both Senecio hybrids but not in either Arabidopsis or maize. For Arabidopsis suecica, gene function data were taken from Wang et al. (2006b), and for maize from an extrapolation of the supplementary data given in Stupar et al. (2007). Sb, S. x baxteri; Sc, S. cambrensis.
As revealed by similar studies in Arabidopsis and maize (Wang et al. 2006b; Stupar et al. 2007), a number of the cDNA clones affected by hybridization in S. x baxteri encode mitochondrial proteins. Of the 18 clones identified, 14 encode subunits of ATP synthase and all displayed upregulation compared with the parental midpoint, with 8 of these displaying transgressive levels of expression. The α-subunit (ATPa) showed the highest degree of upregulation, which was somewhat unexpected, given the findings of Smart et al. (1994) when investigating the role of ATPa in cytoplasmic male sterility in maize hybrids. Smart et al. (1994) showed that misregulation of the maternally inherited mitochondrial genome by the hybrid nucleus results in aberrant RNA editing, resulting in the production of an extended ATPa transcript, which causes pollen to become inviable. The production of this aberrant transcript did not, however, coincide with any increase in ATPa transcript levels: either the aberrant or normal form. Furthermore, where fertility was restored by post-transcriptional elimination of the aberrant transcript, normal ATPa expression was reduced (Smart et al. 1994). Indeed, in our study of gene expression change in resynthesized homoploid S. squalidus, we also observed a reduction in ATPa expression in the hybrids, which was potentially linked to increased levels of a fertility-restoring factor (Hegarty et al. submitted). The high levels of ATP synthase expression in S. x baxteri are therefore intriguing, particularly because ATPa expression levels are reduced in its fully fertile allohexaploid derivative S. cambrensis.
Both S. x baxteri and S. cambrensis differ from their parental taxa in floral morphology, particularly ray flower morphology. We are therefore focusing current studies on a number of floral development gene homologues whose expression was affected in S. x baxteri and S. cambrensis relative to their parents. In S. x baxteri, two highly upregulated genes of particular interest encode putative homologues of FIDDLEHEAD and HOTHEAD involved in floral organ fusion in Arabidopsis thaliana (Lolle & Cheung 1993; Lolle et al. 1998; Krolikowski et al. 2003). FIDDLEHEAD encodes a protein related to β-ketoacyl-CoA synthases (Lolle et al. 1998), while HOTHEAD encodes a protein similar to FAD-containing oxidoreductases (Krolikowski et al. 2003). In Arabidopsis, mutations in either of these genes result in an organ fusion phenotype and allow hydration and germination of wild-type pollen on non-reproductive organs (Lolle & Cheung 1993; Lolle et al. 1998; Krolikowski et al. 2003). Both genes appear to play a role in lipid biosynthesis during cuticle formation and thus act to inhibit signalling between adjoining epidermal cells (Pruitt et al. 2000; Kurdyukov et al. 2006). Expression of the putative Senecio FIDDLEHEAD and HOTHEAD genes was also differential relative to both parents in allohexaploid S. cambrensis, but these expression changes were largely a shift towards the expression seen in S. squalidus (in the case of HOTHEAD) or additive expression (in the case of FIDDLEHEAD). Both hybrids also displayed non-additive expression in a clone encoding CUT1, which is also involved in cuticle formation (Millar et al. 1999). In this particular case, the expression was more similar to S. vulgaris in the S. x baxteri triploid and more like S. squalidus in the S. cambrensis allohexaploid. It will be interesting to determine whether these altered expression patterns are related to particular changes in floral organ development in S. x baxteri and S. cambrensis, for instance, during processes involved in ray flower production in the hybrids that are the product of a ray-forming genome (S. squalidus) and a non-ray genome (S. vulgaris). We are currently investigating this possibility using rayless phenotypes of resynthesized S. cambrensis.
The final ‘interesting’ group of genes affected in both hybrids is putatively involved in defence or stress responses. Among these, we observe downregulation of a gene putatively encoding a protein with significant homology to the lectin-like toxicity protein Hfr2 (Williams et al. 2002). Inheritance of herbivore resistance is not always consistent between polyploid lineages (Nuismer & Thompson 2001); so it is interesting to note that the Hfr2 gene is affected similarly in both S. x baxteri (which was produced under glasshouse conditions) and a wild lineage of S. cambrensis. Other genes potentially involved in response to herbivory were affected differently between the two hybrids, with S. x baxteri showing upregulation of chitinase and lipoxygenase, and S. cambrensis displaying increased expression of a germin-like protein. This is consistent with the findings of Pearse et al. (2006) in Nicotiana allopolyploids, which suggested that different components of anti-herbivore defence mechanisms could be flexibly inherited in neopolyploids. Altered expression of genes involved in response to herbivory has also been demonstrated to vary between two Nicotiana polyploids that share a common ancestor (Qu et al. 2004). Increased resistance to herbivores in polyploids compared with their diploid relatives has been proposed (Qu et al. 2004) as possibly due to an increase in the number of alleles for resistance genes seen in polyploids (Oswald & Nuismer 2007). Recent research in Gossypium (Liu & Adams 2007) suggests that different adaptive phenotypes such as this could also result from modulated expression of homeologous parental gene copies. The ability of allopolyploids to increase their phenotypic range in such a manner may explain the vast evolutionary success of polyploid taxa in the plant kingdom; however, it should be noted that empirical studies of resistance have been performed in only a limited number of genera; so it remains to be seen if this phenomenon is common to all polyploids.
5. Conclusion
Recent molecular genetic studies of hybridization and polyploidy in plants have shown that these key evolutionary processes have profound effects on patterns of gene expression. Importantly, changed patterns of gene expression identified in wild hybrids and allopolyploids can be recapitulated in synthetic hybrids and allopolyploids, indicating that these processes generate an immediate source of genetic variation upon which natural selection can act. Our studies in Senecio have demonstrated that hybridization between the diverged genomes of S. squalidus and S. vulgaris results in a wide range of alterations to gene expression in both the sterile triploid intermediate S. x baxteri and its allohexaploid derivative S. cambrensis, with a relatively high proportion of these changes being non-additive. However, the percentage of genes displaying non-additive expression in S. cambrensis was lower than that in S. x baxteri, confirming our earlier observations that polyploidization has an ameliorating effect on changed levels of gene expression experienced by S. x baxteri (Hegarty et al. 2006). Our analyses have identified non-additively expressed genes (examples shown in table 1) potentially involved in (i) gene regulation or gene silencing, (ii) metabolism, (iii) floral organ development and (iv) defence, which are now the focus of more detailed study to determine the extent of their role in these processes and in phenotypic change during hybrid speciation in Senecio. These results confirm and extend findings from similar studies in other hybrid systems, such as maize (Stupar et al. 2007) and Arabidopsis (Wang et al. 2006b), that hybridization generally results in non-additive gene expression changes. Importantly, the percentages of assayed microarray features displaying non-additive expression are similar between these systems and Senecio, as are many of the affected gene categories (figure 7). In particular, genes involved in nucleotide binding and transcriptional regulation are affected in both Senecio hybrids and also in maize and Arabidopsis hybrids, while those involved in defence or development are within the top five affected categories in one or both Senecio hybrids and either A. suecica or maize. This suggests that hybridization and polyploidy have somewhat predictable consequences in terms of the classes of genes affected, even if the specific genes themselves may differ. The deviation in gene expression in hybrids from a purely additive combination of the expression levels of their parents could contribute to phenotypic phenomena such as hybrid vigour and, importantly, may serve as a source of variation in hybrids upon which selection can act. The most important future goal will be to determine whether non-additive and transgressive changes in gene expression in hybrids contribute directly to local adaptation and speciation.
Table 1.
Non-additively expressed genes in Senecio x baxteri and S. cambrensis. (Genes showing non-additive expression in S. x baxteri and S. cambrensis that are discussed in the text: the number of array features (cDNA clones) displaying non-additive expression is displayed for each gene, together with the direction and level of expression change from the parental midpoint value (MPV), together with its putative functional class.)
| gene product | number of clones | mean expression versus MPV | functional category | ||
|---|---|---|---|---|---|
| S. x baxteri | S. cambrensis | S. x baxteri | S. cambrensis | ||
| pectin methylesterase | 1 | 2 | up (1.63-fold) | up (1.67-fold) | cell wall |
| caffeic acid O-methyltransferase | 1 | 1 | up (1.55-fold) | up (2.07-fold) | cell wall |
| pore-forming toxic-like protein Hfr2 | 5 | 5 | down (1.85-fold) | down (1.3-fold) | defence |
| germin-like protein | — | 3 | — | up (1.89-fold) | defence |
| putative senescence-associated protein | 6 | 0 | up (3.93-fold) | — | defence |
| α-tubulin | 6 | — | up (2.04-fold) | — | development |
| HOTHEAD | 2 | — | up (2.63-fold) | — | fatty acid biosynthesis |
| FIDDLEHEAD | 1 | — | up (2.39-fold) | — | fatty acid biosynthesis |
| CUT1 | — | 1 | — | down (2.14-fold) | fatty acid biosynthesis |
| adenosyl kinase | 2 | — | up (1.64-fold) | — | kinase activity |
| subunit of ATP synthase | 14 | 1 | up (2.73-fold) | up (1.72-fold) | mitochondrion |
| LIM domain transcription factor SF3 | 5 | 2 | down (2.34-fold) | up (1.59-fold) | nucleotide binding |
| OGG1 | 1 | — | up (1.72-fold) | — | nucleotide binding |
| cytidine deaminase | 1 | — | up (3.07-fold) | — | nucleotide binding |
| adenosylhomocysteinase | 1 | — | up (1.61-fold) | — | unknown |
Acknowledgments
We thank Christopher Thorogood and Alexandra Allen for their useful comments on an early version of this manuscript, and Pete Hollingsworth and three anonymous referees for their constructive comments on improving the final version. Work done in S.J.H.'s laboratory is supported by the Natural Environment Research Council (NERC), the Leverhulme Trust and the Lady Emily Smyth Agricultural Research Station, University of Bristol.
Footnotes
One contribution of 12 to a Theme Issue ‘Speciation in plants and animals: pattern and process’.
References
- Abbott, R. J., Brennan, A. C., James, J. K., Forbes, D. G., Hegarty, M. J. & Hiscock, S. J. In press. Recent hybrid origin and invasion of the British Isles by a self-incompatible species, Oxford ragwort (Senecio squalidus L., Asteraceae). Biol. Inv
- Adams S, Vinkenoog R, Spielman M, Dickinson H.G, Scott R.J. Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development. 2000;127:2493–2502. doi: 10.1242/dev.127.11.2493. [DOI] [PubMed] [Google Scholar]
- Adams K.L, Cronn R, Percifield R, Wendel J.F. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl Acad. Sci. USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. doi:10.1073/pnas.0630618100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K.L, Percifield R, Wendel J.F. Organ-specific silencing of genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–2226. doi: 10.1534/genetics.104.033522. doi:10.1534/genetics.104.033522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleman M, Doctor J. Genomic imprinting in plants: observations and evolutionary implications. Plant Mol. Biol. 2000;43:147–161. doi: 10.1023/a:1006419025155. doi:10.1023/A:1006419025155 [DOI] [PubMed] [Google Scholar]
- Baltz R, Evrard J.L, Domon C, Steinmetz A. A LIM motif is present in a pollen-specific protein. Plant Cell. 1992;4:1465–1466. doi: 10.1105/tpc.4.12.1465. doi:10.1105/tpc.4.12.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Martiennssen R.A. Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl Acad. Sci. USA. 1991;88:3502–3506. doi: 10.1073/pnas.88.8.3502. doi:10.1073/pnas.88.8.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.A, Burke J.M. Genetic divergence and hybrid speciation. Evolution. 2007;61:1773–1780. doi: 10.1111/j.1558-5646.2007.00134.x. doi:10.1111/j.1558-5646.2007.00134.x [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi A.P, Winter K, Holmes-Davis R, Reynolds S.H, Stevens Y, Byers B. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–1567. doi: 10.1105/tpc.12.9.1551. doi:10.1105/tpc.12.9.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Madlung A, Josefsson C, Tyagi A. Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Phil. Trans. R. Soc. B. 2003;358:1149–1155. doi: 10.1098/rstb.2003.1305. doi:10.1098/rstb.2003.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy A.A. Allopolyploidy—a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005;109:250–258. doi: 10.1159/000082407. doi:10.1159/000082407 [DOI] [PubMed] [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy A.A, Vega J.M. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homeologous chromosomes. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson D, Sang T. Speciation through homoploid hybridisation between allotetraploids in peonies (Paeonia) Proc. Natl Acad. Sci. USA. 2001;98:3915–3919. doi: 10.1073/pnas.061288698. doi:10.1073/pnas.061288698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R.T, Pires J.C, Iniguez-Luy F, Leon E, Osborn T.C. Genomic changes in resynthesised Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. doi:10.1105/tpc.107.054346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M.D, Devos K.M. Plant comparative genetics after 10 years. Science. 1998a;282:656–659. doi: 10.1126/science.282.5389.656. doi:10.1126/science.282.5389.656 [DOI] [PubMed] [Google Scholar]
- Gale M.D, Devos K.M. Comparative genetics in the grasses. Proc. Natl Acad. Sci. USA. 1998b;95:1971–1974. doi: 10.1073/pnas.95.5.1971. doi:10.1073/pnas.95.5.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortiz M.V, Ariza R.R, Roldán-Arjona T. An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol. Biol. 2001;47:795–804. doi: 10.1023/a:1013644026132. doi:10.1023/A:1013644026132 [DOI] [PubMed] [Google Scholar]
- Godfrey D, Able A.J, Dry I.B. Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: a possible role in defense? Mol. Plant Microbe Interact. 2007;20:1112–1125. doi: 10.1094/MPMI-20-9-1112. doi:10.1094/MPMI-20-9-1112 [DOI] [PubMed] [Google Scholar]
- Grant V. Cytogenetics of the hybrid Gilia millefoliata x achilleaefolia. I. Variations in meiosis and polyploidy rate as affected by nutritional and genetic conditions. Chromosoma. 1952;5:372–390. doi:10.1007/BF01271494 [PubMed] [Google Scholar]
- Grant V. 2nd edn. Columbia University Press; New York, NY: 1981. Plant speciation. [Google Scholar]
- Gucciardo S, Wisniewski J.P, Brewin N.J, Bornemann S. A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J. Exp. Bot. 2007;58:1161–1171. doi: 10.1093/jxb/erl282. doi:10.1093/jxb/erl282 [DOI] [PubMed] [Google Scholar]
- Hammerle B, Ferrus A. Expression of enhancers is altered in Drosophila melanogaster hybrids. Ecol. Dev. 2003;5:221–230. doi: 10.1046/j.1525-142x.2003.03030.x. doi:10.1046/j.1525-142X.2003.03030.x [DOI] [PubMed] [Google Scholar]
- Hegarty M.J, Hiscock S.J. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 2004;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. doi:10.1111/j.1469-8137.2004.01253.x [DOI] [PubMed] [Google Scholar]
- Hegarty M.J, Hiscock S.J. Polyploidy: doubling up for evolutionary success. Curr. Biol. 2007;17:927–929. doi: 10.1016/j.cub.2007.08.060. doi:10.1016/j.cub.2007.08.060 [DOI] [PubMed] [Google Scholar]
- Hegarty M.J, Hiscock S.J. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. doi:10.1016/j.cub.2008.03.043 [DOI] [PubMed] [Google Scholar]
- Hegarty M.J, et al. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol. Ecol. 2005;14:2493–2510. doi: 10.1111/j.1365-294x.2005.02608.x. doi:10.1111/j.1365-294x.2005.02608.x [DOI] [PubMed] [Google Scholar]
- Hegarty M.J, Barker G.L, Wilson I.D, Abbott R.J, Edwards K.J, Hiscock S.J. Transcriptome shock after interspecific hybridisation in Senecio is ameliorated by genome duplication. Curr. Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. doi:10.1016/j.cub.2006.06.071 [DOI] [PubMed] [Google Scholar]
- Hegarty, M. J., Barker, G. L., Brennan, A. C., Edwards, K. J., Abbott, R. J. & Hiscock S. J. Submitted. Extreme changes to gene expression associated with homoploid hybrid speciation. [DOI] [PubMed]
- Ingram R. The genomic relationship of Senecio squalidus L. and S. vulgaris L. and the significance of genomic balance in their hybrid, S. x baxteri Druce. Heredity. 1978;40:459–462. doi:10.1038/hdy.1978.51 [Google Scholar]
- James J.K, Abbott R.J. Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution. 2005;59:2533–2547. doi:10.1111/j.0014-3820.2005.tb00967.x [PubMed] [Google Scholar]
- Jiggins C.D, Salazar C, Linares M, Mavarez J. Hybrid trait speciation and Heliconius butterflies. Phil. Trans. R. Soc. B. 2008;363:3047–3054. doi: 10.1098/rstb.2008.0065. doi:10.1098/rstb.2008.0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.-K, Bennetzen J.L. Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell. 1994;6:1177–1186. doi: 10.1105/tpc.6.8.1177. doi:10.1105/tpc.6.8.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.A. What happens to genes in duplicated genomes. Proc. Natl Acad. Sci. USA. 2003;100:4369–4371. doi: 10.1073/pnas.0831050100. doi:10.1073/pnas.0831050100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Dadejova M, Lim Y.K, Chase M.W, Clarkson J.J, Knapp S, Leitch A.R. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Ann. Bot. 2008;101:815–823. doi: 10.1093/aob/mcn019. doi:10.1093/aob/mcn019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolikowski K.A, Victor J.L, Nussbaum Wagler T, Lolle S.J, Pruitt R.E. Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 2003;35:501–511. doi: 10.1046/j.1365-313x.2003.01824.x. doi:10.1046/j.1365-313X.2003.01824.x [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, et al. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-, omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006;224:315–329. doi: 10.1007/s00425-005-0215-7. doi:10.1007/s00425-005-0215-7 [DOI] [PubMed] [Google Scholar]
- Lai Z, Gross B.L, Zou Y, Andrews J, Rieseberg L.H. Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 2006;15:1213–1227. doi: 10.1111/j.1365-294X.2006.02775.x. doi:10.1111/j.1365-294X.2006.02775.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S, Chen Z.J. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl Acad. Sci. USA. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. doi:10.1073/pnas.121064698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch I.J, Bennett M.D. Polyploidy in angiosperms. Trends Plant Sci. 1997;2:470–476. doi:10.1016/S1360-1385(97)01154-0 [Google Scholar]
- Liu Z, Adams K.L. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr. Biol. 2007;17:1669–1674. doi: 10.1016/j.cub.2007.08.030. doi:10.1016/j.cub.2007.08.030 [DOI] [PubMed] [Google Scholar]
- Lolle S.J, Cheung A.Y. Promiscuous germination and growth of wildtype pollen from Arabidopsis and related species on the shoot of the Arabidopsis mutant, fiddlehead. Dev. Biol. 1993;155:250–258. doi: 10.1006/dbio.1993.1022. doi:10.1006/dbio.1993.1022 [DOI] [PubMed] [Google Scholar]
- Lolle S.J, Hsu W, Pruitt R.E. Genetic analysis of organ fusion in Arabidopsis. Genetics. 1998;149:607–619. doi: 10.1093/genetics/149.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Baldwin T.T. Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol. 2006;140:1126–1136. doi: 10.1104/pp.105.073700. doi:10.1104/pp.105.073700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matya´šek R, Tate J.A, Lim Y.K, Šrubařova´ H, Koh J, Leitch A.R, Soltis D.E, Soltis P.S, Kovařík A. Concerted evolution of rDNA in recently formed Tragopogon allotetraploids is typically associated with an inverse correlation between gene copy number and expression. Genetics. 2007;176:2509–2519. doi: 10.1534/genetics.107.072751. doi:10.1534/genetics.107.072751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A, Matzke A.J.M. Polyploidy and transposons. Trends Ecol. Evol. 1998;13:241. doi: 10.1016/s0169-5347(98)01390-1. doi:10.1016/S0169-5347(98)01390-1 [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. doi:10.1126/science.15739260 [DOI] [PubMed] [Google Scholar]
- Millar A.A, Clemens S, Zachgo S, Giblin E.M, Taylor D.C, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. doi:10.1105/tpc.11.5.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt B.A, et al. Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 2002;128:812–821. doi: 10.1104/pp.010880. doi:10.1104/pp.010880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. Meiosis in allopolyploids: the importance of “Teflon” chromosomes. Trends Genet. 2002;18:456–463. doi: 10.1016/s0168-9525(02)02730-0. doi:10.1016/S0168-9525(02)02730-0 [DOI] [PubMed] [Google Scholar]
- Mull L, Ebbs M.L, Bender J. A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics. 2006;174:1161–1171. doi: 10.1534/genetics.106.063974. doi:10.1534/genetics.106.063974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuismer S.L, Thompson J.N. Plant polyploidy and non-uniform effects on insect herbivores. Proc. R. Soc. B. 2001;268:1937–1940. doi: 10.1098/rspb.2001.1760. doi:10.1098/rspb.2001.1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Springer; Berlin, Germany: 1970. Evolution by gene duplication. [Google Scholar]
- Oswald B.P, Nuismer S.L. Neopolyploidy and pathogen resistance. Proc. R. Soc. B. 2007;274:2393–2397. doi: 10.1098/rspb.2007.0692. doi:10.1098/rspb.2007.0692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P. In polyploids, one plus one does not equal two. Trends Ecol. Evol. 2003;18:431–433. doi:10.1016/S0169-5347(03)00213-1 [Google Scholar]
- Otto S.P, Whitton J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. doi:10.1146/annurev.genet.34.1.401 [DOI] [PubMed] [Google Scholar]
- Pearse I.S, Krügel T, Baldwin I.T. Innovation in anti-herbivore defense systems during neopolypoloidy—the functional consequences of instantaneous speciation. Plant J. 2006;47:196–210. doi: 10.1111/j.1365-313X.2006.02776.x. doi:10.1111/j.1365-313X.2006.02776.x [DOI] [PubMed] [Google Scholar]
- Pereira L.A, Schoor S, Goubet F, Dupree P, Moffatt B.A. Deficiency of adenosine kinase activity affects the degree of pectin methyl-esterification in cell walls of Arabidopsis thaliana. Planta. 2006;224:1401–1414. doi: 10.1007/s00425-006-0323-z. doi:10.1007/s00425-006-0323-z [DOI] [PubMed] [Google Scholar]
- Pikaard C.S. The epigenetics of nucleolar dominance. Trends Genet. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. doi:10.1016/S0168-9525(00)02113-2 [DOI] [PubMed] [Google Scholar]
- Pires J.C, Zhao J.W, Schranz M.E, Leon E.J, Quijada P.A, Lukens L.N, Osborn T.C. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol. J. Linn. Soc. Lond. 2004;82:675–688. doi:10.1111/j.1095-8312.2004.00350.x [Google Scholar]
- Pruitt R.E, Vielle-Calzada J.-P, Ploense S.E, Grossniklaus U, Lolle S.J. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl Acad. Sci. USA. 2000;97:1311–1316. doi: 10.1073/pnas.97.3.1311. doi:10.1073/pnas.97.3.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu N, Schittko U, Baldwin I.T. Consistency of Nicotiana attenuata's herbivore- and jasmonate-induced transcriptional responses in the allotetraploid species Nicotiana quadrivalvis and Nicotiana clevelandii. Plant Physiol. 2004;135:539–548. doi: 10.1104/pp.103.037036. doi:10.1104/pp.103.037036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada M.N, Benito M.I, Walbot V. The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 2001;25:79–91. doi: 10.1046/j.1365-313x.2001.00939.x. doi:10.1046/j.1365-313x.2001.00939.x [DOI] [PubMed] [Google Scholar]
- Ramsey J. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae) Heredity. 2007;98:143–150. doi: 10.1038/sj.hdy.6800912. doi:10.1038/sj.hdy.6800912 [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske D.W. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 2002;29:589–639. doi:10.1146/annurev.ecolsys.33.010802.150437 [Google Scholar]
- Riddle N.C, Birchler J.A. Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 2003;19:597–600. doi: 10.1016/j.tig.2003.09.005. doi:10.1016/j.tig.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 1997;28:359–389. doi:10.1146/annurev.ecolsys.28.1.359 [Google Scholar]
- Rieseberg L.H, Willis J.H. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. doi:10.1126/science.1137729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H, Sinervo B, Linder C.R, Ungerer M.C, Arias D.M. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science. 1996;272:741–745. doi: 10.1126/science.272.5262.741. doi:10.1126/science.272.5262.741 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, Archer M.A, Wayne R.K. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. doi:10.1038/sj.hdy.6886170 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H, Widmer A, Arntz A.M, Burke J.M. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Phil. Trans. R. Soc. B. 2003;358:1141–1147. doi: 10.1098/rstb.2003.1283. doi:10.1098/rstb.2003.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A, Ainouche M.L, Wendel J.F. Genetic and epigenetic consequences of recent hybridisation and polyploidy in Spartina (Poaceae) Mol. Ecol. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. doi:10.1111/j.1365-294X.2005.02488.x [DOI] [PubMed] [Google Scholar]
- Salone V, Rüdinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. doi:10.1016/j.febslet.2007.07.075 [DOI] [PubMed] [Google Scholar]
- Shaked H, Kashkusk K, Ozkan H, Feldman M, Levy A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridisation and allopolyploidy in wheat. Plant Cell. 2001;13:1749–1759. doi: 10.1105/TPC.010083. doi:10.1105/tpc.13.8.1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart C.J, Monéger F, Leaver C.J. Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell. 1994;6:811–825. doi: 10.1105/tpc.6.6.811. doi:10.1105/tpc.6.6.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E, Soltis P.S. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. doi:10.1016/S0169-5347(99)01638-9 [DOI] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn T.C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl Acad. Sci. USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. doi:10.1073/pnas.92.17.7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R.M, Hermanson P.J, Springer N.M. Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. 2007;145:411–425. doi: 10.1104/pp.107.101428. doi:10.1104/pp.107.101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol. 2006;140:879–889. doi: 10.1104/pp.105.073825. doi:10.1104/pp.105.073825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J.A, Soltis D.E, Soltis P.S. Polyploidy in plants. In: Gregory T.R, editor. The evolution of the genome. Elsevier Science & Technology, Academic Press; London, UK: 2005. pp. 371–426. [Google Scholar]
- Ungerer M.C, Baird S.J.E, Pan J, Rieseberg L.H. Rapid hybrid speciation in wild sunflowers. Proc. Natl Acad. Sci. USA. 1998;95:11 757–11 762. doi: 10.1073/pnas.95.20.11757. doi:10.1073/pnas.95.20.11757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, Knowles V.L, Snedden W.A, Plaxton W.C. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 2006;142:1282–1293. doi: 10.1104/pp.106.087171. doi:10.1104/pp.106.087171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee H.-Y, Chen Z.J. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics. 2006a;173:965–974. doi: 10.1534/genetics.106.056580. doi:10.1534/genetics.106.056580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006b;172:507–517. doi: 10.1534/genetics.105.047894. doi:10.1534/genetics.105.047894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C.F, Wessler S.R. Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell. 1993;5:512–522. doi: 10.1105/tpc.5.5.515. doi:10.1105/tpc.5.5.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000;42:225–249. doi:10.1023/A:1006392424384 [PubMed] [Google Scholar]
- Williams C.E, Collier C.C, Nemacheck J.A, Liang C, Cambron S.E. A lectin-like wheat gene responds systemically to attempted feeding by avirulent first-instar Hessian fly larvae. J. Chem. Ecol. 2002;28:1411–1428. doi: 10.1023/a:1016200619766. doi:10.1023/A:1016200619766 [DOI] [PubMed] [Google Scholar]
- Zhu H, Qian W, Lu X, Li D, Liu X, Liu K, Wang D. Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 2005;59:581–594. doi: 10.1007/s11103-005-0183-0. doi:10.1007/s11103-005-0183-0 [DOI] [PubMed] [Google Scholar]