Abstract
New Zealand taxa from the Orthopteran family Anostostomatidae have been shown to consist of three broad groups, Hemiandrus (ground weta), Anisoura/Motuweta (tusked weta) and Hemideina–Deinacrida (tree–giant weta). The family is also present in Australia and New Caledonia, the nearest large land masses to New Zealand. All genera are endemic to their respective countries except Hemiandrus that occurs in New Zealand and Australia. We used nuclear and mitochondrial DNA sequence data to study within genera and among species-level genetic diversity within New Zealand and to examine phylogenetic relationships of taxa in Australasia. We found the Anostostomatidae to be monophyletic within Ensifera, and justifiably distinguished from the Stenopelmatidae among which they were formerly placed. However, the New Zealand Anostostomatidae are not monophyletic with respect to Australian and New Caledonian species in our analyses. Two of the New Zealand groups have closer allies in Australia and one in New Caledonia. We carried out maximum-likelihood and Bayesian analyses to reveal several well supported subgroupings. Our analysis included the most extensive sampling to date of Hemiandrus species and indicate that Australian and New Zealand Hemiandrus are not monophyletic. We used molecular dating approaches to test the plausibility of alternative biogeographic hypotheses for the origin of the New Zealand anostostomatid fauna and found support for divergence of the main clades at, or shortly after, Gondwanan break-up, and dispersal across the Tasman much more recently.
Keywords: Anostostomatidae, biogeography, COI, 18S, Zealandia, New Zealand
1. Introduction
The biology of New Zealand is, unlike that of most Pacific islands, viewed as continental in nature (Cowie & Holland 2006). This is justified geologically because New Zealand is formed from continental rather than oceanic crust (Neall & Trewick 2008). Consequently, the biota of New Zealand is considered to be predominantly ‘Gondwanan’, having its principal affinities in Australasia (Fleming 1979) and the Southern Hemisphere in general (Gibbs 2006). Although the Gondwanan nature of the New Zealand biota is often attributed to the continental (vicariant) history of the land, this explanation has not always been pre-eminent (e.g. Fleming 1962; Caughley 1964). The importance of dispersal is widely recognized, and it is more generally accepted that biogeographical pattern alone does not reveal the process(es) of origination of biota (Waters & Craw 2006). Indeed, the distribution of one former icon of vicariance biogeography in New Zealand (and the Southern Hemisphere), southern beech (Nothofagus) has recently been shown to be best explained by dispersal to New Zealand (Cook & Crisp 2005a; Knapp et al. 2005). Numerous other molecular studies of a range of taxa demonstrate that a substantial (if not predominant) part of the New Zealand biota are products of long-distance dispersal (Waters & Craw 2006 and references therein; Trewick et al. 2007).
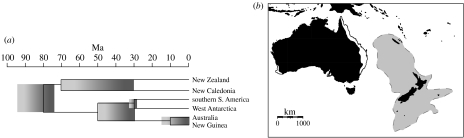
An improving, though far from perfect understanding of the tectonic history in the New Zealand region (Mortimer 2004) is helping to reveal why this continental land does not in fact have a predominantly continental biota. The rifting of the continent Zealandia from Gondwana (including Australia), commenced ca 83 Ma (figure 1a) and the Tasman Sea reached its current width between 63.5 and 55.5 Ma (Veevers & Li 1991; McLoughlin 2001). This was just the start of New Zealand and New Caledonia's story. Zealandia (figure 1b) subsequently sank so that today approximately 93 per cent of the continent is below the surface of the sea (Landis et al. 2008).
Figure 1.
(a) Geological area cladogram, after Cook & Crisp (2005b) showing the accepted Gondwanan break-up sequence. Shaded boxes represent uncertainty about timing of vicariant events. (b) Australasia sampled for anostostomatid weta (Insecta: Orthoptera) in this study (Australia, New Caledonia and New Zealand). Australian species are limited to the east coast of Queensland and New South Wales and one genus in Western Australia (white fill) whereas most parts of New Zealand and New Caledonian support one or more weta species. The dashed outline and light grey shading indicate the approximate boundaries of the submerged continent Zealandia.
This active geological history contrasts with the prolonged geological stability of Australia, the nearest remnant of continental Gondwana. With respect to land area, climate and biotic assemblage, New Zealand (and New Caledonia) has few continental attributes and ample evidence suggests that these islands are biologically more like oceanic islands than southern continents (Goldberg et al. 2008; Grandcolas et al. 2008; Trewick & Morgan-Richards in press).
Evidence for the persistence of land in the New Zealand region throughout the Oligocene has been obscured by the extensive tectonic activity initiated in the early Miocene (Landis et al. 2008). The tectonic upheaval that resulted in the formation of New Zealand (as we know it today) began ca 24 Ma and still continues (Trewick et al. 2007). For example, the major mountain ranges of New Zealand started forming only ca 5 Ma. This, and other local geophysical events, may have been more important in the development of the modern biota than ancient vicariant processes. New Caledonia has a similar geological history with tectonic activity forcing a submerged section of Zealandia (and obducted oceanic ultramafic strata) to the sea surface in the late Eocene (ca 40 Ma Chardon & Chevillotte 2006; Mortimer et al. 2006; Grandcolas et al. 2008; Neall & Trewick 2008).
One of the most interesting components of New Zealand's terrestrial fauna, with both taxonomic and ecological diversity, are insects of the orthopteran family Anostostomatidae, known in New Zealand by their Maori name, weta. Of particular biogeographic interest is the presence of the family on all three major Australasian landmasses: Australia, New Caledonia and New Zealand. The group consists of relatively large insects (20–80 mm) that are nocturnal, predominantly flightless and predatory, with a Gondwanan distribution (also found in Central and South America, South Africa, Madagascar and India). In New Zealand, the family is represented by five genera and approximately 56 species. These five genera fall into three distinct groups: (i) nine (plus approx. 30 undescribed) species of Hemiandrus Ander 1938 (ground weta), (ii) one species of Anisoura Ander 1938 and two species of Motuweta Johns 1997 (tusked weta), and (iii) seven Hemideina White 1846 (tree weta) and 11 Deinacrida White 1842 (giant weta) (Trewick & Morgan-Richards 2004, 2005).
The Hemideina and Deinacrida are unusual among Anostostomatidae in that all species are primarily herbivorous. The diversification of Hemideina–Deinacrida dates to the Miocene, with adaptation to diverse habitats following mountain uplift (ca 5 Ma Trewick & Morgan-Richards 2005). The three tusked weta species (Anisoura/Motuweta), so named owing to the impressive tusk-like structures on the mandibles of mature males, form a monophyletic group among New Zealand taxa (Trewick & Morgan-Richards 2004), although analogous ornamentations are found in some South African species (i.e. Libanasidus vittatus; Field & Deans 2001). Within the Australasian anostostomatid genera, Hemiandrus is the only genus not endemic to a single landmass, being recorded in both Australia and New Zealand (Johns 1997). Of the approximately 40 species from New Zealand (P. M. Johns 2005, personal communication) only nine are described (Johns 1997; Jewell 2007), making them the least well-characterized weta group in this country. Ovipositor length, which appears to be correlated with degree of maternal care (Gwynne 1995; Johns 1997; Gwynne 2004), was in the past the key morphological character distinguishing the genus Zealandosandrus Salmon 1950 from Hemiandrus Ander 1838. However, Johns (1997) synonymized Zealandosandrus, retaining Hemiandrus by precedent. The Australian anostostomatid fauna is poorly characterized with just 13 described species but probably comprises nine genera with approximately 60 species (Johns 1997; G. Monteith 1999, personal communication). Australia's fauna includes three genera with winged species, one of which (Transaevum) is considered to be the most ‘ancestral’ extant member of the group (Monteith & Field 2001). Intriguingly, fossil Orthoptera that are putatively ‘weta’ are reported from 190 Ma deposits in Queensland, Australia (Meads 1990) but have not been formally described. The New Caledonian weta fauna consists of two genera, Aistus (three species with several undescribed; Johns 1997) and Carcinopsis (six species; Johns 1997).
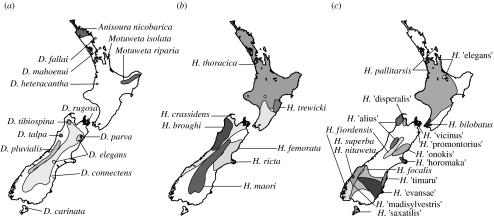
Anostostomatids occupy a variety of environments across their geographical range in Australasia. In New Caledonia, they are tropical forest inhabitants; in Australia, almost all are found in the wet tropical forests of Queensland with one genus endemic to coastal Western Australia (Monteith & Field 2001; figure 1b). The New Zealand weta live mostly in temperate forest and subalpine environments, with the majority of species in South Island (figure 2). Curiously, anostostomatids are absent from comparable temperate forest in southern Australia (Victoria and Tasmania).
Figure 2.
Approximate distributions of New Zealand Anostostomatidae taxa: (a) Deinacrida (giant weta) and Anisoura/Motuweta (tusked weta) species; (b) Hemideina (tree weta) (based on Trewick & Morgan-Richards in press); (c) Hemiandrus (ground weta) based on Johns (2001), Jewell (2007) and P. M. Johns (2005, personal communication). In addition to the species shown, H. maculifrons is widely distributed throughout both the North and South Islands. Additional undescribed taxa appear to have local distributions and predominate in South Island (P. M. Johns 2005, personal communication).
We have undertaken sampling across New Zealand, New Caledonian and Australia to explore the evolution of the New Zealand Anostostomatidae. Representatives of all nine Australian genera, both New Caledonian genera and the five New Zealand genera were included in the present study. We used molecular phylogenetics to recover support for the relationships of the three New Zealand anostostomatid groups. We include representatives of New Zealand Hemiandrus species diversity, which have been absent from previous work, in order to explore diversity within this group and test support for monophyly of the genus. We apply relaxed molecular clock methods, to estimate the likely age and origin of New Zealand weta lineages.
2. Material and methods
(a) Sampling
The majority of sampling was undertaken by the authors in New Zealand and New Caledonia. In addition, samples of Hemiandrus were supplied by Darryl Gwynne (University of Toronto, Canada). The New Zealand sampling included all three tusked weta species (Anisoura/Motuweta), representatives of the Hemideina and Deinacrida (previously shown to be monophyletic; Trewick & Morgan-Richards 2005) and representatives within the taxonomic diversity of Hemiandrus including putative and new species (see the electronic supplementary material). Assistance with identification and sampling of undescribed species was provided by P. M. Johns. Material from Australia was supplied by Geoff Monteith (Brisbane Museum, Australia) and Dave Rentz (CSIRO, Australia). This sampling, although not exhaustive, includes at least one representative of each genus in the region and is the most complete dataset to date.
(b) Molecular methods
Whole genomic DNA was extracted from hind leg muscle following the salting-out method (Sunnucks & Hale 1996) and resuspended in 50 μl TE buffer (0.1 mM EDTA, 10 mM Tris) or water. Polymerase chain reactions (PCRs) were performed in 10 μl volume using ABgene Red Hot Taq. Products were visualized on 1 per cent agarose gels stained with SYBRSafe (Invitrogen). Thermal cycling PCR was carried out on an MJ Research PTC-200 thermal cycler and consisted of initial denaturation of 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 48–50°C for 30 s and 72°C for 1 min 30 s with a final extension of 72°C for 3 min. PCR products were purified with Shrimp Alkaline Phosphatase and Exonuclease I following manufacturer's recommendations (USB Corporation). Sequencing used BigDye Terminator v. 3.1 chemistry and an ABI3730XL Genetic Analyzer (Applied Biosystems; Foster City, CA). DNA sequences were deposited on NCBI GenBank (see the electronic supplementary material; EU676657–EU676800 and EU713453–EU13461). Primers used for PCR were the following: mtDNA COI: LCO1490, HCO2198 (Folmer et al. 1994), CI-J-2195 and L2-N-3014 (Simon et al. 1994); mtDNA third domain 12S rRNA: SR-N-14588 and LR-J-13417 (Simon et al. 1994); nuclear rRNA 18S: 18S-S22, 18S-A1984 (Vawter 1991), 18S_1F (gac gaa aaa taa cga tac ggg) and 18S_1R (ctc aat ctg tca atc ctt cca) (this study); and 28S: 28SrD1.2a, 28SrD3.2a, 28SrD4.8a, 28SA, 28SrD4.2b, 28SrD5b, 28SrD7bl and 28SB (Whiting 2002a).
(c) Phylogenetic analysis
Individual sequence reads were checked against ABI trace files using Sequencher v. 4.70 (Gene Codes Corp. Ann Arbour, MI) and aligned using Se-Al v2.0a11 (Rambaut 1996). The protein-coding gene COI was translated into amino acids to ensure correct reading frame and to detect evidence of nuclear copies (which were subsequently removed). Ribosomal RNA genes were checked for indels. In cases where missing data were included, they were coded as N in analyses. In order to evaluate individual genes and concatenated data, we divided the datasets into the following: I—18S Ensifera; II—18S Australasian Anostostomatidae; III—combined 18S and 28S Australasian Anostostomatidae; IV—COI–RY-coded Australasian Anostostomatidae; V—combined COI–RY and 12S Australasian Anostostomatidae; and VI—COI Hemiandrus only.
The COI data were partitioned into three character sets according to the codon position, first, second and third. In order to maximize third codon information, we treated it in three different ways: as four nucleotides (A, G, T, C), Y-coded (Y, A, G) or RY-coded (A and G=R, T and C=Y). In order to avoid potential tree estimation bias due to nucleotide composition or saturation, we used Y or RY coding on the third codon position nucleotides for COI sequences in dataset IV and V. Recoding of this sort has been shown to greatly improve consistency in phylogenetic resolution by reducing bias from differences in nucleotide composition (Phillips & Penny 2003), which is useful when looking at deeper divergences. To assist with tree rooting and thus confirm ingroup status of our sample, we used published Ensifera DNA sequences from both EMBL and NCBI GenBank (see the electronic supplementary material).
Models of DNA evolution were optimized separately for each dataset using Modeltest v. 3.7 (Posada & Crandall 1998) and Akaike Information Criterion was preferred to the hierarchical likelihood ratio test (Posada & Buckley 2004). Maximum-likelihood (ML) analyses were implemented using the programs PAUP* (Swofford 2003), GARLI v. 0.951 (Zwickl 2006) and PhyML (Guindon & Gascuel 2003). Model parameters from Modeltest were implemented using a general time-reversible model with invariable sites and a gamma distribution for variable rate sites (GTR+I+G) model with a heuristic search under the likelihood criterion with trees obtained from stepwise addition.
Bayesian analyses were implemented using MrBayes v. 3.1 (Huelsenbeck & Ronquist 2001). We specified nst=2 (HKY) and nst=6 (GTR) with a proportion of invariant sites and gamma distribution of rate variation. Analyses of datasets III (18S+28S), IV (COI) and V (COI +12S) were undertaken with (parameters unlinked) and without character set partitions. We used two runs of four Markov chains (each with one cold chain) with 1–10×106 generations and default priors, sampling every thousandth tree. A ‘burn-in’ of 10 per cent was removed after examination of log-likelihood scores and average standard deviation of the split frequencies. Trees saved below the burn-in generation were discarded and a majority rule consensus of the remaining trees was calculated. Multiple replicates of the Bayesian runs were carried out to insure convergence of the posteriors.
(d) Tree comparisons
We assessed the degree of conflict between our phylogenetic estimates by using tree comparison tests, to see if one topology was significantly better at explaining the molecular data than alternative phylogenies. We used the SH tests (Shimodaira & Hasegawa 1999) implementing a RELL distribution derived from 1000 bootstrap replicates as executed in PAUP*. For dataset IV (COI), we carried out multiple analyses manipulating the third codon position so that it was; four states, Y-coded and RY coded. To observe the effect of this simple noise reduction technique, we compared ML topologies obtained from PhyML for each state using either a simple model (HKY85) or a parameter-rich model (GTR +I+G). We also used constraint analysis to test the likelihood of alternative tree topologies for the monophyly of New Zealand taxa and the genus Hemiandrus (New Zealand and Australia).
(e) Divergence time estimation
We compared the likelihood scores obtained from ML analysis both with and without the implementation of a molecular clock in PAUP* for dataset II (18S Australasia) and dataset IV (COI–RY-coded Australasia). This was carried out both with and without the inclusion of taxa we suspected of having a rate shift due to long branches observed in initial analyses. SH tests were applied to resulting trees to determine whether there was rate heterogeneity and therefore if the data were acting in a clock-like manner to determine whether to use a strict or relaxed molecular clock in BEAST v. 1.4.6 (Drummond & Rambaut 2007).
As there are no suitable fossils for molecular dating, we used geological events as points of reference to test the plausibility of vicariant versus dispersal explanations for the New Zealand weta diversification (figure 1a). In order to explicitly examine the alternative hypotheses for patterns of diversity, we calibrated trees using initial separation of Zealandia from Gondwana (less than 82 Ma as applied by avian evolutionists, see Ericson et al. 2002; Baker et al. 2005) and emergence of New Caledonian (less than 40 Ma). The two dating constraints were separately applied to the nuclear dataset II (18S Australasia) and mitochondrial dataset IV (COI–RY-coded Australasia). We removed a clade of five taxa (clade A plus New Caledonian taxa), shown by initial analyses to have long branches and a substantially elevated rate of molecular evolution (indicated by BEAST rates). First, if Zealandia and Australia parted ca 82 Ma, we assumed vicariance and constrained the most basal split of Anostostomatidae to more than 82 Ma (BEAST parameters; relaxed lognormal clock, lognormal distribution, mean=4.74; s.d.=0.2, run for 100 million repetitions sampling trees every 10 000). Second, we constrained the same point on the tree as above, but assuming Zealandia submerged completely and land resurfaced ca 40 Ma (BEAST parameters; normal distribution: mean=36, s.d.=0.2, initial value=35). We used a root value for the trees (BEAST parameters; uniform distribution: lower limit=85, upper limit=250, initial=100) as unpublished fossils have been dated from 190–200 Ma from Queensland that have been attributed to the Anostostomatidae and fossil Ensifera are dated back to ca 250 Ma (fig. 7.18, p. 202, Grimaldi & Engel 2005). Calibration points were implemented in BEAST using a relaxed uncorrelated lognormal molecular clock (Drummond et al. 2006). Resulting trees were analysed using software distributed with BEAST–TreeAnnotator v. 1.4.6 where the burn-in (1000 trees) was removed and a maximum credibility tree constructed. Trees were then viewed in FigTree v. 1.1.2. Details of the XML files are available on request from the authors.
3. Results
A summary of all sequence data collected can be viewed in the electronic supplementary material along with locality information and accession numbers. NCBI BLAST searches returned matches for previously published Orthopteran sequences. The alignment used in each analysis is available as a Nexus file from the authors on request.
(a) Phylogeny
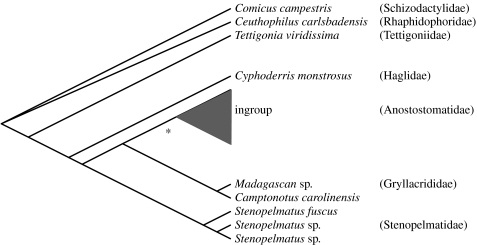
Dataset I (18S Ensifera) consisted of 1863 bp after removal of a 37 bp hypervariable indel region between bp 719 and 756 from our original alignment to accommodate the diverse range of taxa, which included seven ingroup taxa and nine outgroup Ensifera (Flook et al. 1999; Terry & Whiting 2005; Jost & Shaw 2006). We confirm the monophyly of Anostostomatidae in our sample and found the Gryllacrididae to be sister to Anostostomatidae with Stenopelmatidae sister to the Anostostomatidae–Gryllacrididae clade. Both of these families have previously been suggested as close relatives to Anostostomatidae (figure 3).
Figure 3.
Bayesian cladogram for 1863 bp of nuclear 18S Ensifera data including seven ingroup taxa (EU676721, EU713453, EU713454, EU713455, EU713458, EU713459 and Z97570) and published taxa (Flook et al. 1999; Terry & Whiting 2005; Jost & Shaw 2006) used to determine appropriate outgroup (see the electronic supplementary material). The hypervariable indel region between bp 719 and 756 was removed due to alignment uncertainty. The asterisk node indicates Bayesian posterior probability of 0.96.
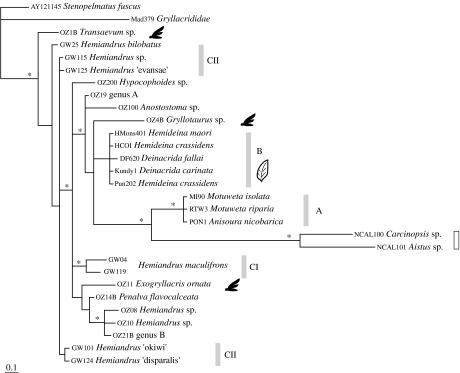
Dataset II-18S Australasian Anostostomatidae: After establishing support for monophyly of the Anostostomatidae, we turned our focus to the relationships within the family. We included more representatives from the Australasian region and a slightly shorter fragment of 18S (29 taxa, 1746 bp), again excluding the problematic indel region. Bayesian and ML analyses yielded similar topologies (figure 4). We observed that the New Zealand tusked weta (Anisoura/Motuweta; clade A) and New Caledonian taxa (Aistus and Carcinopsis) formed long branches in the phylogeny. Long branches like these can result in misleading results even without rate differences (Hendy & Penny 1989) that affect all further tree selection criteria. We explored the effect of these long branches by subjecting the dataset to identical analyses with the inclusion or exclusion of either or both the New Caledonian taxa (Aistus and Carcinopsis), or the tusked weta (Anisoura/Motuweta) sequences. The exclusion of Anisoura/Motuweta resulted in Aistus and Carcinopsis together being placed as sister to the rest of the Anostostomatidae, from which we infer long branch attraction, resulting from lineage-specific rate increases. When Aistus and Carcinopsis were removed, the Anisoura/Motuweta (clade A; figure 4) were sister to the Hemideina–Deinacrida (clade B; figure 4). In no instances were Hemiandrus (New Zealand and Australian) found to be monophyletic, a finding consistent in the following analyses with nuclear and mitochondrial sequences. We identified two clades within the New Zealand Hemiandrus (CI and CII; figure 4) although not monophyletic in every analysis, both were separate from the Australian Hemiandrus species. Dataset III (concatenated 18S+28S) Australasian anostostomatids (35 taxa, 1549 bp) drew on data published by Jost & Shaw (2006) and to allow for direct comparison with that study, we cut our 18S sequences to 1204 bp and included 344 bp of 28S data for a subset of our taxa. We again failed to find the monophyly of Australian taxa or New Zealand taxa but the grouping of Anisoura/Motuweta (clade A) with Aistus and Carcinopsis as sister to Hemideina–Deinacrida (clade B) was supported (tree not shown).
Figure 4.
Bayesian phylogram for 1746 bp of nuclear genes 18S including representatives of the Australasian anostostomatids. The grey and white bars indicate New Zealand and New Caledonian taxa, respectively. Clades of New Zealand taxa are indicated as A, B, CI and CII. The single asterisks at nodes indicate Bayesian posterior probabilities above 0.9.
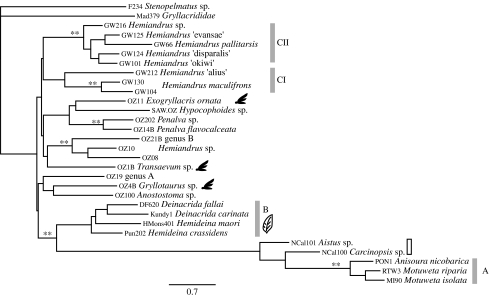
Dataset IV (COI sequences from Australasian taxa), 1225 bp aligned DNA sequence data for 28 ingroup taxa, plus two outgroup taxa (one Madagascan gryllacridid (Mad 379) and one South African stenopelmatid (F234); see the electronic supplementary material). With RY-coded mitochondrial COI data, our phylogenetic analyses returned clades consistent with those observed in analysis of 18S data (figure 4), including strong support for multiple, distinct weta lineages in New Zealand (clades A, B, CI, CII; figure 5). This result was strongly supported by the SH test constraining New Zealand taxa to form a monophyletic lineage (p<0.0001). Anisoura/Motuweta (clade A) was again found to be sister to Aistus and Carcinopsis and collectively were sister to the Hemideina/Deinacrida (clade B; figure 5). We examined the translated amino acid sequence and observed 10 changes shared by Aistus, Carcinopsis and Anisoura/Motuweta (out of 409) clearly separating them from the rest of Anostostomatidae. Analysis of dataset V, a reduced set of taxa with concatenated sequences, consisted of COI (1225 bp) and 12S (435 bp) for Australasian Anostostomatids (25 taxa, 1860 bp). Analyses returned a similar topology with the same taxon subgroupings and no support for the New Zealand monophyly as with previous datasets (not shown).
Figure 5.
ML phylogram for 1225 bp of the mitochondrial COI gene representing Australasian anostostomatids. Sequences were RY coded on the third codon position to reduce noise while retaining phylogenetic signal. Winged silhouette indicates winged species and the leaf symbol indicates herbivorous species. The grey and open bars indicate New Zealand and New Caledonian taxa, respectively. Clades of New Zealand taxa are indicated as A, B, CI and CII. The double asterisks at nodes indicate Bayesian posterior probabilities of 0.99 and above.
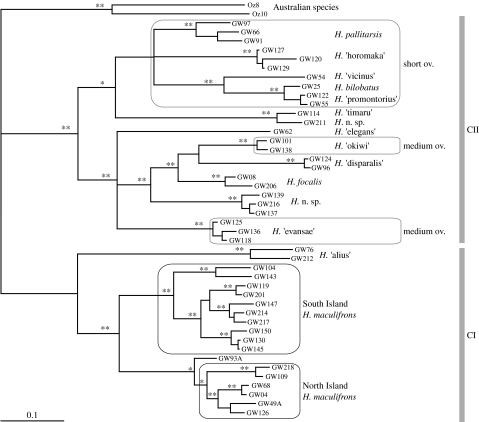
Analysis of dataset VI (COI Hemiandrus only), for 46 individuals from at least 15 New Zealand species and two Australian Hemiandrus species (848 bp), revealed high genetic diversity (mean genetic distance, 6.5 and 7.2% for the North and South Islands, respectively) among samples of the widespread New Zealand taxon Hemiandrus maculifrons. Samples of H. maculifrons formed two main clades, one from South Island and one from North Island (clade CI, figure 6), with a specimen from northern South Island (sample GW93A from Pelorus Bridge) sister to the North Island clade. By contrast, most other New Zealand Hemiandrus species were closely related to one another (e.g. 3.9% between H. bilobatus and H. ‘promontorius’), and these species tend to have narrow geographical ranges. All ground weta (whether North Island or South Island) species with extremely reduced/short ovipositors form a monophyletic group with long ovipositor species paraphyletic to this. Two taxa with medium ovipositors (H. ‘okiwi’ and H. ‘evansae’) each are sister to long ovipositor species, and not together. Other anostostomatids, including the sister Australian ‘Hemiandrus’, have long ovipositors and therefore the reduction of ovipositor length (along with the unusual maternal care that appears to correlate with these conditions) appears to have evolved at least three times in New Zealand. Investigation into monophyly of the genus using the SH test on dataset IV revealed that constraining all Hemiandrus (Australian and New Zealand) to be monophyletic resulted in a tree with a significantly worse likelihood score (p<0.0001).
Figure 6.
Bayesian phylogram of the genus Hemiandrus for 848 bp of the mitochondrial COI gene. Species with short or medium-length ovipositor are indicated; all other species have long ovipositors. The two New Zealand clades CI and CII are indicated. The single and double asterisks at nodes indicate Bayesian posterior probabilities above 0.9 and 0.99, respectively.
(b) Summary of phylogenetics
New Zealand weta are not monophyletic, instead forming three (or four) clades. The New Zealand tusked weta (clade A) genera are more closely related to the New Caledonian taxa (Aistus sp. and Carcinopsis sp.; figures 4 and 5) with no apparent close Australian relative. New Zealand's large herbivorous tree and giant weta (Hemideina and Deinacrida) form a distinct clade that is strongly supported with less than 1 per cent sequence divergence in the nuclear rDNA gene 18S (clade B, figure 4). All our analyses returned a clade consisting of Aistus, Carcinopsis, Anisoura/Motuweta (clade A) and Hemideina–Deinacrida (clade B). However, long branches due to an apparent rate shift in the Aistus, Carcinopsis and Anisoura/Motuweta (clade A) lead to us remove these taxa from further analyses, so this relationship is not fully understood. The third New Zealand clade, the Hemiandrus, consists of two lineages (clades CI and CII; figures 4–6) that may not be sisters. One lineage consists of only H. maculifrons and H. ‘alius’ while the other includes 11+ species sampled (figure 6). Finally, the nine Australian taxa are not monophyletic but form four clades throughout the tree, sister to New Zealand clades; however, there is little BPP support for these nodes. This is consistent with the short branch lengths obtained at the base of the tree.
Dataset IV (COI–RY-coded Australasia; figure 5) returned three Australian clades of interest: (i) winged Transaevum sister to the non-winged Australian Hemiandrus and genus B, (ii) winged Exogryllacris sister to the non-winged Hypocophoides and Penalva, and (iii) winged Gryllotaurus sister to the non-winged Anostostoma and genus A. These three Australian clades were not resolved in the analysis of dataset II (18S Australasia; figure 4) but the three winged species were never monophyletic and Transaevum was sister to all the other Anostostomatidae.
(c) Divergence estimation
Edge lengths on branches suggest that Aistus and Carcinopsis along with Anisoura/Motuweta (clade A; figures 4 and 5) may have an increased rate of mutation. In order to test for this, we compared ML trees with and without a clock enforced and found evidence in both 18S and COI datasets for a significant deviation from a clock-like model of evolution. By removing Aistus, Carcinopsis and Anisoura/Motuweta and running the ML analysis again, the significance of the SH test decreased.
We carried out relaxed-clock analyses on 18S (dataset II) and COI (dataset IV). Our calibration points assumed either existence of all anostostomatid weta lineages prior to the separation of Zealandia (82 Ma) or emergence of New Caledonia (40 Ma). However, due to the observed rate change in Aistus, Carcinopsis and Anisoura/Motuweta, we removed this clade (five sequences) from the dating analysis.
Estimates of the rate of evolution of COI vary a great deal, primarily due to the disparity between population- and species-level rates (time-dependent rates or J-shape curve; Ho et al. 2005; Penny 2005). Here, we restrict our comparisons to recent studies that examine family-level divergences of insects, which estimate the rate of evolution for COI and COII of insects at approximately 0.007–0.012 substitutions per site per million years (Zakharov et al. 2004 and references therein). In our relaxed molecular clock phylogenetic analysis, constraining the base of the Anostostomatidae clade to 82 Ma, our estimated mutation rates for COI were between 0.0097 and 0.0376 [95% CI], compared with a mutation rate of 0.0223–0.1219 when applying the 40 Ma constraint. The mutation rate derived using the 82 Ma constraint is more similar to published data for other insects (Zakharov et al. 2004), and therefore supports divergences within Anostostomatidae of more than 80 Ma. Our relaxed-clock analyses of COI data suggest that both New Zealand Hemiandrus and Hemideina–Deinacrida lineages may have diverged from Australian relatives more than 82 Ma. Analysis of our 18S sequences with the 82 Ma constraint estimates divergence for New Zealand Hemiandrus clades that extend to before continental break-up (45–120 Ma and 11–103 Ma), but the Hemideina–Deinacrida clade was estimated to have diverged post-Gondwanan break-up (3–38 Ma). Thus, our sequence data support the idea that some weta lineages may have diverged before the separation of Zealandia from Australia but also that dispersal has since occurred. The phylogenetic placement of Aistus, Carcinopsis and Anisoura/Motuweta as sister to Hemideina–Deinacrida suggests genetic exchange between New Zealand and New Caledonia after separation of Zealandia.
4. Discussion
Despite comprehensive morphological studies, phylogenetic relationships within the Ensifera are poorly understood (Gwynne 1995; Whiting 2002b; Desutter-Grandcolas 2003). Johns (1997) removed taxa from Stenopelmatidae to form Anostostomatidae, a separation subsequently supported by molecular analyses (Jost & Shaw 2006). Although we are not concerned here with deeper Ensiferan relationships, it is important to know that our taxon set comprises a true ingroup. We found support for the monophyly of Anostostomatidae in our analysis (0.96 BPP) and for the close relationship with the Gryllacrididae and Stenopelmatidae (figure 3), supporting previous inferences (Jost & Shaw 2006; P. M. Johns 2007, personal communication). However, we did not find evidence of a sister relationship of Deinacridinae (Hemideina and Deinacrida) and Anostostomatidae (rest of the family; Johns 1997; Gorochov 2001).
For the first time, we have shown that members of the family Anostostomatidae are not monophyletic in New Zealand or Australia. To explain the phylogenetic diversity of the New Zealand weta by vicariance requires that at least four distinct clades of Anostostomatidae were already present in Gondwana before Zealandia split from Australia, and that some of these subsequently went extinct in Australia. On the face of it, this seems an unlikely scenario, given the small size and geological activity of New Zealand compared with Australia, and indeed this has been shown to be a poor explanation for the distribution of Nothofagus beech in the region (Cook & Crisp 2005a). Although we found some variation in node dates inferred from COI and 18S data, we have to reject the hypothesis that all New Zealand lineages arose before continental break-up (ca 82 Ma). However, relaxed molecular clock calibrated phylogenies do suggest that some New Zealand clades may have formed before continental separation. These inferred early splits are consistent with a vicariant origin and survival of some Anostostomatidae lineages on Zealandia throughout the Oligocene marine transgression. Taxa missing from analyses (owing to extinction) will always result in long unbroken branches in phylogenetic trees and thus the inference of great age since common ancestors (Cook & Crisp 2005b) whereas recent splits (short branches) cannot be made older by the inclusion of ‘missing taxa’.
Colonization of New Zealand from the Australian biota, which includes three separate winged lineages, might have been facilitated by increasing land area after the Oligocene (less than 22 Ma). Dispersal events continue today, and include the establishment of an Australian Gryllacridid in recent years (Green & Ramsay 2003). The current study suggests that the two New Caledonia genera are more closely related to one of the New Zealand lineages but not to any Australian taxa. This is despite the comparatively close physical proximity and more similar climate of New Caledonia and Queensland, Australia. Despite evidence of an elevated substitution rate in both nuclear and mitochondrial genes, we observed clear phylogenetic evidence, supported by amino acid substitutions, for the sister relationship of the Aistus, Carcinopsis and Anisoura/Motuweta. Weta must have colonized New Caledonia after it emerged from the sea ca 40 Ma (Grandcolas et al. 2008), but our current taxon sampling is not sufficient to prove whether those weta ancestors came to or from New Zealand. Hemideina and Deinacrida were found to form a monophyletic group corresponding to the subfamily Deinacridinae Karny 1932. The lack of variation at the 18S gene among 18 species in these two genera is indicative of a recent radiation. Interestingly, Johns (1997) suggested that one of the tusked weta genera, Anisoura, should be included in the Deinacridinae; however, if Deinacridinae is to be valid, it should include all tusked weta genera plus the New Caledonian genera. Further morphological study is required to test this.
Our analysis of Australian and New Zealand Hemiandrus shows clearly that they represent separate lineages (figure 4). Furthermore, Hemiandrus in New Zealand consists of two distinct clades that may not be monophyletic. It is clear that taxonomic revision of the genus is required, with Hemiandrus being retained only for New Zealand taxa. Our data are the first to illustrate the depth of diversity within the New Zealand Hemiandrus that may amount to some 40 species (Johns 2001), using a broad range of habitats from alpine to lowland forest. We note relatively high genetic diversity in the most widespread species (H. maculifrons) that might justify separation as two or more allopatric species, but similar levels of sequence divergence are reported from other widespread weta with wide geographical ranges, e.g. Deinacrida connectens (Trewick et al. 2000) and Hemideina thoracica (Morgan-Richards et al. 2001). There is a stark contrast between the wide geographical range of H. maculifrons and the small ranges of the numerous, genetically similar, local endemics of South Island (figures 2 and 6). This pattern implies recent diversification in South Island, perhaps in response to habitat diversification since the Pliocene, which is also observed in the giant weta (figure 2) and many other taxa (Wagstaff & Garnock-Jones 1998; Lockhart et al. 2001; Trewick 2001; Chinn & Gemmell 2004; Trewick 2008). By contrast, much of the North Island is young (less than 1 Ma) and relatively homogeneous in terms of habitat (Trewick & Morgan-Richards in press).
In terms of geological history, Zealandia bears little resemblance to other larger, more stable southern continents that together originated from Gondwana. Instead, its history is more like that of many oceanic islands, undergoing extensive geological transformations that mark significant changes in climate and topology, and ultimately shaping the biota. The lack of monophyly within the Anostostomatidae fauna is not entirely unexpected, but the idea that some lineages represent more ancient links between Australia and New Zealand is exciting, as is the apparent exchange between New Zealand and the other Zealandian island, New Caledonia. This pattern might be indicative of the New Zealand biota as a whole, where some old lineages surviving from Zealandia, are all but overshadowed by more recent biotic exchange.
Acknowledgments
A special thank you to all those involved in collecting over the years: Geoff Monteith, Peter Johns, George Gibbs, Darryl Gwynne, Scott Dunavan. Special thanks to Matt Phillips and Simon Hills for guidance and assistance with the inner workings of BEAST. Thanks to Andrew Clarke, Sheralee Cleland, Barbara Holland, Manda Jost, David Penny and Michael Whiting for their constructive comments on the manuscript. This research was supported by funding from the Marsden Fund granted to David Penny and the Allan Wilson Centre for Molecular Ecology and Evolution. The Orthoptera Society grant was awarded to R.C.P. facilitating collaboration with Peter Johns. New Zealand Department of Conservation provided one Motuweta and two Deinacrida species.
Footnotes
One contribution of 15 to a Theme Issue ‘Evolution on Pacific islands: Darwin's legacy’.
Supplementary Material
References
- Baker A.J, Huynen L.J, Haddrath O, Millar C.D, Lambert D.M. Reconstructing the tempo and mode of evolution in an extinct clade of birds with ancient DNA: the giant moas of New Zealand. Proc. Natl Acad. Sci. USA. 2005;102:8257–8262. doi: 10.1073/pnas.0409435102. doi:10.1073/pnas.0409435102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughley G. Does the New Zealand vertebrate fauna conform to zoogeographic principles? Tuatara. 1964;12:50–57. [Google Scholar]
- Chardon D, Chevillotte V. Morphotectonic evolution of the New Caledonia ridge (Pacific Southwest) from post-obduction tectonosedimentary record. Tectonophysics. 2006;420:473–491. doi:10.1016/j.tecto.2006.04.004 [Google Scholar]
- Chinn W.G, Gemmell N.J. Adaptive radiation within New Zealand endemic species of the cockroach genus Celatoblatta Johns (Blattidae): a response to Plio-Pleistocene mountain building and climate change. Mol. Ecol. 2004;13:1507–1518. doi: 10.1111/j.1365-294X.2004.02160.x. doi:10.1111/j.1365-294X.2004.02160.x [DOI] [PubMed] [Google Scholar]
- Cook L.G, Crisp M.D. Not so ancient: the extant crown group of Nothofagus represents a post-Gondwanan radiation. Proc. R. Soc. B. 2005a;272:2535–2544. doi: 10.1098/rspb.2005.3219. doi:10.1098/rspb.2005.3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook L.G, Crisp M.D. Directional asymmetry of long-distance dispersal and colonization could mislead reconstructions of biogeography. J. Biogeogr. 2005b;32:741–754. doi:10.1111/j.1365-2699.2005.01261.x [Google Scholar]
- Cowie R.H, Holland B.S. Dispersal is fundamental to biogeography and the evolution of biodiversity on oceanic islands. J. Biogeogr. 2006;33:193–198. doi:10.1111/j.1365-2699.2005.01383.x [Google Scholar]
- Desutter-Grandcolas L. Phylogeny and evolution of acoustic communication in extant Ensifera (Insecta, Orthoptera) Zool. Scr. 2003;32:525–561. doi:10.1046/j.1463-6409.2003.00142.x [Google Scholar]
- Drummond A.J, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. doi:10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J, Ho S.Y.W, Phillips M.J, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson P.G.P, Christidis L, Cooper A, Irestedt M, Jackson J, Johansson U.S, Norman J.A. A Gondwanan origin of passerine birds supported by DNA sequences of the endemic New Zealand wrens. Proc. R. Soc. B. 2002;269:235–241. doi: 10.1098/rspb.2001.1877. doi:10.1098/rspb.2001.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field L.H, Deans N.A. Sexual selection and secondary sexual characters of wetas and king crickets. In: Field L.H, editor. The biology of wetas, king crickets and their allies. CABI Publishing; Wallingford, UK: 2001. pp. 179–204. [Google Scholar]
- Fleming C.A. New Zealand biogeography—a palaeontologist's approach. Tuatara. 1962;10:53–108. [Google Scholar]
- Fleming C.A. Oxford University Press; Auckland, New Zealand: 1979. The geological history of New Zealand and its life. [Google Scholar]
- Flook P.K, Klee S, Rowell C.H.F. Combined molecular phylogenetic analysis of the Orthoptera (Arthropoda, Insecta) and implications for their higher systematics. Syst. Biol. 1999;48:233–253. doi: 10.1080/106351599260274. doi:10.1080/106351599260274 [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Gibbs G. Craig Potton Publishing; Nelson, New Zealand: 2006. Ghosts of Gondwana: the history of life in New Zealand. [Google Scholar]
- Goldberg J, Trewick S.A, Paterson A.M. Evolution of New Zealand's terrestrial fauna: a review of molecular evidence. Phil. Trans. R. Soc. B. 2008;363:3319–3334. doi: 10.1098/rstb.2008.0114. doi:10.1098/rstb.2008.0114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorochov A.V. The higher classification, phylogeny and evolution of the superfamily Stenopelmatiodea. In: Field L.H, editor. The biology of wetas, king crickets and their allies. CABI Publishing; Wallingford, UK: 2001. pp. 3–33. [Google Scholar]
- Grandcolas P, Murienne J, Robillard T, Desutter-Grandcolas L, Jourdan H, Guilbert E, Deharveng L. New Caledonia: a very old Darwinian island? Phil. Trans. R. Soc. B. 2008;363:3309–3317. doi: 10.1098/rstb.2008.0122. doi:10.1098/rstb.2008.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green O.R, Ramsay G.W. A winged weta, Pterapotrechus (Orthoptera: Gryllacrididae), established in New Zealand. New Zeal. Entomol. 2003;26:75–77. [Google Scholar]
- Grimaldi D, Engel M.S. Cambridge University Press; New York, NY: 2005. Evolution of insects. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Gwynne D.T. Phylogeny of the Ensifera (Orthoptera): a hypothesis supporting multiple origins of acoustical signalling, complex spermatophores and maternal care in crickets, katydids, and weta. J. Orthopt. Res. 1995;4:203–218. doi:10.2307/3503478 [Google Scholar]
- Gwynne D.T. Reproductive behaviour of ground weta (Orthoptera: Anostostomatidae): drumming behaviour, nuptial feeding, post-copulatory guarding and maternal care. J. Kans. Entomol. Soc. 2004;77:414–428. doi:10.2317/E-34.1 [Google Scholar]
- Hendy M.D, Penny D. A framework for the quantitative study of evolutionary trees. Syst. Biol. 1989;38:297–309. [Google Scholar]
- Ho S.Y.W, Phillips M.J, Cooper A, Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. doi:10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jewell T. Two new species of Hemiandrus (Orthoptera: Anostostomatidae) from Fiordland National Park. New Zealand. Zootaxa. 2007;1542:49–57. [Google Scholar]
- Johns P.M. The Gondwanaland weta: family Anostostomatidae (formerly in Stenopelmatidae, Henicidae or Mimmermidae): nomenclatural problems, world checklist, new genera and species. J. Orthopt. Res. 1997;6:125–138. doi:10.2307/3503546 [Google Scholar]
- Johns P.M. Department of Conservation; Wellington, New Zealand: 2001. Distribution and conservation status of ground weta, Hemiandrus species (Orthoptera: Anostostomatidae) [Google Scholar]
- Jost M.C, Shaw K.L. Phylogeny of Ensifera (Hexapoda: Orthoptera) using three ribosomal loci, with implications for the evolution of acoustic communication. Mol. Phylogenet. Evol. 2006;38:510–530. doi: 10.1016/j.ympev.2005.10.004. doi:10.1016/j.ympev.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Knapp M, Stöckler K, Havell D, Delsuc F, Sebastiani F, Lockhart P.J. Relaxed molecular clock provides evidence for long distance dispersal of Nothofagus (Southern Beech) PLoS Biol. 2005;3:e14. doi: 10.1371/journal.pbio.0030014. doi:10.1371/journal.pbio.0030014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis C.A, Campbell H.J, Begg J.G, Paterson A.M, Trewick S.A. The Waipounamu erosion surface: questioning the antiquity of the New Zealand land surface and terrestrial fauna and flora. Geol. Mag. 2008;145:1–25. doi:10.1017/S0016756807004268 [Google Scholar]
- Lockhart P.J, McLenachan P.A, Havell D, Glenny D, Huson D, Jensen U. Phylogeny, radiation and transoceanic dispersal of New Zealand alpine buttercups: molecular evidence under split decomposition. Ann. Miss. Bot. Gard. 2001;88:458–477. doi:10.2307/3298586 [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Aust. J. Bot. 2001;49:271–300. doi:10.1071/BT00023 [Google Scholar]
- Meads M.J. DSIR Land Resources; Lower Hutt, New Zealand: 1990. The weta book: a guide to the identification of wetas. [Google Scholar]
- Monteith G.B, Field L.H. Australian king crickets: distribution, habitats and biology (Orthoptera: Anostostomatidae) In: Field L.H, editor. The biology of wetas, king crickets and their allies. CABI Publishing; Wallingford, UK: 2001. pp. 79–94. [Google Scholar]
- Morgan-Richards M, Trewick S.A, Wallis G. Chromosome races with Pliocene origins: evidence from mtDNA. Heredity. 2001;86:303–312. doi: 10.1046/j.1365-2540.2001.00828.x. doi:10.1046/j.1365-2540.2001.00828.x [DOI] [PubMed] [Google Scholar]
- Mortimer N. New Zealand's geological foundations. Gondw. Res. 2004;7:261–272. doi:10.1016/S1342-937X(05)70324-5 [Google Scholar]
- Mortimer N, Hoernle K, Hauff F, Palin J.M, Dunlap W.J, Werner R, Faure K. New constraints on the age and evolution of the Wishbone Ridge, southwest Pacific Cretaceous microplates, and Zealandia-West Antarctica breakup. Geology. 2006;34:185–188. doi:10.1130/G22168.1 [Google Scholar]
- Neall V.E, Trewick S.A. The age and origin of the Pacific Islands: a geological overview. Phil. Trans. R. Soc. B. 2008;363:3293–3308. doi: 10.1098/rstb.2008.0119. doi:10.1098/rstb.2008.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny D. Relativity for molecular clocks. Nature. 2005;436:183–184. doi: 10.1038/436183a. doi:10.1038/436183a [DOI] [PubMed] [Google Scholar]
- Phillips M.J, Penny D. The root of the mammalian tree inferred from whole mitochondrial genomes. Mol. Phylogenet. Evol. 2003;28:171–185. doi: 10.1016/s1055-7903(03)00057-5. doi:10.1016/S1055-7903(03)00057-5 [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley T.R. Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over Likelihood Ratio tests. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. doi:10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut, A. 1996 Se-Al: sequence alignment editor. See http://evolve.zoo.ox.ac.uk
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Simon C, Frati F, Bechenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Sunnucks P, Hale D.F. Numerous transposed sequences of mitochondrial cytochrome oxidase I–II in aphids of the genus Sitobion (Hemiptera: Aphididae) Mol. Biol. Evol. 1996;13:510–524. doi: 10.1093/oxfordjournals.molbev.a025612. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2003 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. Sunderland, MA: Sinauer Associates.
- Terry M.D, Whiting M.F. Mantophasmatodea and phylogeny of the lower neopterous insects. Cladistics. 2005;21:240–257. doi:10.1111/j.1096-0031.2005.00062.x [Google Scholar]
- Trewick S.A. Identity of an endangered grasshopper (Acididae: Brachaspis): taxonomy, molecules and conservation. Conserv. Genet. 2001;2:233–243. doi:10.1023/A:1012263717279 [Google Scholar]
- Trewick S.A. DNA barcoding is not enough: mismatch of taxonomy and genealogy in New Zealand grasshoppers (Orthoptera: Acrididae) Cladistics. 2008;24:240–254. doi:10.1111/j.1096-0031.2007.00174.x [Google Scholar]
- Trewick S.A, Morgan-Richards M. Phylogenetics of New Zealand's tree, giant and tusked weta (Orthoptera: Anostostomatidae): evidence from mitochondrial DNA. J. Orthopt. Res. 2004;13:185–196. doi:10.1665/1082-6467(2004)013[0185:PONZTG]2.0.CO;2 [Google Scholar]
- Trewick S.A, Morgan-Richards M. After the deluge: mitochondrial DNA indicates Miocene radiation and Pliocene adaptation of tree and giant weta (Orthoptera: Anostostomatidae) J. Biogeogr. 2005;31:295–309. doi:10.1111/j.1365-2699.2004.01179.x [Google Scholar]
- Trewick, S. A. & Morgan-Richards, M. In press. Biology of New Zealand. Encyclopedia of Islands Berkeley, CA: University of California Press.
- Trewick S.A, Wallis G.P, Morgan-Richards M. Phylogeographical pattern correlates with Pliocene mountain building in the alpine scree weta (Orthoptera, Anostostomatidae) Mol. Ecol. 2000;9:657–666. doi: 10.1046/j.1365-294x.2000.00905.x. doi:10.1046/j.1365-294x.2000.00905.x [DOI] [PubMed] [Google Scholar]
- Trewick S.A, Paterson A.M, Campbell H.J. Hello New Zealand. J. Biogeogr. 2007;34:1–6. doi:10.1111/j.1365-2699.2006.01643.x [Google Scholar]
- Vawter, L. 1991 Evolution of blattoid insects and of the small subunit ribosomal gene. PhD dissertation, University of Michigan, Ann Arbor.
- Veevers J.J, Li Z.X. Review of seafloor spreading around Australia. II. Marine magnetic anomaly modelling. Aust. J. Earth Sci. 1991;38:391–408. doi:10.1080/08120099108727980 [Google Scholar]
- Wagstaff S.J, Garnock-Jones P.J. Evolution and biogeography of the Hebe complex (Scrophulariaceae) inferred from ITS sequences. New Zeal. J. Bot. 1998;36:425–437. [Google Scholar]
- Waters J.M, Craw D. Goodbye Gondwana? New Zealand biogeography, geology and the problem of circularity. Syst. Biol. 2006;55:351–356. doi: 10.1080/10635150600681659. doi:10.1080/10635150600681659 [DOI] [PubMed] [Google Scholar]
- Whiting M.F. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zool. Scr. 2002a;31:93–104. doi:10.1046/j.0300-3256.2001.00095.x [Google Scholar]
- Whiting M.F. Phylogeny of the holometabolous insect orders: molecular evidence. Zool. Scr. 2002b;31:3–15. doi:10.1046/j.0300-3256.2001.00093.x [Google Scholar]
- Zakharov E, Caterino M, Sperling F. Molecular phylogeny, historical biogeography and divergence time estimates for swallowtail butterflies of the genus Papilio (Lepidoptera: Papilionidae) Syst. Biol. 2004;54:193–215. doi: 10.1080/10635150490423403. doi:10.1080/10635150490423403 [DOI] [PubMed] [Google Scholar]
- Zwickl, D. J. 2006 Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, The University of Texas, Austin.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.