Abstract
The endemic moth genus Hyposmocoma (Lepidoptera: Cosmopterigidae) may be one of the most speciose and ecologically diverse genera in Hawaii. Among this diversity is the Hyposmocoma saccophora clade with previously unrecorded aquatic larvae. I present a molecular phylogeny based on 773 base pairs (bp) of the mitochondrial gene cytochrome c oxidase subunit I and 762 bp of the nuclear gene elongation factor 1-α. Topologies were constructed from data using maximum-parsimony, maximum-likelihood and Bayesian search criteria. Results strongly support the monophyly of the H. saccophora clade and the monophyly of the genus Hyposmocoma. The H. saccophora clade has single-island endemic species on Oahu, Molokai and West Maui. By contrast, there are three species endemic to Kauai, two being sympatric. The H. saccophora clade appears to follow the progression rule, with more basal species on older islands, including the most basal species on 11 Myr-old Necker Island, one of the Northwestern Hawaiian Islands. Aquatic behaviour either evolved recently in the species on the main Hawaiian Islands or was secondarily lost on the arid northwestern Necker Island. The phylogeny suggests that Hyposmocoma is older than any of the current main islands, which may, in part, explain Hyposmocoma's remarkable diversity.
Keywords: Hyposmocoma, Cosmopterigidae, aquatic caterpillar, progression rule, Lepidoptera, Hawaii
1. Introduction
The Hawaiian archipelago has sponsored remarkable diversification in many groups of organisms and could rightly be called another ‘Galápagos’. There is little doubt that if the Beagle had landed in Hawaii instead of the Galápagos, Darwin would have found himself equally inspired by Hawaii's remarkable diversity. Although there are many spectacular radiations in Hawaii, few are as diverse as the species of the endemic case-bearing moth genus Hyposmocoma (Cosmopterigidae). Hyposmocoma contains more than 350 recognized species (Zimmerman 1978), but the ecological diversity of the genus is so broad, and rates of endemism so high, that discovery of many more species is anticipated in the course of future research. Hyposmocoma larvae occur from alpine moss mats at altitudes of over 3000 m down to rocks just above the high-tide splash zone. Larvae are remarkably ecologically diverse, occurring in habitats as different as lichen-covered crevices of still-steaming lava flows to pristine isolated remnants of rainforest. Hyposmocoma includes a species with the only known molluscivorous caterpillar (Rubinoff & Haines 2005). Different species occur in all native plant communities on all the islands, though very few are widespread; more than 90 per cent of recognized Hyposmocoma species are single-island endemics (Zimmerman 1978).
The origins of Hyposmocoma are completely unknown, probably obscured by tremendous diversification and time; the genus may be a very early colonist in the Hawaiian archipelago and only distantly related to extant continental taxa. The distribution of Hyposmocoma across the ancient Northwestern Hawaiian Islands, the number of species in the genus and their ecological diversity suggest a long period of isolation, perhaps over 10 Ma, similar to what has been estimated for Hawaiian drosophilid flies (Bonacum et al. 2005) and Megalagrion damselflies (Jordan et al. 2003). Understanding the origin and relationships of Hyposmocoma is further challenged by the poor state of knowledge of the systematics of the Cosmopterigidae. This globally distributed family contains 106 genera and 1628 recognized species (Hodges 1999), but these numbers are probably a severe underestimate since the moths are small and frequently cryptic, discouraging casual study. Perhaps as a result of this diversity and obscurity, research is still in the alpha taxonomy stage and there is no systematic framework for Cosmopterigidae as a whole, making phylogenetic placement of Hyposmocoma within the family challenging. This is a question worth long-term pursuit since an understanding of the origins of such a diverse genus will have applications to broader themes of evolutionary pattern and process. More immediate systematics research within Hyposmocoma is also essential to these macroevolutionary issues, and this paper is an effort to understand a particularly remarkable part of Hyposmocoma's diversity in a phylogeographic context.
Some species of Hyposmocoma have aquatic larvae, reflecting a dramatic ecological shift in life history that was inexplicably unrecorded by previous researchers. These larvae feed on algae growing on rocks at and below the water's surface in fast-moving streams. Such streams are extremely prone to flash floods and larvae may be submerged suddenly in torrents that can last for days when storms pass. This requires the larvae not only to be able to breath underwater but also to enact specific behaviour to secure themselves to inundated rocks to avoid being washed out to sea. Thus, the riparian habitat presents several challenges not faced by terrestrial species of Hyposmocoma and suggests an interesting evolutionary history for these populations. Although one aquatic species, Hyposmocoma saccophora, was described by Walsingham (1907), the aquatic behaviour of its larvae was never recognized. While the adult moth is neither aquatic nor particularly unusual in its behaviour, H. saccophora larvae are easily distinguished by their unique cone-shaped larval case, constructed largely of silk and diatoms (figure 1). Hyposmocoma saccophora, or related species, occur in fast-flowing streams on the main Hawaiian Islands of Kauai, Oahu, Molokai and Maui.
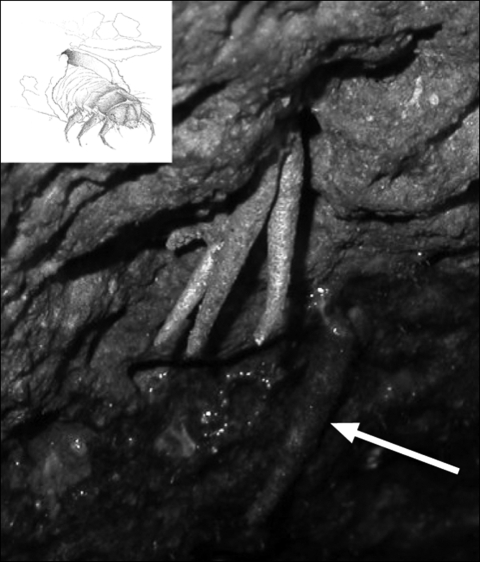
Figure 1.
Hyposmocoma saccophora larvae. Note the three larvae sheltering in one hole just above the waterline. Just below them (arrow) is a pupated larva in a case below the waterline, where it will remain for several weeks before the adult moth emerges. Inset: a sketch of a larva emerging from its case, the lid of which is retractable.
The discovery of multiple populations of an aquatic Hyposmocoma with a distinct case shape and ecology fostered many questions about their evolution and phylogeography, including the following. Is H. saccophora a single, widespread species or are there many species of cone-cased Hyposmocoma? What is the scale of intra- and inter-island genetic diversity and are populations isolated by island or drainage? Does the H. saccophora clade follow the progression rule (Funk & Wagner 1995) with monophyly and more basal lineages on older islands, including the much older Northwestern Hawaiian Islands? Alternatively, is the pattern of diversification more complex and inconsistent with this colonization pattern, suggesting repeated independent derivation of the cone case and aquatic habit as part of separate radiations (Gillespie et al. 1997; Hormiga et al. 2003)? A progression rule pattern might indicate that Hyposmocoma as a genus is older than the current main islands, as has been suggested for Drosophila (Bonacum et al. 2005) and Megalagrion damselflies (Jordan et al. 2003), but that is generally thought to be a rare phenomenon with most extant lineages colonizing the archipelago after the current main islands arose (Price & Clague 2002). Within Hyposmocoma, are these cone-cased species monophyletic or does the cone case type reoccur in paraphyly with other case shape lineages? Finally, what are the conservation implications of the phylogenetics? The answers to these questions are best addressed through the use of DNA-based phylogenetics, tools that Darwin would have no doubt greatly appreciated in his efforts to understand the evolution of unusual organisms on isolated archipelagos.
2. Material and methods
(a) Sample collection and moth rearing
Accessible fast-moving streams on all main islands were searched for cone-cased Hyposmocoma. Repeated surveys revealed no stream-dwelling Hyposmocoma in low-elevation areas heavily modified by agriculture and urbanization, so searching was focused on higher reaches of streams. Larvae were collected from the surfaces and undersides of rocks in and around streams and returned to the laboratory for rearing to adulthood. Larvae were maintained in standard Petri dishes and fed a mixture of carrot, stream algae (various species) and various commercial fish food products. The time from larval collection to the emergence of adult moths varied from a few weeks to nearly six months, presumably depending on the population and developmental stage at the time of collection. Adult moths were harvested daily and frozen at −80°C to preserve DNA for sequencing. In addition to the cone-cased Hyposmocoma collected from the younger main islands (table 1), I acquired cone-cased larvae from the Northwestern Hawaiian Island of Necker, an 11-Ma remnant on which there are no streams and where there may not have been any streams for perhaps several hundred thousand years or more as the island has eroded (Price & Clague 2002).
Table 1.
Hyposmocoma and outgroup taxa sequenced. (Species groupings are based on the phylogenies (figure 2).)
| taxon | locality | voucher numbers | case type | larval habitat |
|---|---|---|---|---|
| H. saccophora | Oahu | cp20, cp24, cp33, cp43, cp53, cp107 | cone | aquatic |
| jw12, jw13, jw14, tl3h, cp107 | ||||
| H. saccophora species group | Molokai | cp75, cp82, cp116, cp117 | cone | aquatic |
| West Maui | cp86, cp122, cp123 | cone | aquatic | |
| cp128, cp158, cp160 | ||||
| Kauai | cp111, cp138, cp146 | cone | aquatic | |
| Kauai | cp63, cp64, cp118, cp119 | cone | aquatic | |
| Kauai | cp84, cp120, cp121 | cone | aquatic | |
| Necker | cp85, cp94 | cone | terrestrial | |
| terrestrial burrito case | Hawaii | cp144, cp145 | burrito | terrestrial |
| Kauai | cp141, cp142 | burrito | terrestrial | |
| aquatic burrito case | East Maui | cp45, cp55, cp56, cp68 | burrito | aquatic |
| Oahu | cp66, cp67 | burrito | aquatic | |
| Hawaii | cp105 | burrito | aquatic | |
| West Maui | cp87, cp88, cp90, cp91 | burrito | aquatic | |
| cp161, cp162 | ||||
| outgroups | ||||
| Cosmopterix sp. | Taiwan | tl8h | — | terrestrial |
| Chrysopleinae | Taiwan | tl1h, tl7h | — | terrestrial |
| Walshia sp. | California | tl2h | — | terrestrial |
A broad assortment of ‘burrito-cased’ Hyposmocoma, both aquatic and terrestrial species (largely undescribed), was added to the analysis as outgroup taxa. These larvae were reared in the same manner as the cone-cased Hyposmocoma. For both Oahu and West Maui, I used aquatic burrito-cased species found in the same streams as their cone-cased counterparts to assess the possibility that these geographically proximate but morphologically different species were not the products of an intra-island adaptive radiation as found, for instance, in Hawaiian spiders (Blackledge & Gillespie 2004; Gillespie 2004). If this were the case, then the cone case of Hyposmocoma would be the result of convergent evolution of similar phenotypes in unrelated radiations under similar conditions on different islands. This convergence would manifest itself as paraphyly of the cone-cased species with respect to burrito-cased species, especially aquatic burrito-cased species from the same island. To assess the monophyly of Hyposmocoma as a genus more broadly, three other cosmopterigid taxa were included. I selected these non-Hyposmocoma taxa from two of the continental land masses closest to Hawaii to assess most effectively the possibility of multiple colonizations and paraphyly in Hyposmocoma. Included were two taxa from Taiwan, a species of Cosmopterix and an unidentified species in the subfamily Chrysopleinae, and one from California, a species of Walshia.
In total, I collected and reared populations of cone-cased Hyposmocoma from streams on Kauai, Oahu, Molokai and West Maui, resulting in 53 samples, including outgroup taxa (table 1). Despite extensive searches on East Maui and Hawaii, no aquatic cone-cased Hyposmocoma larvae were found. All vouchers are deposited in the University of Hawaii Insect Museum (table 1).
(b) DNA extraction and sequencing
The thorax of frozen moths was dissected from the body and processed following the manufacturer's protocols for DNeasy extraction kits (Qiagen, Inc.). Using the mitochondrial gene cytochrome c oxidase subunit I (COI) alone for phylogenetic analyses is unlikely to provide the most robust or accurate results (Rubinoff & Holland 2005; Cognato 2006; Rubinoff et al. 2006), nor is apparent incongruence of datasets likely to impair combined resolution (Bull et al. 1993; Cunningham 1997; Rubinoff & Sperling 2002). Therefore, both COI and a nuclear gene were sequenced for analysis in a combined dataset. In total, 1535 base pairs (bp) were sequenced from COI (773 bp) and the nuclear gene elongation factor 1-α (EF1-α, 762 bp). Primers used for polymerase chain reaction (PCR) amplification of COI were sense primer Jerry (CAACAT TTA TTT TGA TTT TTT GG), beginning at base pair 2183, and antisense primer Pat (TCC AAT GCA CTA ATC TGC CAT ATT A), beginning at base pair 3020. EF1-α primers were designed by C. Pong specifically for Hyposmocoma: Coma (GGC CCA GGA AAT GGG CAA AGG) beginning at base pair 95 and Toes (GGA GTC WCC AGC KAC GTA ACC) beginning at base pair 1045. Each 50 μl reaction contained 20 μl HotMasterMix (HotMaster Taq DNA polymerase, 0.3 U; 2.5× HotMaster Taq Buffer pH 8.5, 45 mM KCl and 2.5 mM MgCl2; 2 μl of each dNTP (Brinkman Instruments, Inc.)), 25 pmol of each primer and approximately 5 ng of total genomic DNA (see Le Roux et al. (2007) for more details). The PCR thermal profile for COI was 94°C for 2 min initial denaturation, and then 94°C for 1 min, 51°C for 1 min, 72°C for 2 min for 34 cycles, followed by a 72°C extension for 12 min. For EF1-α, the thermal profile was 94°C for 2 min initial denaturation, and then 94°C for 1 min, 53°C for 1 min, 72°C for 2 min for 34 cycles, followed by a 72°C extension for 12 min. Cycle sequencing and DNA sequencing were performed on both sense and antisense strands. Sequences are available from GenBank (accession numbers in table 2).
Table 2.
COI and EF1-α GenBank accession numbers for vouchers.
| GenBank reference numbers | ||
|---|---|---|
| voucher number | COI | EF1-α |
| cp55 | EU697277 | EU697330 |
| cp56 | EU697278 | EU697331 |
| cp161 | EU697279 | EU697332 |
| cp162 | EU697280 | EU697333 |
| cp158 | EU697281 | EU697334 |
| cp160 | EU697282 | EU697335 |
| cp87 | EU697283 | EU697336 |
| cp88 | EU697284 | EU697337 |
| cp86 | EU697285 | EU697338 |
| cp122 | EU697286 | EU697339 |
| cp123 | EU697287 | EU697340 |
| cp128 | EU697288 | EU697341 |
| cp90 | EU697289 | EU697342 |
| cp91 | EU697290 | EU697343 |
| cp33 | EU697291 | EU697344 |
| cp43 | EU697292 | EU697345 |
| cp66 | EU697293 | EU697346 |
| cp67 | EU697294 | EU697347 |
| cp75 | EU697295 | EU697348 |
| cp82 | EU697296 | EU697349 |
| cp116 | EU697297 | EU697350 |
| cp117 | EU697298 | EU697351 |
| cp138 | EU697299 | EU697352 |
| cp111 | EU697300 | EU697353 |
| cp145 | EU697301 | EU697354 |
| cp146 | EU697302 | EU697355 |
| cp45 | EU697303 | EU697356 |
| cp68 | EU697304 | EU697357 |
| cp144 | EU697305 | EU697358 |
| cp24 | EU697306 | EU697359 |
| cp20 | EU697307 | EU697360 |
| cp141 | EU697308 | EU697361 |
| cp142 | EU697309 | EU697362 |
| jw14 | EU697310 | EU697363 |
| cp85 | EU697311 | EU697364 |
| cp94 | EU697312 | EU697365 |
| cp107 | EU697313 | EU697366 |
| cp53 | EU697314 | EU697367 |
| cp63 | EU697315 | EU697368 |
| cp64 | EU697316 | EU697369 |
| cp84 | EU697317 | EU697370 |
| cp118 | EU697318 | EU697371 |
| cp119 | EU697319 | EU697372 |
| cp120 | EU697320 | EU697373 |
| cp121 | EU697321 | EU697374 |
| jw12 | EU697322 | EU697375 |
| jw13 | EU697323 | EU697376 |
| cp105 | EU697324 | EU697377 |
| tl3h | EU697325 | EU697378 |
| tl1h | EU697326 | EU697379 |
| tl2h | EU697327 | EU697380 |
| tl7h | EU697328 | EU697381 |
| tl8h | EU697329 | EU697382 |
(c) Phylogenetic analysis
DNA sequences were aligned manually using forward and reverse sequenced strands for independent confirmation. Sequences from both genes were analysed as separate data partitions whenever possible. Maximum-parsimony (MP) and maximum-likelihood (ML) models were used as implemented in PAUP (Swofford 2003). Maximum parsimony was conducted using a heuristic search and all standard defaults (e.g. unordered, equal-weight characters, stepwise addition, tree bisection–reconnection). Partitioned decay index values (Bremer support) were calculated using TreeRot v. 2 (Sorenson 1999). The ML model was optimized first using Modeltest (Posada & Crandall 1998), which selected the GTR+I+G model (general time reversible with estimated proportion of invariable sites and rate heterogeneity) as optimum for the molecular dataset. MrBayes (Huelsenbeck & Ronquist 2001) was used for Bayesian posterior probability analysis, with all model parameters estimated by the program. For the analysis, COI and EF1-α were designated as independent character sets, and two simultaneous runs were performed with four chains (one cold, three hot) with a sample frequency of every 10 generations and a burn-in of 2000. Three million generations for each chain were run for the combined COI and EF1-α dataset. Confidence in tree topologies was assessed as posterior probabilities calculated for each node as implemented in MrBayes.
3. Results
Initial morphological analysis revealed relatively low levels of variation among populations on different islands (species descriptions are in preparation). By contrast, DNA sequence analysis revealed clear, well-supported genetic breaks between populations on each island commensurate with species-level divergence (figure 2).
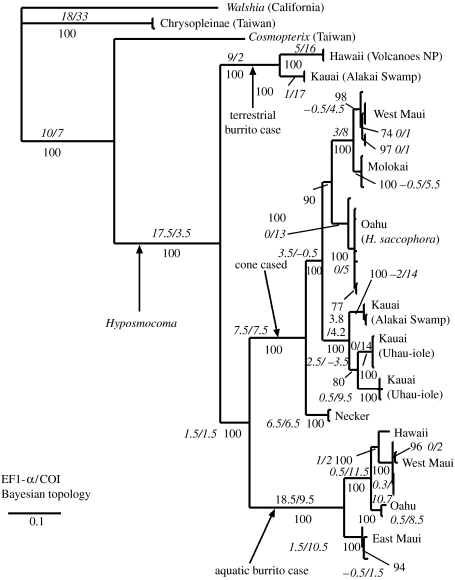
Figure 2.
EF1-α and COI combined Bayesian phylogram. Numbers below the branches are posterior probabilities. Parsimony-based decay index values are in italics adjacent to corresponding nodes that were also supported by MP analysis. Decay index values are partitioned by gene: EF1-α/COI. Negative decay index values indicate support for an alternative topology. Cone-cased and aquatic burrito-cased clades are indicated. Note strongly supported monophyly of Hyposmocoma and case-bearing Hyposmocoma. Burrito-cased species are rendered paraphyletic by the cone-cased clade. NP, National Park.
ML analysis using the optimized model gave a tree with a −ln likelihood score of −ln 6173.7646. The Bayesian analysis resulted in a standard deviation between split frequencies of 0.0055, well below the 0.01 suggested to ensure that the searches were converging on an optimal topology (Huelsenbeck & Ronquist 2001). Both Bayesian and ML models generated the same topology except that resolution between three cryptic species on Kauai differed; under Bayesian analysis, the two populations from Uhau-iole Stream were monophyletic (but still quite divergent), while under ML, the species from Kawaikoi Stream in the Alakai Swamp population is placed between the two Uhau-iole species. Support for either resolution between these three Kauai taxa was poor. The maximum parsimony analysis generated 6775 most parsimonious trees of 788 steps, with a consistency index of 0.627 and retention index of 0.896. The strict consensus of the MP trees agreed with the structure of both the Bayesian and ML trees, though it was more poorly resolved (probably as a result of the strict consensus). Partitioned decay index values strongly supported all but the most recently derived nodes. In a few cases, the two genes disagreed, but this was generally weak disagreement overwhelmed by much stronger support for the dominant topology in the other gene tree and in trees reconstructed using other methods. The only time the two genes disagreed with nearly equal support was in the resolution of Kauai Uhau-iole and Alakai Swamp cone-cased species relationships. There was slightly stronger support in COI (decay index 3.5) for a relationship not in agreement with EF1-α (decay index 2.5) or Bayesian- and ML-based reconstructions (figure 2). Bayesian branch support was above 0.90 for nearly all branches, the only three places it was below 0.90 being for within-population structure in the West Maui and Oahu species, and the aforementioned Kauaian Uhau-iole and Alakai Swamp species trinity.
MP, ML and Bayesian search criteria all suggest that the H. saccophora species group consists of six species, one per island on Oahu, Molokai and West Maui, and three species, two of which are sympatric, on Kauai. These cone-cased Hyposmocoma are monophyletic, including the terrestrial population of cone-cased Hyposmocoma from Necker, which is basal to the high island aquatic species. Burrito-cased Hyposmocoma were rendered paraphyletic by the cone-cased H. saccophora species group, with aquatic burrito-cased species being more closely related to aquatic cone-cased species. Hyposmocoma as a genus was strongly monophyletic with respect to the cosmopterigid outgroups, Cosmopterix from Taiwan being the closest.
4. Discussion
The H. saccophora species group appears to follow the progression rule with basal and, presumably, older lineages originating on older islands, while more derived lineages result from dispersal to younger islands where they speciate (figure 3) but thus far have not radiated into new habitats. Even on Kauai, where three species occur, there has been no apparent change in ecology. Understanding the reasons why some lineages such as certain spiders (Gillespie et al. 1997; Hormiga et al. 2003; Gillespie 2004) might support adaptive radiations, while others follow the progression rule (Liebherr & Zimmerman 1998) as do cone-cased Hyposmocoma and Megalagrion (Jordan et al. 2003), will shed light on the constraints of evolution operating on different lineages, and comparisons between the two kinds of radiations may be particularly valuable in understanding this aspect of island biogeography.
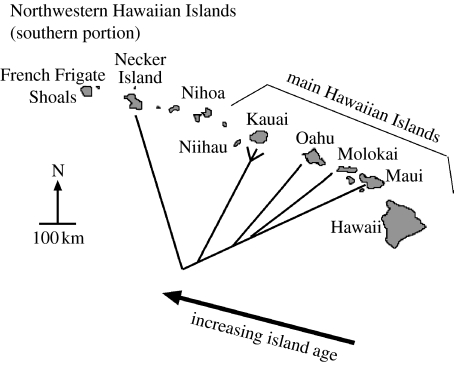
Figure 3.
Cone-cased Hyposmocoma phylogeny mapped onto the Hawaiian Islands. Note the concordance of the species relationships with the age of the islands. As predicted by the progression rule, older islands sustain more basal species than younger islands.
The rarity or absence of cone-cased aquatic Hyposmocoma on the youngest islands of East Maui and Hawaii is probably not an artefact of collecting technique as the Hyposmocoma species group was abundant and easily found in the appropriate stream habitats on the islands where collections were successful, suggesting that the searching technique was appropriate and productive. Instead, this absence may be related to island age since all older islands support unique species. Most remarkably, the terrestrial species on arid Necker Island is strongly supported as the basal member of the H. saccophora species group, but well within the genus as a whole (figure 2). This suggests that the H. saccophora lineage has been extant at least as long as 11-Ma Necker Island, and that the ancestor of Hyposmocoma may have arrived in the Hawaiian Islands even earlier, a pattern supported in other groups of endemic insects (Jordan et al. 2003; Bonacum et al. 2005). Furthermore, the strictly terrestrial nature of Necker Island's cone-cased Hyposmocoma suggests that either aquatic adaptations first occurred when the clade colonized Kauai or the Necker species was able to secondarily revert to a dry terrestrial existence. Considering the physiological challenges of making aquatic/terrestrial shifts and their global rarity in Lepidoptera, either scenario is remarkable.
The apparent decoupling of morphological and genetic variation in cone-cased Hyposmocoma is a typical example of cryptic speciation, wherein genetically isolated species do not manifest their evolutionary independence with divergent morphology (Henry et al. 1999; Parsons & Shaw 2001; Molbo et al. 2003). This lack of morphological divergence in the face of significant genetic divergence may be a result of stabilizing selection on both the larvae and adults and related to the peculiar demands of an aquatic and streamside life history.
Patterns of genetic diversity in the H. saccophora species group are inconsistent across the islands. Among the Oahu, Molokai and West Maui species, there is a relatively low genetic divergence of 1 per cent or less. Using COI as a benchmark (Brower 1994), there is approximately 2.5 per cent divergence in COI between the West Maui and Molokai species and 5.5 per cent between those species and the Oahu species. Although delimiting species based on per cent divergence is counterproductive (Cognato 2006; Rubinoff 2006), such measures are useful in understanding gene flow. The pattern suggested by both genes is of panmictic populations on the three islands, but isolation among the species on different islands. On Kauai, not only are there three species, but also two occur in the same stream, and, with divergence levels of 4–6% for COI for the closest species pair, genetic distances are as large as or larger than those between the species on the other islands. Although the relationships between the three species on Kauai are poorly resolved, the level of divergence between them is equivalent to inter-island species divergence on other islands, suggesting that the Kauai species are apparently not the result of recent divergence. This deeper divergence indicates, as evidenced by the hard polytomy, that the three species arose from an older and nearly simultaneous cladogenesis, leaving very few synapomorphies supporting resolution for whichever is the slightly younger clade.
Equally remarkable is the contrast in the rate of speciation on Kauai compared with the other islands. Perhaps the greater age of Kauai has permitted intra-island speciation, whereas the cone-cased Hyposmocoma on the younger islands of Oahu, Molokai and West Maui have simply not had enough time to generate intra-island diversity. This supposition may help in understanding the relative importance of inter- and intra-island isolation and age in dictating rates of genetic divergence and speciation in Hawaii and elsewhere. While a simple comparison between island age and isolation may answer this question, further research may reveal a more complex association between age, habitat and topology that has promoted a higher level of speciation on Kauai than on the other three islands.
The conservation implications for the H. saccophora species group are complex. Hawaii's riparian systems have been heavily impacted by development, diversion and introduced species (Polhemus 1997). While the loss of a few streams on Oahu, Molokai or West Maui is unlikely to lead to extinction of those islands' endemic species of cone-cased Hyposmocoma, the spatial scale of speciation on Kauai suggests that more localized species may be vulnerable to the loss of the sole drainage they inhabit. As with plants (Price 2004) and Megalagrion damselflies (Jordan et al. 2003), Kauai appears to sustain a higher number of species, and endemic species, than the larger, but younger islands, suggesting that, at least for the H. saccophora group within Hyposmocoma, island age is more important than size in generating endemic diversity (see also Cowie 1995). Because Hawaii lacks most of the aquatic insect orders used to assess water quality in continental systems, aquatic Hyposmocoma may offer a surrogate measure for stream conservation and habitat quality. Restricted to relatively unmodified stretches of flowing water, the presence of these endemic insects may be useful in assessing ecosystem health.
Cone-cased aquatic Hyposmocoma have accomplished a remarkable invasion of the treacherous aquatic habitats in Hawaiian streams. Understanding why and how a particular clade in the genus Hyposmocoma has been successful in this dramatic ecological diversification, while the vast majority of lepidopteran lineages remain solely terrestrial, will be a fruitful goal for future research. Establishing a systematic framework for these aquatic cone-cased Hyposmocoma is an essential first step in understanding the diversification of a remarkable and globally rare life history. Further investigations into the relationships between these aquatic species and the rest of Hyposmocoma, the vast majority of which are terrestrial, will be essential to understanding not only the remarkable evolution of aquatic moths, but also the diversification of one of the most speciose endemic island lineages anywhere.
Acknowledgments
This research would not have been possible without permits and/or assistance from the following: Hawaii Division of Forestry and Wildlife (B. Gagné, G. Kawakami and J. S. Cumming); US Fish and Wildlife Service (D. Horvath and C. Rehkemper); Maui Land and Pineapple Co., Inc. (R. Bartlett); Hawaii Division of Aquatic Resources (D. Polhemus); Hawaii Volcanoes National Park (R. Loh); and Haleakala National Park (E. Gordon). I also thank the following for assistance with fieldwork: C. Buddenhagen; M. Caterino; J. Cooper; J. Eiben; R. Englund; W. Haines; G. Hansen; M. Heddle; B. Holland; R. Kaholoa‘a; C. King; T. Lee; S. Montgomery; S. Myers; C. Pong; M. Shin; P. Welton; J. Winhall-Rice; and many others. I thank S. Yen for outgroup specimens from Taiwan and J. Powell for those from California. J. J. Le Roux commented on the manuscript and assisted in the field. Comments by R. Cowie and two anonymous reviewers greatly improved the quality of the paper. B. Holland helped with the illustrations. Pre-sequencing molecular work was performed by C. Pong, T. Lee and J. Winhall-Rice, and B. Holland and M. San Jose assisted with data entry. S. Myers provided the drawing in figure 1. Funding was provided by the National Geographic Society's Committee for Research and Exploration and by the State of Hawaii's US Fish and Wildlife Service State Wildlife grant (T-3-P).
Footnotes
One contribution of 15 to a Theme Issue ‘Evolution on Pacific islands: Darwin's legacy’.
References
- Blackledge T.A, Gillespie R.G. Convergent evolution of behavior in an adaptive radiation of Hawaiian web-building spiders. Proc. Natl Acad. Sci. USA. 2004;101:16 228–16 233. doi: 10.1073/pnas.0407395101. doi:10.1073/pnas.0407395101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacum J, O'Grady P.M, Kambysellis M, DeSalle R. Phylogeny and age of diversification of the planitibia species group of the Hawaiian Drosophila. Mol. Phylogenet. Evol. 2005;37:73–82. doi: 10.1016/j.ympev.2005.03.008. doi:10.1016/j.ympev.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Brower A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. doi:10.1073/pnas.91.14.6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J.J, Huelsenbeck J.P, Cunningham C.W, Swofford D.L, Waddell P.J. Partitioning and combining data in phylogenetic analysis. Syst. Biol. 1993;42:384–397. doi:10.2307/2992473 [Google Scholar]
- Cognato A. Standard percent DNA sequence difference for insects does not predict species boundaries. J. Econ. Entomol. 2006;99:1037–1045. doi: 10.1603/0022-0493-99.4.1037. [DOI] [PubMed] [Google Scholar]
- Cowie R.H. Variation in species diversity and shell shape in Hawaiian land snails: in situ speciation and ecological relationships. Evolution. 1995;49:1191–1202. doi: 10.1111/j.1558-5646.1995.tb04446.x. doi:10.2307/2410444 [DOI] [PubMed] [Google Scholar]
- Cunningham C.W. Is congruence between data partitions a reliable predictor of phylogenetic accuracy? Empirically testing an iterative procedure for choosing among phylogenetic methods. Syst. Biol. 1997;46:464–478. doi: 10.1093/sysbio/46.3.464. doi:10.2307/2413692 [DOI] [PubMed] [Google Scholar]
- Funk V.A, Wagner W.L. Biogeographic patterns in the Hawaiian Islands. In: Wagner W.L, Funk V.A, editors. Hawaiian biogeography. Smithsonian Institution Press; Washington, DC: 1995. pp. 379–419. [Google Scholar]
- Gillespie R.G. Community assembly through adaptive radiation in Hawaiian spiders. Science. 2004;303:356–359. doi: 10.1126/science.1091875. doi:10.1126/science.1091875 [DOI] [PubMed] [Google Scholar]
- Gillespie R.G, Croom H.B, Hasty G.L. Phylogenetic relationships and adaptive shifts among major clades of Tetragnatha spiders (Araneae: Tetragnathidae) in Hawaii. Pac. Sci. 1997;51:380–394. [Google Scholar]
- Henry C.S, Wells M.L.M, Simon C.M. Convergent evolution of courtship songs among cryptic species of the carnea group of green lacewings (Neuroptera: Chrysopidae: Chrysoperla) Evolution. 1999;53:1165–1179. doi: 10.1111/j.1558-5646.1999.tb04530.x. doi:10.2307/2640820 [DOI] [PubMed] [Google Scholar]
- Hodges, R. W. 1999 The Gelechoidea. In Lepidoptera: moths and butterflies. Handbook of zoology, vol. VI (ed. N. P. Kristensen), pp. 131–158. New York, NY: W. de Gruyter.
- Hormiga G, Arnedo M.A, Gillespie R.G. Speciation on a conveyor belt: sequential colonization of the Hawaiian Islands by Orsonwelles spiders (Araneae: Linyphiidae) Syst. Biol. 2003;52:70–88. doi: 10.1080/10635150390132786. doi:10.1080/10635150390132786 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jordan S, Simon C, Polhemus D. Molecular systematics and adaptive radiation of Hawaii's endemic damselfly genus Megalagrion (Odonata: Coenagrionidae) Syst. Biol. 2003;52:89–109. doi: 10.1080/10635150390132803. doi:10.1080/10635150390132803 [DOI] [PubMed] [Google Scholar]
- Le Roux J.J, Wieczorek A.M, Wright M.G, Tran C.T. Super-genotype: global monoclonality defies the odds of nature. PLoS ONE. 2007;2:e590. doi: 10.1371/journal.pone.0000590. doi:10.1371/journal.pone.0000590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebherr J.K, Zimmerman E.C. Cladistic analysis, phylogeny and biogeography of the Hawaiian Platynini (Coleoptera: Carabidae) Syst. Entomol. 1998;23:137–172. doi:10.1046/j.1365-3113.1998.00044.x [Google Scholar]
- Molbo D.C, Machado A, Sevenster J.G, Keller L, Herre E.A. Cryptic species of fig-pollinating wasps: implications for the evolution of the fig-wasp mutualism, sex allocation, and precision of adaptation. Proc. Natl Acad. Sci. USA. 2003;100:5867–5872. doi: 10.1073/pnas.0930903100. doi:10.1073/pnas.0930903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons Y.M, Shaw K.L. Species boundaries and genetic diversity among Hawaiian crickets of the genus Laupala identified using amplified fragment length polymorphism. Mol. Ecol. 2001;10:1765–1772. doi: 10.1046/j.1365-294x.2001.01318.x. doi:10.1046/j.1365-294X.2001.01318.x [DOI] [PubMed] [Google Scholar]
- Polhemus D.A. Phylogenetic analysis of the Hawaiian damselfly genus Megalagrion (Odonata: Coenagrionidae): implications for biogeography, ecology, and conservation biology. Pac. Sci. 1997;51:395–412. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Price J.P. Floristic biogeography of the Hawaiian Islands: influences of area, environment and paleogeography. J. Biogeogr. 2004;31:487–500. [Google Scholar]
- Price J.P, Clague D.A. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc. R. Soc. B. 2002;269:2429–2435. doi: 10.1098/rspb.2002.2175. doi:10.1098/rspb.2002.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinoff D. Utility of mitochondrial DNA barcodes in species conservation. Conserv. Biol. 2006;20:1026–1033. doi: 10.1111/j.1523-1739.2006.00372.x. doi:10.1111/j.1523-1739.2006.00542.x [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Haines W.P. Web-spinning caterpillar stalks snails. Science. 2005;309:575. doi: 10.1126/science.1110397. doi:10.1126/science.1110397 [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Holland B.S. Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst. Biol. 2005;54:952–961. doi: 10.1080/10635150500234674. doi:10.1080/10635150500234674 [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Sperling F.A.H. Evolution of ecological traits and wing morphology in Hemileuca (Saturniidae) based on a two gene phylogeny. Mol. Phylogenet. Evol. 2002;25:70–86. doi: 10.1016/s1055-7903(02)00213-0. doi:10.1016/S1055-7903(02)00213-0 [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Cameron S, Will K. Are plant DNA barcodes a search for the Holy Grail? Trends Ecol. Evol. 2006;21:1–2. doi: 10.1016/j.tree.2005.10.019. doi:10.1016/j.tree.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Sorenson, M. D. 1999 TreeRot, v. 2. Boston, MA: Boston University.
- Swofford, D. L. 2003 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. Sunderland, MA: Sinauer Associates.
- Walsingham T. de G., Lord Microlepidoptera. Fauna Hawaiiensis. 1907;1:469–759. pls. 10–25. [Google Scholar]
- Zimmerman E.C. Microlepidoptera. Insects of Hawaii. 1978;9:i–xviii. See also 1–1903. [Google Scholar]