Abstract
To gain insight into the early stages of speciation, we reconstructed a DNA-based phylogeny, using combined mitochondrial (cytochrome c oxidase subunits I and II: 1008 bp) and nuclear (elongation factor 1-α and wingless: 1062 bp) markers of populations of the moth Galagete darwini endemic to the Galápagos, which belongs to an insular radiation similar in size to that of Darwin's finches. Adults of G. darwini were collected in the arid lowlands of 11 of the Galápagos Islands (Baltra, Española, Fernandina, Floreana, Isabela, Pinta, Pinzón, San Cristobal, Santa Cruz, Santiago and Seymour) and the humid highlands of a subset of 5 of them (Fernandina, Floreana, Isabela, Santa Cruz and Santiago). The combined phylogeographic analysis surprisingly revealed that G. darwini populations at higher elevation on the western islands (Fernandina, Isabela and Santiago) represent a distinct lineage from the one in the low arid zones of these same islands. This is the first reported case in the archipelago of genetic cryptic differentiation correlated with elevation on the western Galápagos volcanoes.
Keywords: micromoth, Galagete darwini, cryptic differentiation, Galápagos Islands, speciation
1. Introduction
Since the advent of inexpensive and rapid DNA sequencing, the recognition and description of cryptic species, or sibling species, has increased drastically (Bickford et al. 2007). Yet this trend has not been especially evident for terrestrial organisms on volcanic archipelagos like the Canary, Hawaiian and Galápagos Islands, where only a few cryptic species have been reported based on the use of molecular (DNA) tools. Following Bickford et al. (2007), we consider a taxon cryptic if it has been classified as a single nominal species, principally as a result of the difficulty of distinguishing it based on morphological characters.
For example, in the Canary Islands, cryptic species were discovered in the beetle genus Nesotes (Coleoptera: Tenebrionidae), in which species described morphologically that were thought to occur on a number of different islands in fact represented various single island endemics that are genetically distinct (Rees et al. 2001). Similarly, in Liparthum bark beetles, genetic evidence highlighted three cases of cryptic speciation in one polyphyletic species associated with different host plants (Jordal et al. 2004).
In the Hawaiian Islands, only two possible cryptic species were detected in the endemic katydids in the genus Banza (Shapiro et al. 2006). Among Hawaiian Laupala crickets, molecular studies showed some discrepancies in the phylogeny (Shaw 2002) that could have been attributed to the presence of cryptic species, but further investigation recognized the different populations as belonging to a single species (Mendelson et al. 2004; Rubinoff & Holland 2005).
In the Galápagos Islands, the surprise came from the well-documented Darwin's finches with the discovery of two allopatric species within the Warbler finch that showed considerable genetic differences (Freeland & Boag 1999; Petren et al. 1999). Also, three divergent lineages were found representing at least two cryptic taxa in the giant tortoise population of Santa Cruz (Russello et al. 2005). But no evidence of cryptic taxa has been revealed by genetic studies in insects in the archipelago (Finston & Peck 1997; Sequeira et al. 2000, 2008).
Because volcanic oceanic islands show considerable habitat diversity as a result of gradients in topology and humidity, which, combined with their isolation, result in lower competition and empty ecological niches (Whittaker 2002), they represent an ideal microcosm for studying evolution in terrestrial arthropods (Roderick & Gillespie 1998; Gillespie & Roderick 2002). In the Galápagos, as observed in plants (Porter 1979), the greatest insect diversity and endemism is in the arid lowlands (Peck 2001), the largest ecological zone in the archipelago. This zone is predominant on the low islands, and on the larger islands it extends inland to an elevation of over 120 m on the southern slopes, and up to 300 m or higher on the northern sides, where it is replaced by more humid vegetation zones at higher elevations (Wiggins & Porter 1971). In insect genera that have undergone extensive speciation in the Galápagos there is a general pattern in which the ancestral species were lowland colonists and evolutionary radiations within lineages took place in the arid, lowland regions, e.g. Galapaganus weevils (Sequeira et al. 2000), Calosoma carabid beetles, Neoryctes scarab beetles and Gryllus crickets (Peck 2001). Such a scenario is consistent with the Galápagos having been drier than they are currently, with the vegetated areas of the wetter upland forests diminished or even absent (Colinvaux 1972).
The micromoth Galagete darwini belongs to the largest endemic Galápagos radiation of Lepidoptera, and the genus is comparable in species numbers with Darwin's finches (Landry 2002; Schmitz & Landry 2005). Although their feeding habits are poorly known, larvae appear to be scavengers of dead plant material (Landry 2002; Schmitz & Landry 2007). Nested phylogenetically inside the Galagete insular radiation, G. darwini is the most widespread species of the genus, occurring on each major island at almost all elevations (Schmitz et al. 2007).
In this paper, we present a detailed phylogeographic analysis of G. darwini, based on individuals collected on different islands and at different altitudes, and reveal the differentiation of a possible cryptic taxon linked to elevation. Our results provide a phylogenetic context for future studies of this interesting micromoth.
2. Material and methods
(a) Samples and DNA methods
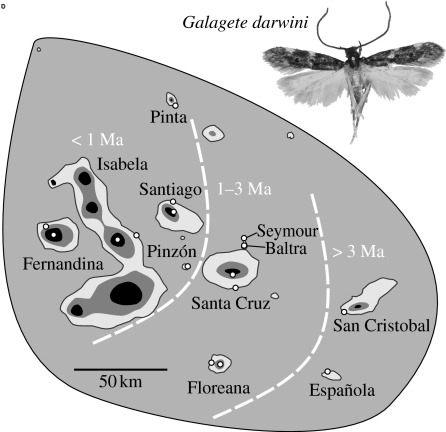
A light trap was used to collect adult G. darwini in the arid lowlands of 11 of the Galápagos Islands (Baltra, Española, Fernandina, Floreana, Isabela, Pinta, Pinzón, San Cristobal, Santa Cruz, Santiago and Seymour) and the humid highlands of a subset of 5 of them (Fernandina, Floreana, Isabela, Santa Cruz and Santiago; figure 1). A single specimen of each of the 16 populations was selected for analysis and all genitalia were dissected for proper identification. Elevations of collecting localities were recorded with a global positioning system (Garmin, Geko 301). Methods for DNA extraction, polymerase chain reaction amplification and sequencing of the mitochondrial DNA (mtDNA) markers cytochrome c oxidase subunits I and II (COI and COII), and nuclear DNA (nDNA) elongation factor 1-α (EF1-α) and wingless (WG) followed Schmitz et al. (2007). The resulting sequences encompass 555, 453, 711 and 351 bp fragments of COI, COII, EF1-α and WG, respectively, for a total of 2070 bp. The new sequences obtained for G. darwini are deposited in GenBank (accession numbers EU780561–EU780580). We used the 11 described Galagete species as out-groups (see Schmitz et al. 2007).
Figure 1.
Distribution of Galagete darwini in the Galápagos archipelago (map modified from Tonnis et al. (2005)). Shaded area encompasses islands where G. darwini occurs. White circles represent the 16 sampling localities. The 300 and 600 m elevation contours are shown. Approximate ages of the islands are indicated by dashed lines.
(b) Data analysis
Sequences were easily assembled manually, edited and aligned with Bioedit v. 7.0.5 (Hall 1999), and alignments were unambiguous. To ensure that we were not getting conflicting topologies, we analysed the combined COI and COII fragments (analysed together since mtDNA acts as a single locus) and the EF1-α and WG nDNA fragments separately. Concatenated alignments were used in the final phylogenetic analysis. The presence of stop codons or indels, which could reveal pseudogene sequences, was checked using MEGA v. 3.1 (Kumar et al. 2004). Hierarchical likelihood ratio tests performed with Modeltest v. 3.04 (Posada & Crandall 1998) determined that a general time-reversible model with rate variation among sites and a proportion of invariable sites (GTR+Γ8+I) was the best-fit model of nucleotide substitution for the combined datasets. A maximum-likelihood (ML) analysis was then performed with a heuristic search and random addition of sequences as implemented in PAUP* v. 4.0.b10 (Swofford 2003), with the starting tree obtained via stepwise addition of taxa and then swapped using the tree-bisection-reconnection algorithm. The reliability of internal branches was assessed using the bootstrap (BS) method (Felsenstein 1985) with 1000 replicates by using the program PHYML 2.4.4 (Guindon & Gascuel 2003). Bayesian posterior probabilities (PP) were calculated using a Metropolis-coupled, Markov chain Monte Carlo sampling approach, as implemented in MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2003), using the same GTR+Γ8+I model. Four simultaneous Markov chains were run in parallel twice for one million generations with trees sampled every 10 generations. Stationarity was evaluated graphically and the first 250 000 initial trees were discarded as burn-in. MrBayes was set to estimate model parameters independently and simultaneously for each gene partition. All analyses were run through the Bioportal web-based service platform for phylogenomic analysis at the University of Oslo, Norway.
3. Results
All haplotypes for each locus were unique, except for the specimens from Pinzón and Seymour, which shared their mtDNA haplotype. Overall sequence divergences were 4.1, 1.3 and 1.5 per cent for mtDNA, EF1-α and WG, respectively. The topologies recovered from the individual locus analyses did not differ from each other (results not shown). The final concatenated alignment of DNA was 2070 bp long, including 1008 bp mtDNA (COI and COII) and 1062 bp nDNA (EF1-α and WG). The A/T ratio was 74 and 44 per cent for mtDNA and nDNA, respectively. No indels or stop codons were found and no pseudogenes were suspected for the mitochondrial genes. No incongruencies between the mtDNA and nDNA datasets were detected using the partition homogeneity test (p>0.05; Farris et al. 1995). Therefore, the combined mitochondrial and nuclear dataset was used in further phylogenetic analyses.
The ML tree of the combined mitochondrial and nuclear dataset is shown in figure 2. Nodes were considered fully resolved if BS support and PP were greater than 0.95 (Hillis & Bull 1993) or greater than 75 per cent (Huelsenbeck & Ronquist 2003), respectively. The tree topologies obtained with MrBayes and PAUP* for the combined analysis were identical.
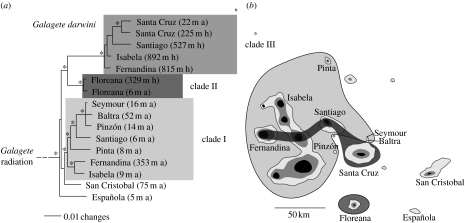
Figure 2.
(a) The ML tree and population relationships of Galagete darwini populations based on a combined molecular analysis of mtDNA and nDNA, and (b) the geographical locations of the specimens analysed. Horizontal branch lengths are proportional to inferred evolutionary time. Asterisks indicate nodes for which PP and BS were greater than or equal to 95 and 75%, respectively. The three clades are indicated and elevation and collecting zone (a, arid zone; h, humid zone) were identified for each specimen. The map shows the 300 and 600 m elevation contours.
All sequenced specimens of G. darwini form a monophyletic clade in both reconstructions (1.00 PP, 88% BS). This clade is subdivided into three monophyletic lineages: clade I (0.96 PP, 77% BS), which includes all specimens from the small dry islands of Baltra, Pinzón and Seymour, the island of Pinta, and from the arid lowlands of the larger western islands of Fernandina, Isabela and Santiago; clade II (1.00 PP, 100% BS), which includes specimens from both the lowlands and highlands of Floreana; and clade III (1.00 PP, 100% BS), which includes the specimens from the highlands of the western islands of Fernandina, Isabela and Santiago, and from Santa Cruz. The relationship between the specimens from Española and San Cristobal and the three other clades is unresolved and therefore represents a basal polytomy. An interesting feature of the combined data tree is the strong genetic differentiation between specimens from the humid highlands and those found in the arid lowlands on the western islands. By contrast, no differences appear between highland and lowland moths on Floreana and Santa Cruz.
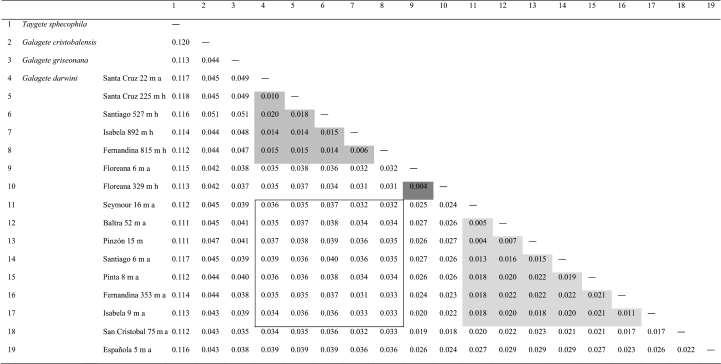
Uncorrected intra- and interspecific genetic distances for the combined dataset are given in table 1. Average intraspecific divergence is 2.7 per cent for G. darwini compared with 4.2 per cent interspecific divergence with G. griseonana, 4.4 per cent with G. cristobalensis and 11.4 per cent with Taygete sphecophila, the closest known relative of the genus Galagete (Schmitz et al. 2007). Within clades I and III, average divergence is 1.7 and 1.4 per cent, respectively, and within clade II it is 0.4 per cent (one comparison only). The average divergence between clades I and II is 2.5 per cent, between clades I and III is 3.6 per cent and between clades II and III is 3.4 per cent. The average divergence between lowland and highland populations on Floreana and Santa Cruz is 0.4 and 1.0 per cent, respectively.
Table 1.
Intra- and interspecific genetic distances (uncorrected; combined COI, COII, EF1-α and WG) among Galagete darwini specimens and three out-groups (two other Galagete species were chosen for comparison, and the closest known relative, Taygete sphecophila). Shaded areas indicate divergences inside clades and bordered area indicates divergences between the three clades.
 |
4. Discussion
The combined mtDNA and nDNA phylogeographic analysis surprisingly revealed that G. darwini populations at higher elevation on the western islands (Fernandina, Isabela and Santiago) represent a distinct lineage from the one in the low arid zones of these same islands (figure 2). This result supports one clear differentiation of a cryptic lineage, at a different elevation on these islands, and suggests a geographical isolation process that is correlated with elevation.
The placement of the specimens of G. darwini from the oldest islands of Española and San Cristobal and those from clade I in a basal polytomy is consistent with the phylogenetic reconstruction, in that the basal taxa are found in coastal arid habitats whereas the more derived taxa occur in the higher and more humid zones. A similar trend is observed also in other Galápagos insects, such as the flightless Galapaganus weevils (Sequeira et al. 2000). This biogeographic pattern suggests that colonization of the higher elevations, or a possible niche shift into humid highland habitat on the western islands of Fernandina, Isabela and Santiago by an arid lowland ancestor resulted in clearly differentiated populations from those in the lowlands.
On the oldest islands of Española and San Cristobal (figure 1), the prevailing ecological conditions are similar to those on all the small islands and on the coasts of the larger islands (clade I). The intermediate aged islands of Floreana and Santa Cruz support individuals that show little genetic divergence between lowlands and highlands, although the colonization scenario could also have been hidden by anthropological factors owing to high human impacts on these two islands, which, for example, could have influenced a secondary colonization of lowlands from highlands or vice versa. Populations clearly differentiated according to elevation are found exclusively on the younger western islands, which emerged less than 1 Ma. One explanation for this pattern is that the western islands, owing to their young age, are higher than the rest of the islands in the archipelago, especially Fernandina and Isabela, which probably offered good opportunities for populations to become isolated and to differentiate in new ecological niches.
The recent and historical volcanic activity of the young western islands of Fernandina, Isabela and Santiago (White et al. 1993) could also have influenced the colonization of the higher elevations by clade III, with eruptions causing damage to high vegetation zones, thereby delaying colonization or extinguishing established populations, as has occurred in tortoise populations on the volcano Alcedo on Isabela (Beheregaray et al. 2003). The long branch of clade III, contrasting with the two other clades, may reflect this recent colonization.
Although cryptic taxa have been found in other terrestrial groups in the Galápagos (Russello et al. 2005; Tonnis et al. 2005), this is the first reported case in the archipelago of cryptic genetic differentiation correlated with elevation. These populations are not yet sufficiently differentiated to be considered cryptic species, as shown by their genetic divergences (table 1), but they are probably in the early stages of divergence. Our data support the idea that these small-winged insects are vagile taxa, whose speciation may be driven by other mechanisms than geographical barriers to dispersal alone, these having long been generally accepted to play the key roles in species formation and maintenance (Lack 1947).
However, limited sampling could have contributed to the appearance of phylogeographic divisions. Increased sampling across the range of G. darwini and along altitudinal gradients will be needed to assess the evolutionary reality of these phylogeographic breaks that could then be portrayed by a minimum spanning network approach. Also a detailed study of the ecological distribution of G. darwini should be carried out to test for a link between lineages and microhabitat types.
Acknowledgements
We thank the Galápagos National Park Service for permission to conduct this research, and the Charles Darwin Research Station and the Charles Darwin Foundation for logistical support. We are also thankful for the assistance and companionship provided to B.L. and P.S. in the field by Stefania Bertoli, Novarino Castillo, Bill Conner, Valentina Cruz, Sarah Garrett, Henri Herrera, Jill Key, Jose Loaiza, Ana Maria Ortega and Lazaro Roque-Albelo. We thank Ken Petren and Dan Rubinoff for their comments on early drafts of the manuscript. This project was supported by Foundations Claraz, Lombard, Schmidheiny, the Swiss Academy of Sciences, the Baslerstiftung für biologische Forschung and the Département des Affaires culturelles of the City of Geneva.
Footnotes
One contribution of 15 to a Theme Issue ‘Evolution on Pacific islands: Darwin's legacy’.
References
- Beheregaray L.B, Ciofi C, Geist D, Gibbs J.P, Caccone A, Powell J.R. Genes record a prehistoric volcano eruption in the Galápagos. Science. 2003;302:75. doi: 10.1126/science.1087486. doi:10.1126/science.1087486 [DOI] [PubMed] [Google Scholar]
- Bickford D, Lohman D.J, Sodhi N.S, Ng P.K.L, Meier R, Winker K, Ingram K.K, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. doi:10.1016/j.tree.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Colinvaux P.A. Climate and the Galápagos Islands. Nature. 1972;240:17–20. doi:10.1038/240017a0 [Google Scholar]
- Farris J.S, Källersjö M, Kluge A.G, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. doi:10.1111/j.1096-0031.1994.tb00181.x [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. doi:10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Finston T.L, Peck S.B. Genetic differentiation and speciation in Stomion (Coleoptera: Tenebrionidae): flightless beetles of the Galápagos Islands, Ecuador. Biol. J. Linn. Soc. 1997;61:183–200. [Google Scholar]
- Freeland J.R, Boag P.T. Phylogenetics of Darwin's finches: paraphyly in the tree-finches, and two divergent lineages in the warbler finch. Auk. 1999;116:577–588. [Google Scholar]
- Gillespie R.G, Roderick G.K. Arthropods on islands: colonization, speciation, and conservation. Annu. Rev. Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. doi:10.1146/annurev.ento.47.091201.145244 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hall T.A. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hillis D.M, Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993;42:182–192. doi:10.2307/2992540 [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Jordal B.H, Kirkendall L.R, Harkestad K. Phylogeny of a Macaronesian radiation: host-plant use and possible cryptic speciation in Liparthrum bark beetles. Mol. Phylogenet. Evol. 2004;31:554–571. doi: 10.1016/j.ympev.2003.09.008. doi:10.1016/j.ympev.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lack D. Cambridge University Press; Cambridge, MA: 1947. Darwin's finches. [Google Scholar]
- Landry B. Galagete, a new genus of Autostichidae representing the first case of an extensive radiation of endemic Lepidoptera in the Galápagos Islands. Rev. Suisse Zool. 2002;109:813–868. [Google Scholar]
- Mendelson T.C, Siegel A.M, Shaw K.L. Testing geographical pathways of speciation in a recent island radiation. Mol. Ecol. 2004;13:3787–3796. doi: 10.1111/j.1365-294X.2004.02375.x. doi:10.1111/j.1365-294X.2004.02375.x [DOI] [PubMed] [Google Scholar]
- Peck S.B. NRC Research Press; Ottawa, ON: 2001. Smaller orders of insects of the Galápagos Islands, Ecuador: evolution, ecology, and diversity. [Google Scholar]
- Petren K, Grant B.R, Grant P.R. A phylogeny of Darwin's finches based on microsatellite DNA length variation. Proc. R. Soc. B. 1999;266:321–329. doi:10.1098/rspb.1999.0641 [Google Scholar]
- Porter D.M. Endemism and evolution in Galápagos Islands vascular plants. In: Bramwell D, editor. Plants and islands. Academic Press; London, UK: 1979. pp. 225–256. [Google Scholar]
- Posada D, Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rees D.J, Emerson B.C, Oromi P, Hewitt G.M. Mitochondrial DNA, ecology and morphology: interpreting the phylogeography of the Nesotes (Coleoptera: Tenebrionidae) of Gran Canaria (Canary Islands) Mol. Ecol. 2001;10:427–434. doi: 10.1046/j.1365-294x.2001.01227.x. doi:10.1046/j.1365-294x.2001.01227.x [DOI] [PubMed] [Google Scholar]
- Roderick G.K, Gillespie R.G. Speciation and phylogeography of Hawaiian terrestrial arthropods. Mol. Ecol. 1998;7:519–531. doi: 10.1046/j.1365-294x.1998.00309.x. doi:10.1046/j.1365-294x.1998.00309.x [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Holland B.S. Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst. Biol. 2005;54:952–961. doi: 10.1080/10635150500234674. doi:10.1080/10635150500234674 [DOI] [PubMed] [Google Scholar]
- Russello M.A, Glaberman S, Gibbs J.P, Marquez C, Powell J.R, Caccone A. A cryptic taxon of Galápagos tortoise in conservation peril. Biol. Lett. 2005;1:287–290. doi: 10.1098/rsbl.2005.0317. doi:10.1098/rsbl.2005.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz P, Landry B. Two new taxa of Galagete (Lepidoptera, Autostichidae) from the Galápagos Islands, Ecuador. Rev. Suisse Zool. 2005;112:511–517. [Google Scholar]
- Schmitz P, Landry B. Immature stages of Galagete protozona (Lepidoptera: Gelechioidea: Autostichidae) from the Galápagos Islands: description and notes on biology. Can. Entomol. 2007;139:201–208. [Google Scholar]
- Schmitz P, Cibois A, Landry B. Molecular phylogeny and dating of an insular endemic moth radiation inferred from mitochondrial and nuclear DNA genes: the genus Galagete (Lepidoptera: Autostichidae) of the Galápagos Islands. Mol. Phylogenet. Evol. 2007;45:180–192. doi: 10.1016/j.ympev.2007.05.010. doi:10.1016/j.ympev.2007.05.010 [DOI] [PubMed] [Google Scholar]
- Sequeira A.S, Lanteri A.A, Roque Albelo L, Bhattacharya S, Sijapati M. Colonization history, ecological shifts and diversification in the evolution of endemic Galápagos weevils. Mol. Ecol. 2008;17:1089–1107. doi: 10.1111/j.1365-294X.2007.03642.x. doi:10.1111/j.1365-294X.2007.03642.x [DOI] [PubMed] [Google Scholar]
- Sequeira A.S, Lanteri A.A, Scataglini M.A, Confalonieri V.A, Farrell B.D. Are flightless Galapaganus weevils older than the Galápagos Islands they inhabit? Heredity. 2000;85:20–29. doi: 10.1046/j.1365-2540.2000.00690.x. doi:10.1046/j.1365-2540.2000.00690.x [DOI] [PubMed] [Google Scholar]
- Shapiro L.H, Strazanac J.S, Roderick G.K. Molecular phylogeny of Banza (Orthoptera: Tettigoniidae), the endemic katydids of the Hawaiian archipelago. Mol. Phylogenet. Evol. 2006;41:53–63. doi: 10.1016/j.ympev.2006.04.006. doi:10.1016/j.ympev.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Shaw K.L. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc. Natl Acad. Sci. USA. 2002;99:16 122–16 127. doi: 10.1073/pnas.242585899. doi:10.1073/pnas.242585899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. [Google Scholar]
- Tonnis B, Grant P.R, Grant B.R, Petren K. Habitat selection and ecological speciation in Galápagos warbler finches (Certhidea olivacea and Certhidea fusca) Proc. R. Soc. B. 2005;272:819–826. doi: 10.1098/rspb.2004.3030. doi:10.1098/rspb.2004.3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White W.M, McBirney A.R, Duncan R.A. Petrology and geochemistry of the Galápagos Islands: portrait of a pathological mantle plume. J. Geophys. Res. 1993;98:19 533–19 563. doi:10.1029/93JB02018 [Google Scholar]
- Whittaker R.J. Oxford University Press; Oxford, UK: 2002. Island biogeography: ecology, evolution and conservation. [Google Scholar]
- Wiggins I.L, Porter D.M. Stanford University Press; Stanford, CA: 1971. Flora of the Galápagos Islands. [Google Scholar]