Abstract
The Pacific iguanas of the Fijian and Tongan archipelagos are a biogeographic enigma in that their closest relatives are found only in the New World. They currently comprise two genera and four species of extinct and extant taxa. The two extant species, Brachylophus fasciatus from Fiji, Tonga, and Vanuatu and Brachylophus vitiensis from western Fiji, are of considerable conservation concern with B. vitiensis listed as critically endangered. A recent molecular study has shown that Brachylophus comprised three evolutionarily significant units. To test these conclusions and to reevaluate the phylogenetic and biogeographic relationships within Brachylophus, we generated an mtDNA dataset consisting of 1462 base pairs for 61 individuals from 13 islands, representing both Brachylophus species. Unweighted parsimony analyses and Bayesian analyses produced a well-resolved phylogenetic hypothesis supported by high bootstrap values and posterior probabilities within Brachylophus. Our data reject the monophyly of specimens previously believed to comprise B. fasciatus. Instead, our data demonstrate that living Brachylophus comprise three robust and well-supported clades that do not correspond to current taxonomy. One of these clades comprises B. fasciatus from the Lau group of Fiji and Tonga (type locality for B. fasciatus), while a second comprises putative B. fasciatus from the central regions of Fiji, which we refer to here as B. n. sp. Animals in this clade form the sister group to B. vitiensis rather than other B. fasciatus. We herein describe this clade as a new species of Brachylophus based on molecular and morphological data. With only one exception, every island is home to one or more unique haplotypes. We discuss alternative biogeographic hypotheses to explain their distribution in the Pacific and the difficulties of distinguishing these. Together, our molecular and taxonomic results have important implications for future conservation initiatives for the Pacific iguanas.
Keywords: Pacific biogeography, island biogeography, molecular phylogeny, speciation, Iguaninae, Brachylophus bulabula
1. Introduction
The occurrence of true iguanas in the western Pacific has long fascinated biogeographers because their closest relatives occur exclusively in the Americas. The origin of the clade that comprises the iconic Fijian iguanas (Brachylophus) and their extinct relatives from Fiji and Tonga (Brachylophus, Lapitiguana) is often presented as the product of one of the longest overwater dispersal events inferred for a terrestrial vertebrate—over 8000 km of ocean separates them from their nearest relatives (Gibbons 1981). Although there is debate about how and when such a dispersal event happened (Cogger 1974; Gibbons 1981, 1985), or if it transpired (Gorham 1965; Worthy 2000; Burns et al. 2006), there is no dispute that the Pacific Island iguanas comprise part of the monophyletic Iguaninae and are a divergent sister group to other Iguaninae based on both morphological and molecular data (Etheridge & De Queiroz 1988; Frost & Etheridge 1989; Wiens & Hollingsworth 2000; Schulte et al. 2003).
The Pacific Island iguanas currently comprise two genera and four species. Lapitiguana impensa was a gigantic (500 mm snout–vent length; SVL), presumably ground-dwelling, species described from Late Quaternary fossil deposits from Viti Levu in Fiji (Pregill & Worthy 2003). The genus Brachylophus comprises the now extinct Brachylophus gibbonsi, another large species (350 mm SVL) described from Late Holocene fossil deposits from Tonga (Pregill & Steadman 2004), and two extant species. The Fijian banded iguana, Brachylophus fasciatus (Brongniart 1800), is known historically from the largest Fijian Islands of Viti Levu and Vanua Levu as well as many islands east and south of these two including Kadavu, Ovalau, Gau, Taveuni and the many Lau Group Islands in the east of the Fijian Republic (Gibbons 1981). The Fijian crested iguana, B. vitiensis (Gibbons 1981), is known from the Yasawa, Mamanuca, Yadua and Macuata Islands in the western portion of the Fijian Republic (Gibbons 1981; Alberts 1991; Harlow et al. 2007). There are two additional populations of B. fasciatus whose status has been controversial. There is good evidence that B. fasciatus animals on Vanuatu have recently been introduced from Fiji (Gibbons 1981), but the origin of these animals is unknown. More difficult is the distribution of B. fasciatus known from four islands in Tonga. Based on morphological differences, these animals were described as a new taxon, B. brevicephalus, by Avery & Tanner (1970), which was later synonymized with B. fasciatus by Gibbons (1981). There is disagreement over whether the Tongan animals are endemic to Tonga (Avery & Tanner 1970; Gibbons 1981; Burns et al. 2006) or have recently been introduced from Fiji following the extinction of B. gibbonsi after human arrival in Tonga (Pregill & Steadman 2004; Steadman 2006).
All four Pacific iguanas have undergone a dramatic decline since humans arrived in these islands. The gigantic L. impensa in Fiji and B. gibbonsi in Tonga were both eaten to extinction after the arrival of humans ca 2800 years ago (Steadman et al. 2002; Pregill & Worthy 2003; Pregill & Steadman 2004). The two living species are of particular conservation concern. Brachylophus vitiensis has been heavily impacted by human-mediated habitat loss and the introduction of feral cats, rats and goats, which have severely reduced its numbers across the entire Yasawa and Mamanuca Group Islands (Harlow et al. 2007). It is listed as Critically Endangered on the IUCN Red List with only a single secure population remaining on the island of Yadua Taba (Harlow & Biciloa 2001). The status of B. fasciatus in the wild is largely unknown, but large populations still exist on the two Aiwa islands in the Lau group and Kadavu (Cogger 1974; Harlow 2003).
There have been two previous molecular evaluations of Brachylophus, both of which sought to provide some clarification of the evolutionary relationships among living populations, which in turn could be used to help clarify conservation priorities. Based on a small number of samples (15), Colgan & Da Costa (1997) used allozymes and sequence data from a short (300 base pair; bp) fragment of cytochrome b to show that B. fasciatus and B. vitiensis are indeed genetically divergent. More recently, Burns et al. (2006) used five polymorphic nuclear microsatellites and the same short cytochrome b fragment on a larger number of samples (35) to produce a molecular phylogeny for the group. Based on their mtDNA phylogeny, Burns et al. (2006) identified three major clades; they suggested that B. fasciatus and B. vitiensis as currently understood are not reciprocally monophyletic, and that B. fasciatus animals from Tonga should be recognized as a separate species. Then they went on to identify their three clades as evolutionarily significant units (ESUs) for conservation (Burns et al. 2006).
Unfortunately, Burns et al. (2006) were not able to include the animals from the Lau group in eastern Fiji, which means that the interpretation of their dataset is necessarily limited with regard to both testing the taxonomic status of Tongan B. fasciatus and clearly defining ESUs. Once defined, ESUs often become the focus for management action and investment of limited resources. As this genus is endangered and continues to decline, taxonomic confusion and resultant management actions may result in further loss of diversity, as was seen in the New Zealand tuatara during much of the last century (Daugherty et al. 1990). Currently, enough confusion still exists about the systematics and the distribution of extant Brachylophus species to warrant a reappraisal of these iconic animals.
Based on greater sampling and a substantially larger molecular dataset, we generate a comprehensive molecular phylogeny to test the primary results of Burns et al. (2006) with a particular focus on the status of the Tongan B. fasciatus animals and the number, composition, distribution and description of ESUs. We build on their and our results with a detailed evaluation of morphological variation among the major identified clades. We combine our molecular data with the published data for other members of the Iguaninae for a comparison of the levels of molecular differentiation, and also discuss the alternative biogeographic scenarios provided to explain the Pacific distribution of this clade and the weight of evidence required to test these scenarios. Based on our molecular, morphological and distributional data, we herein describe a new species of Fijian iguana and discuss the conservation implications of our findings.
2. Material and methods
(a) Taxonomic sampling for molecular data
Sequence data were obtained for a total of 61 individuals representing 13 islands, which include much of the current known distribution of these critically endangered animals (figure 1; electronic supplementary material, table 1). Field-collected tissue samples consisted of either blood samples, scale or tail–tip clips, which were stored in 100 per cent ethanol (no animals were killed) or muscle/liver was collected from limited voucher specimens. These samples were collected and exported under CITES permit EP 8/33/2 and through pre-CITES permits from the Republics of Vanuatu and Fiji. Several samples were obtained from museum collections (electronic supplementary material, table 1). We were able to obtain the most samples of B. vitiensis because these islands have been recently surveyed for iguanas (Harlow et al. 2007). The recent survey for iguanas in the eastern Lau group of islands covered only the island of Aiwa Lailai, so our sampling in this region is poor. We also have no samples from the two largest Fijian islands (Viti Levu and Vanua Levu) because iguanas are extremely rare on these islands, probably due to the introduced mongoose, Herpestes javanicus, but we were able to include B. vitiensis animals from the island of Macuata, which is less than 1 km off the north coast of Viti Levu. We were able to include Brachylophus samples from Tonga and Vanuatu to test the origins and affinities of these two potentially introduced populations. Importantly, our sampling includes animals from the type localities for both B. vitiensis (Yadua Taba) and B. fasciatus (Tonga).
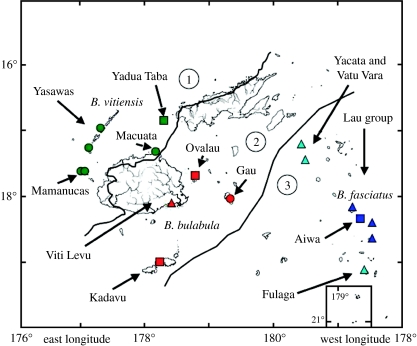
Figure 1.
Species distributions for Brachylophus iguanas in Fiji based on genetic and morphological analyses with B. bulabula sp. nov. in red, B. fasciatus in blue and B. vitiensis in green. Turquoise triangles represent populations that were assessed morphologically but not assigned to a species due to the limited number of samples. Most island names mentioned in the text are noted. Numbers represent proposed biotic provinces for terrestrial herpetofauna as defined by Olson et al. (modified from Olson et al. in press). 1, Fiji dry forest; 2, Fiji moist forest; 3, Lau group. Squares, both genetics and morphology; circles, only genetics; triangles, only morphology. Background map courtesy of the Smithsonian Institution.
(b) Choice of phylogenetic markers
We generated sequence data for the mitochondrial DNA genes ND4 and cytochrome b because they have proved useful in the phylogenetic studies of numerous lizard groups and because a large number of sequences were available on GenBank for both genes for other iguanid lizards. For ND4, we targeted an approximately 900 bp DNA fragment that included the 3′ half of the ND4 gene and most of the tRNA cluster containing the histidine, serine and leucine tRNA genes. This region has proved particularly useful at comparable taxonomic levels in numerous squamate reptile groups, including other iguanas (Malone et al. 2000). For cytochrome b, we targeted an approximately 900 bp DNA fragment that has proved very useful in the phylogenetic studies of many squamate groups, including iguanids (Petren & Case 1997). Because phylogenetic studies comprising multiple unlinked loci can provide independent corroboration of conclusions drawn from single-gene phylogenies, we also generated nuclear DNA data for the commonly used genes RAG-1 and Rhodopsin but they were invariant within Brachylophus (data not shown). Therefore, we generated a morphological dataset to complement the mtDNA dataset (see below).
(c) DNA extraction and polymerase chain reaction
Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) protocol, suspended in TE and stored at −20°C. Two mitochondrial fragments were targeted. An ND4 fragment was amplified using a modified primer pair ND4—TGACTACCAAAAGCTCATGTAGAAGC; and tRNA-Leu—TRCTTTTACTTGGATTTGCACCA (Arévalo et al. 1994). The target cytochrome b fragment was amplified using a modified primer pair rGlu-1L—GAAAAACCRCCGTTGTWATTCAACTA; and rCytb-1H—GCGTAGGCRAATAGGAAGTATCA. Targeted fragments were amplified in 50 μl reactions using GoTaq Green Master Mix (Promega) using approximately 100 ng of genomic template DNA and 10 pmol of each primer. Polymerase chain reaction (PCR) amplification was done using a step-down cycling profile (94°C, 5 min; followed by a series of touchdown cycles of 94°C, 30 s; 65–45°C, 20 s; 72°C, 90 s each repeated twice; with a final cycle of 94°C, 30 s; 40°C, 30 s; 72°C, 45 s repeated 40 times; then held at 72°C, 4 min repeated once; finishing at 4°C, 1 min) on a Corbett PC-960C cooled thermal cycler. All PCR products were run out on a 2 per cent agarose gel and purified using UltraClean 15 DNA purification kit (MoBio laboratories Inc.) as specified by the manufacturer. Cycle-sequencing reactions were performed using BigDye (Applied Biosystems) and approximately 100 ng of purified PCR product according to the manufacturer's specifications. Cleaned reactions were resuspended in HiDi formamide and run on an ABI 3100 autosequencer. Sequences were edited and assembled using Sequencher v. 3.0 (Genes Codes Corporation) and easily aligned manually. Protein-coding regions were translated into amino acid sequences using the mammalian mitochondrial genetic code to ensure an open reading frame, and were compared to various other squamate reptile translations on GenBank to check for stop codons and frame shifts.
(d) Phylogenetic analyses
In order to examine the phylogenetic affinities of Brachylophus within the Iguaninae, we included GenBank data for all available members of the family. A number of short fragments (300 bp) for Brachylophus were available on GenBank from the studies of Colgan & Da Costa (1997) and Burns et al. (2006), but we did not include them because our sampling included these same animals for which we generated complete sequences for both genes. From GenBank we did include a number of sequences of other members of the Iguaninae for comparison. In total, we obtained 31 additional sequences of the mtDNA gene ND4 and 13 additional sequences of the mtDNA gene cytochrome b, which include at least one species of all described genera in the Iguaninae (Amblyrhynchus, Conolophus, Ctenosaurus, Cyclura, Dipsosaurus, Iguana, Sauromalus). Liolaemus kriegi was used as the out-group for all analyses based on its sister group relationship to the Iguaninae (Schulte et al. 2003).
We ran two sets of phylogenetic analyses, one set with the individual genes and the other with the combined data. For the individual datasets, we ran heuristic parsimony searches with the computer program PAUP* (Swofford 2002) to get an assessment of phylogenetic resolution provided by each dataset alone. We then performed a partition-homogeneity test in PAUP* to determine whether the two individual datasets were heterogeneous with regard to the phylogenetic signal. For the combined dataset, we used parsimony and Bayesian approaches. We used TBR branch swapping and ran the parsimony analyses five times from random starting points and with random sequence addition to confirm that overall tree space was well searched. Bayesian analyses were run with the computer program MrBayes (v. 3.0b4; Huelsenbeck & Ronquist 2001) and we partitioned the data into the ND4 and cytochrome b components. We allowed all parameters to be estimated from the data during the run. We used the default value of four Markov chains per run and also ran the full analysis four times to make sure that the overall tree space was well sampled and to avoid getting trapped in local optima. We ran each analysis for a total of 5 000 000 generations and sampled the chain every 100 generations, resulting in 50 000 sampled trees. We discarded the first 5000 trees and used the last 45 000 trees to estimate Bayesian posterior probabilities.
For both sets of analyses, we generated 10 000 ‘fast’ unweighted non-parametric parsimony bootstrap replicates to assess branch support and for the combined dataset we also had Bayesian posterior probabilities. In keeping with the commonly used convention, we consider branches supported by parsimony bootstrap values greater than or equal to 70 per cent and posterior probability values greater than or equal to 95 per cent to be worthy of discussion.
(e) Morphology
We reviewed the detailed morphological studies of Avery & Tanner (1970) and Gibbons (1981) and examined museum specimens and living specimens in the wild and captivity to determine the usefulness of characters described in determining differences between species (or major haplotype clades). We evaluated 37 characters for their usefulness in discriminating the species (14 presented in previous studies and 23 additional characters). Unfortunately, there are few museum specimens and most of these do not have reliable geographical data associated with them. Some of these were acquired by the museums from zoological institutions and many have facial scarring from being in captivity, which makes scoring certain characters suspect. Others in museums are captive offspring from wild-caught animals. Therefore, to be conservative we only examined and scored specimens with accurate locality data (electronic supplementary material, table 1). We grouped specimens into their presumptive haplotype clade based on geography. Thus, we consider specimens from Kadavu, Nairai, Ovalau and Viti Levu to represent Brachylophus n. sp., and specimens from Lakeba, Aiwa, Oneata, Moce and Tonga (type locality of B. fasciatus) to represent B. fasciatus. Specimens from Vanua Levu, Yacata, Vatu Vara and Fulanga were not put in either group pending analysis of larger sample sizes because only one specimen was available from each island and we had no tissue samples from these islands. Brachylophus vitiensis is represented only in museums by specimens collected from Yadua Taba, most of which were included in the original description. Data for this species come from Gibbons (1981) and the few wild-caught paratypes at the Museum of Comparative Zoology, Harvard, and one former captive specimen from the United States National Museum of Natural History. Measurements of live animals were made for some characteristics and coloration representing the different species as follows: Brachylophus n. sp. from live animals at San Diego Zoo sourced from the Orchid Island Cultural Centre; B. fasciatus from live animals from Aiwa Lailai Island; and B. vitiensis from live animals from Yadua Taba Island.
Recent skeletal material with known localities is very rare for this group so we use only external characteristics for the description and to discriminate the three living species. Future work with internal morphology may yield additional differences between the living species, and a careful analysis of skeletal material will be necessary to resolve the relationships of the three living species to the extinct larger allopatric B. gibbonsi (Pregill & Steadman 2004) or the gigantic monotypic sympatric genus L. impensa (Pregill & Worthy 2003). Museum acronyms follow Leviton et al. (1985).
3. Results
(a) Molecular phylogenetics
The edited alignment of the combined dataset comprised 1462 characters (700 ND4+tRNA; 762 cytochrome b), and of these 442 (30.2%) were variable and parsimony informative (202 ND4+tRNA; 240 cytochrome b) across the entire Iguaninae (excluding the out-group L. kriegi) and 136 (9.3%) were variable and parsimony informative (58 ND4+tRNA; 78 cytochrome b) within Brachylophus. Following alignment, protein-coding mtDNA sequences were translated into amino acid sequences using the vertebrate mitochondrial genetic code. No premature stop codons were observed or any other sign of paralogous sequences; therefore, we concluded that all sequences obtained were mitochondrial in origin. Each individual dataset produced well-resolved and well-supported phylogenetic trees and virtually identical topologies both within the Iguaninae as a whole and within Brachylophus based on parsimony (electronic supplementary material, figure 1). A partition-homogeneity test in PAUP* could not reject the null hypothesis that the data were homogeneous with regard to overall phylogenetic signal (p>0.01). Therefore the rest of the results and our discussion are based on analysis of the combined data, which are summarized in the phylogenies shown in figure 2.
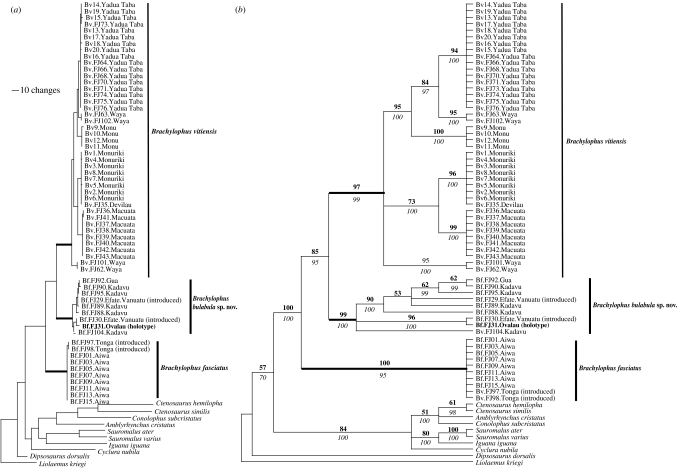
Figure 2.
(a) Parsimony phylogram of the Iguaninae based on combined ND4 and cytochrome b mitochondrial DNA sequences. (b) Parsimony bootstrap consensus tree. Numbers in bold represent bootstraps and numbers in italics represent posterior probabilities. See text for details and the electronic supplementary material, figure 1, for individual gene trees. For both figures, non-Brachylophus individuals include only species for which both genes were available on GenBank (accession numbers in figure 1 in the electronic supplementary material). All Brachylophus samples are from Fiji except the introduced populations in Vanuatu and Tonga. The holotype of B. bulabula sp. nov. (CAS 172524) from Ovalau Island is indicated in bold and the type localities for B. vitiensis (Yadua Taba) and B. fasciatus (Tonga) were included in our sampling.
The unweighted parsimony analyses and the Bayesian analyses of the combined dataset produced virtually identical topologies. Figure 2a shows a representative phylogram of the most parsimonious trees from the unweighted parsimony analysis, and figure 2b shows parsimony bootstrap support values and Bayesian posterior probabilities for the combined dataset. The phylogeny is well resolved and supported by high bootstrap values and posterior probabilities within Brachylophus but poor support for intra-generic relationships (figure 2). Within Brachylophus, our molecular dataset rejects the monophyly of specimens currently identified as B. fasciatus. Instead, our data strongly demonstrate that living Brachylophus comprise three robust and well-supported clades. One clade represents B. vitiensis, primarily from the Fijian dry forest (figure 1; Olson et al. in press) and including samples from the type locality (Yadua Taba). The other two clades comprise animals that have traditionally been allocated to B. fasciatus. One of these clades comprises true B. fasciatus as represented by animals from the Lau group of Fijian islands and Tonga (type locality for B. fasciatus), while the other clade comprises animals from the central regions of Fiji on Kadavu, Gau and Ovalau (Fiji moist forest; Olson et al. in press), which we refer to here as Brachylophus n. sp. Animals in this clade form the sister group to B. vitiensis rather than B. fasciatus. These three clades were also strongly supported in the individual gene trees (electronic supplementary material, figure 1). Genetic distances between the three clades are similar to or larger than that between species within a number of other iguanid genera (electronic supplementary material, table 2).
Intra-specific genetic distances and haplotype diversity were extremely low in B. fasciatus with only two haplotypes identified, one from Aiwa and the other from Tonga, but we have samples from only these two localities (electronic supplementary material, table 2). Genetic distances and haplotype diversity within B. vitiensis was much higher (electronic supplementary material, table 2) and, with only one exception, each island supports at least one unique haplotype and represents a clade in our phylogeny (figure 2). Yadua Taba, the island that is home to the only remaining large population of B. vitiensis, and for which we have the best sampling, displays the most haplotypes with five, Monu and Monuriki each have three haplotypes and Macuata has two. The only haplotype shared between islands was between Devilau and Monuriki. The four samples from Waya yielded two unique haplotypes. One of these haplotypes is closely allied with samples from Yadua Taba. Genetic distances (electronic supplementary material, table 2) and haplotype diversity within Brachylophus n. sp. were intermediate between B. fasciatus and B. vitiensis. We had five samples from the large island of Kadavu and each had a unique haplotype. Each of our single samples from Gau and Ovalau also displayed unique haplotypes.
We included animals from two putative introduced populations in order to examine their possible place of origin. The samples from Tonga are clearly B. fasciatus and extremely close to the Aiwa animals from the Lau group (1 bp difference). The samples from Vanuatu are clearly Brachylophus n. sp. One sample shares a haplotype with an animal from Kadavu and the other is close to the animal from Ovalau.
(b) Morphology
We found several differences in the literature and our analysis that are diagnostic for the three taxa—Brachylophus sp. nov. (figure 3a,b); B. fasciatus (figure 3c,d); and B. vitiensis (figure 3e,f), which we summarize in table 1. We highlight the traits with the greatest consistency within our population samples. First, the maximum size: SVL of up to 235 mm in B. vitiensis (S. Morrison 2007, unpublished data); to 193 mm in B. sp. nov. (Gibbons 1981; listed as B. fasciatus in his table 2); and to 182 mm in B. fasciatus (our data, n=40). Second, the nasal scale shape and colour (figure 4): B. sp. nov. has a circular nasal scale with a round or slightly elliptical opening in the centre and the entire scale is yellow; the nasal scale in B. vitiensis is ‘elliptical with opening often off-centre’ (Gibbons 1981) and we observed that it is off-centre in a frontal–ventral direction, the nasal yellow-orange with the colour extending to adjacent scales; and B. fasciatus has a nasal scale that is slightly elongate/elliptical with an often slit-like opening in the centre, which is offset dorsally, the nasal is orange and the colour does not cover the entire scale (figure 4). In preserved B. sp. nov. specimens over 100 years old, the nasal scale colour is still quite distinctive (ANSP 8316; BMNH 1946.8.3.84). Third, all male B. sp. nov. were observed to have a nuchal stripe extending post-aural over the shoulder, forming a ‘U’ shape over the back neck of the animals, although some have a break in the stripe near the vertebral crest. This character can be easily seen in the first published illustrations of B. sp. nov. (as B. fasciatus) by Girard (1858, Plate XVIII, fig. 8) and in Günther (1862, Plate XXV; BMNH 1946.8.3.84; see the electronic supplementary material, figure 2). Brachylophus vitiensis has this same stripe but it is often thinner and more ‘V’ shaped over the shoulders from a dorsal view. Brachylophus fasciatus lacks this stripe and has nuchal spotting instead, sometimes with small bars but never a more continuous stripe (Brongniart 1805; figure 4). Fourth, in preserved material both B. sp. nov. and B. vitiensis have enlarged, elongated and often coloured parietal eyes, whereas B. fasciatus often has smaller and nondescript parietal eye. Fifth, there is a gradient in the numbers of femoral pores (counting enlarged and smaller ones), with B. vitiensis generally having 12–40 but with the vast majority more than 30 (X=32.7, Gibbons 1981; X=31.23, n=123 our study), followed by B. sp. nov. having 21–39 (X=27.2, Gibbons 1981; X=30, n=19 our study), then B. fasciatus having 14–27 (X=23.7; n=13 our study). Sixth, there are differences in the dorsal crest maximum height, with B. vitiensis spines averaging 6.5 mm (P. S. Harlow 2005–2007, unpublished data) with a maximum length exceeding 10 mm (Gibbons 1981; P. S. Harlow 2005–2007, unpublished data), B. sp. nov. spines averaging 2.1 mm with a maximum not exceeding 4 mm (Gibbons 1981) and B. fasciatus spines averaging 1.4 mm and not exceeding 2 mm in height. Seventh, B. sp. nov. and B. fasciatus display striking sexual dimorphism in colour patterns with the females generally lacking all banding patterns, whereas B. vitiensis females display striping similar to the males. Eighth, eye colour differs between species, with B. sp. nov. having a distinctive red colour iris, B. vitiensis having a tan–gold colour and B. fasciatus having a more orange-red coloured iris.
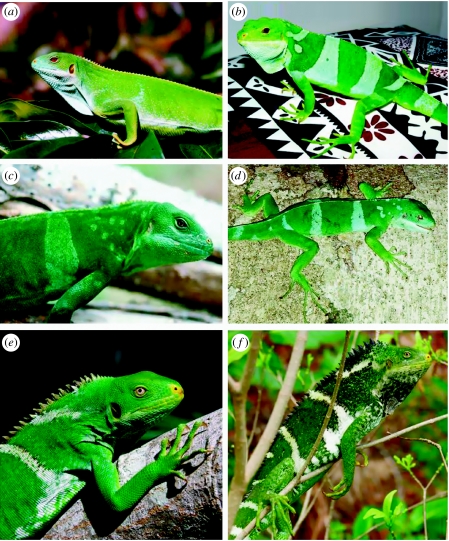
Figure 3.
(a) Female B. bulabula sp. nov. from Kadavu Island, Fiji; photo by Peter Harlow. (b) Male B. bulabula sp. nov. from Kadavu Island, Fiji; photo by Paddy Ryan. Note the sexual dimorphism in colour pattern and diagnostic nuchal stripe, yellow nostril and red eye colour in B. bulabula sp. nov. (c) Male B. fasciatus from Eueiki Island, Tonga, representing the introduced populations that are type location (country) for B. fasciatus; photo by Harold Cogger. (d) Male B. fasciatus from Aiwa Levu Island, Fiji, representing the native populations of this species from the Lau Group Islands; photo by Greg Pregill. Note the diagnostic B. fasciatus features of nuchal spotting, reduced nostril coloration and golden eye colour. (e) Female B. vitiensis from Yadua Taba Island, Fiji. (f) Male B. vitiensis from Yadua Taba Island, Fiji. Both photos by Suzanne Morrison. Brachylophus vitiensis is highly variable in coloration and pattern. Note the prominent crest that is both larger and darker than B. bulabula or B. fasciatus, lack of colour pattern dimorphism, the nuchal stripe that it shares with B. bulabula and the increased yellow nasal coloration and golden eye colour.
Table 1.
Summary of the distinguishing morphological characteristics between the three Brachylophus species.
| B. bulabula | B. fasciatus | B. vitiensis | |
|---|---|---|---|
| maximum body size | 193 mm | 182 mm | 235 mm |
| nasal shape | circular | elongate/elliptical | elliptical |
| nasal colour | yellow | orange but not covering entire scale | yellow-orange with colour extending to surrounding scales |
| nostril shape | round or slightly elliptical and in the centre of nasal scale | slit like and in the centre of nasal scale, offset dorsally | elliptical and off-centre in a frontal–ventral direction in nasal scale |
| nuchal stripe | wide and U shaped | spotting rather than a stripe | thinner and V shaped |
| parietal eye | enlarged, elongated and coloured | small, nondescript | enlarged, elongated and coloured |
| mean no. femoral pores | X=30, s.e.=1.3 | X=24, s.e.=1.0 | X=31, s.e.=1.0 |
| crest height | X=2.1 mm, max=4 mm | X=1.4 mm, max=2 mm | X=6.5 mm, max=10 mm |
| colour pattern sexual dimorphism | extreme with males displaying striking bands and females lacking bands | extreme with males displaying striking bands and females lacking bands | very subtle to non-existent with both sexes displaying striking bands |
| eye colour | red iris | orange-red iris | tan–gold iris |
Figure 4.
Nasal scales of (a) B. bulabula, (b) B. fasciatus and (c) B. vitiensis. Note the difference in shape and colour. The nasal scale of B. bulabula is circular with a central placement of the nostril and yellow pigmentation. The nasal scale of B. fasciatus is elliptical with a more dorsal placement of the slit-like nostril and reduced orange pigmentation that does not cover the entire nasal. The nasal scale of B. vitiensis is elliptical with an elliptical nostril and yellow-orange coloration that extends to the surrounding scales.
We searched the literature for possible synonyms for B. fasciatus, which might have been available as names for this unnamed taxon and could find none available. Thus, we describe the third haplotype clade as a new species.
Systematics.
Reptilia
Lepidosauria
Squamata
Iguanidae
Iguaninae
Brachylophus Cuvier
Brachylophus bulabula sp. nov. Fisher, Harlow, Edwards & Keogh.
Holotype. California Academy of Sciences (CAS) 172524 (Field number CAS 9205), adult male; Navuloa Village, Ovalau Island, Republic of Fiji (17°42′05.95″ S, 178°45′42.12″ E); collected by R. N. Fisher, D. T. Bolger & T. J. Case, 8 October 1988.
Paratypes. Kadavu Island, Republic of Fiji: CAS 54664, adult female; USNM 230303, adult male; AMNH R-40474, sub-adult male; BMNH 1882.8.29.70–71; MCZ R-15010, adult male; MCZ R-15011. Ovalau Island, Republic of Fiji: MCZ R-6458; MCZ R-147303, adult male. Viti Levu Island, Republic of Fiji: BPBM 1824, adult male; AMNH 12827, sub-adult male; USNM 51000, adult male; MCZ R-15006, sub-adult male. Island not specified, Republic of Fiji: BMNH 1946.8.3.84, adult male (Günther 1862; plate XXV).
Diagnosis. Brachylophus bulabula is distinguished from both B. fasciatus and B. vitiensis by its intermediate maximum size, intermediate crest height, circular and yellow nasal scale, rounded nostril, wide and U-shaped nuchal stripe and red iris. These and additional morphological characters that distinguish B. bulabula from B. fasciatus and B. vitiensis are summarized in table 1.
Description of Holotype. Adult male. Meristics are as follows: snout–vent length 165 mm, tail length 475 mm, head length 30 mm, head width 19 mm, head height 22 mm and jaw length 23 mm. The jaw has 21 elongated upper labials (11 left, 10 right) and 20 elongated lower labials (9 left, 11 right). Rostral enlarged but otherwise unremarkable, mental is incised half-way anteriorly. There are six paired post-mental scales. Tympanum translucent and unpigmented, vertical and oval in shape with the same height as the eye is wide. Nasal scale is slightly oval with the opening at the centre and entirely yellow. Parietal eye large and opaque, equivalent to the length of nostril opening. Scale containing parietal eye is larger than most adjacent scales. Supraocular head scales are the smallest on the head, other head scales are larger and polygonal in shape. The dewlap is medium in size, V shaped in outline and has black spots. Poorly defined gular pouch covered with small scales. One nuchal band originating at the tympanum and continuing posteriorly to the nuchal crest, three dorsal body bands with the widest being 20 mm, 1 axial band, 11 tail bands and the banding fades posteriorly into background tail coloration. The dorso-nuchal crest is well defined with spike-shaped spines up to 2 mm in height and 2 mm in width on the nuchal region. The crest on the back region has spines angled backwards. Dorsal scales are small and conical shaped. Ventral scales are much larger than dorsal scales and elongated and strongly keeled, with posterior ends pointed and elevated. Legs are long and thin and covered with keeled scales similar in size to ventral scales. Digits are elongate and palms are covered with small keeled scales. Third digit of hind foot contains a denticulate comb on proximate phalanx with some fusion of scales. Toenails are very long and not worn down. Total of 38 femoral pores (20 left, 18 right) counting all differentiated scales with pore opening (not just ones with obvious exudate). Larger medial pores have orange waxy exudate. Tail is laterally compressed in cross section and weakly crested. Anterior dorsal scales on tail are smaller than ventral scales. All scales are keeled and posteriorly all scales become similar in size and larger.
Colour in life. Head and body background colour lime-green extending around both dorsal and ventral surfaces. Three wide bands cross from ventral edge over dorsum, not crossing ventral surface. These bands are 15–20 mm in width at the widest points. Another thinner band crosses tail at axilla, and a thin nuchal band starts behind external ear opening crossing the back of neck above shoulders. Banding continues on tail with 11 bands present, before fading into background posteriorly. All bands and patches of spots on background are light pale blue colour. Eye is red-orange. Nasal is bright yellow. Underside of throat is paler whitish-blue. Some blue spotting on legs, especially hind limbs and shoulders. Tail becomes darker and brown to black towards tip.
Colour in alcohol. Overall dorsal colour is greatly darkened with banding obscured. Ventral colour maintains lime-green to blue coloration. Dewlap contains black spotting pattern. Nasal scale retains yellow colour. Parietal eye opaque. Tympanum translucent. Tail becomes brownish and bands become less obvious towards tip.
Variation. We have described above some of the variability in this species for the main characteristics that distinguish it from the other living forms. Additional variability includes paired post-mental scales occurring in 60 per cent of the individuals versus B. fasciatus that never has paired post-mental scales. Post-nasal and preocular scales tend to be large with fewer scales in 87 per cent of the individuals, whereas both B. fasciatus and B. vitiensis tend to have smaller scales. Larger octagonal-shaped head scales occur in 83 per cent of specimens, and dorsal crest scales vary in numbers between 55 and 88, with an average of 67.5.
Etymology. The specific epithet is from the Fijian word bula, an all-purpose greeting that literally means ‘to live’, but is most commonly used for ‘hello’, ‘how are you?’ or even just ‘good’. The double bula is a more enthusiastic greeting, and would be used with a close friend. Thus we use bulabula to designate the more familiar banded iguana or ‘Vokai’ of the main Fijian islands, the iguana that is more often depicted as the ‘typical’ banded form. Used as a noun in apposition.
Distribution. The distribution of B. bulabula as currently understood is limited to the larger northwestern islands of the Fijian Islands (Viti group). Specifically, we include iguanas from Ovalau, Gau, Kadavu and Viti Levu in this species based on their relationships from mtDNA and morphology. Further morphological assessment of georeferenced specimens is required to determine more accurately this species' range. Animals from the Lau group (Lakeba, Aiwa, Oneata and Moce) are included in B. fasciatus. While animals from the northern islands (i.e. Vanua Levu, Taveuni, Yacata, Vatu Vara) and southern islands (i.e. Fulaga) have been examined, we have not assigned them to either species due to small sample sizes. Brachylophus bulabula is introduced onto Vanuatu on Efate Island (Gibbons 1981) and this species identification is confirmed in our study with both genetic and morphological data (specimens CAS 172419-20).
4. Discussion
We have demonstrated with a large mtDNA sequence dataset that living Brachylophus comprise three monophyletic clades that do not correspond to current taxonomy. Our molecular findings are corroborated by morphological and distributional data and we herein describe a previously unrecognized species of Brachylophus from the central Fiji Islands. Our molecular data identify the likely origin of the introduced Brachylophus on Vanuatu, and our interpretation of the molecular, fossil and anthropological evidence is that B. fasciatus on Tonga are most likely recent transports from the Lau Group Islands in eastern Fiji. With only one exception, every island is home to one or more unique haplotype. Together, our molecular and taxonomic results have important implications for future conservation initiatives for the Fijian iguanas.
(a) Phylogeny and taxa
Our primary result is that the genus Brachylophus comprises three divergent species-level extant lineages rather than two (figure 2): B. fasciatus (Brongniart 1800) from eastern Fiji and Tonga, B. vitiensis (Gibbons 1981) from western Fiji and B. bulabula sp. nov. from central Fiji (figure 1). This result is supported by morphological and distributional data and is broadly consistent with the published allozyme, microsatellite and sequence data (Colgan & Da Costa 1997; Burns et al. 2006). Each of the three species differs by 3.3–5.6% (ND4) or 4.2–6.5% (cytb) uncorrected sequence divergence from the other two, divergences that are similar to or larger than for a number of other iguana species (see comparisons in the electronic supplementary material, table 2). Based on a simplistic molecular clock of 1–2%/million years for mtDNA, the divergences date to 3.3–13 Ma. The highest islands of Fiji are over 1000 m above sea level, were emergent during the Late Oligocene to Middle Miocene and have been continuously above sea level for at least the last 16 million years (Chase 1971; Rodda 1994). It is likely that extinct and extant members of the Pacific iguana radiation have been on the islands for much of this time.
Our topology is similar in a broad sense to the sequence-based topology of Burns et al. (2006) who identified three ‘ESUs’ that largely correspond to our clades, but our results and interpretations differ in several ways. Burns et al. (2006) suggested that B. vitiensis and B. fasciatus were not reciprocally monophyletic. Their result rested entirely on the phylogenetic placement within the B. vitiensis clade of two B. fasciatus individuals sampled from captive individuals (Kula Eco Park, Fiji) presumed to have been collected on Ovalau. However, in our dataset the placement of an additional wild-collected Ovalau specimen in the B. bulabula clade (holotype of B. bulabula) strongly contradicts their result. All three species are reciprocally monophyletic and very well supported in our dataset. Importantly, the microsatellite data presented by Burns et al. (2006) also suggest that there may be a problem with the Ovalau sample used in their study. If this animal is excluded, their independent nuclear microsatellite data provide additional independent support for three ESUs (fig. 2 of Burns et al. 2006). The most likely explanation for the Burns et al. (2006) result is that there was either a labelling error at the time the tissue was collected (many B. vitiensis in the collection at Kula Eco Park were also sampled at this time) or that the two captive animals were not from Ovalau.
Burns et al. (2006) also showed that B. fasciatus in Tonga comprise a divergent third clade of Brachylophus. Based on the morphological differences, these animals were described as a new taxon, B. brevicephalus, by Avery & Tanner (1970), but it was synonymized with B. fasciatus by Gibbons (1981) who considered them one end of a cline. Burns et al. (2006) interpreted their molecular data as being consistent with the resurrection of B. brevicephalus for Tongan animals, but they did not have any animals from the Lau Group Islands with which to adequately test this hypothesis. The presence of the now extinct congener B. gibbonsi in Tonga suggests that it may be reasonable to assume that the living populations of B. fasciatus in Tonga are endemic. However, several pieces of evidence strongly contradict this hypothesis. While alone it does not provide information on the directionality of movements, the molecular data clearly show that the Tonga specimens are genetically almost identical to Fijian Lau island group B. fasciatus, as predicted by Steadman (2006) based on the lack of any fossil evidence of B. fasciatus on Tonga. Given the level of genetic differentiation among island samples of the two other Brachylophus species, we would expect sizable genetic differences between Fiji and Tonga if the Tongan animals were endemic. Ample evidence exists of trade between Fiji and Tonga prior to European discovery. For example, when Captain Cook visited Tonga in the 1770s, he collected the red shining parrot, which was subsequently described as a Tongan species (Prosopeia tabuensis). The parrot's bright red feathers had long been a valuable trade item for Tongan traders visiting Fiji, as they were used to adorn fine mats worn by Tongan chiefs. Prior to Cook's visit, live individuals of this Fijian endemic had also been exported to Tonga (Watling 1986). The complete lack of any sub-fossil material for B. fasciatus from Tonga (Steadman 2006), but widespread sub-fossil evidence of B. fasciatus from the Lau group (G. K. Pregill & D. W. Steadman 2000–2002, unpublished data), is consistent with the idea that the Tongan animals were introduced from the Lau Group Islands, possibly as an alternative food source after B. gibbonsi was eaten to extinction some 2800 years ago (Pregill & Dye 1989; Pregill & Steadman 2004).
Iguanas on Vanuatu have been introduced fairly recently by people working in the pet trade (Gibbons 1981), but their origin was not known. Our molecular data clearly show that these animals are genetically part of the B. bulabula clade, but the two animals we were able to sample have two different haplotypes. One is more closely allied with the animals from Kadavu and Gua and the other is most closely allied to an animal from Ovalau. This suggests that the animals introduced into Vanuatu possibly came from multiple sources, or these haplotypes are broadly distributed across the range of B. bulabula.
The distribution of the three species matches well with a recent evaluation of the major biotic provinces in Fijian forests, which divides Fijian Islands into three broad groups (figure 1). Brachylophus vitiensis occurs in Fijian dry forests in the northwest, B. bulabula inhabits Fijian wet forest and B. fasciatus occurs in the Lau group to the southeast, which are comparatively dry (Harlow 2003). This pattern is in broad agreement with the habitat preferences identified previously (Gibbons 1981; Harlow 2003; Harlow et al. 2007). However, our molecular and morphological sampling does not include most of the islands from which Brachylophus were once known and so our inferences about the historical distribution of these three species are limited. We are confident that B. vitiensis is (was) distributed across the Yasawas and Mamanucas (Gibbons 1985), and is also found on Macuata, 1 km from the Viti Levu coast, but it is very difficult to comment on the wider distribution of B. vitiensis or B. bulabula on either Viti Levu or Vanua Levu based on samples for which we had access.
Models of sea-level changes offer one possible explanation for the current distribution of clades and unique haplotypes on the islands. At the last glacial maximum, the large island of Viti Levu, most of the Yasawa and Mamanuca group islands as well as Ovalau and Gua comprised a single large land mass; Yadua Taba, today the last stronghold of B. vitiensis, was continuous with the present-day Vanua Levu; and the Lau Group Islands and Kadavu were then, as is now, separated by deep ocean trenches (Gibbons & Clunie 1986). Gibbons (1981, 1985) discussed a possible hybrid zone across both of the larger islands that may have been present at times of low sea level as recently as the last glacial maximum, and he suggested that the southern Mamanuca island of Malolo comprises morphologically intermediate individuals that may be remnants of such a contact zone. Unfortunately, we were not able to obtain any samples from the two large islands or from Malolo to test these hypotheses. However, the present-day distribution of our clades, when considered in the light of our phylogenetic hypothesis, means that at least B. vitiensis and B. bulabula must have been in broad contact over some part of their evolutionary history, as suggested by Gibbons (1981, 1985). When sea levels rose, many new islands were created which isolated populations of iguanas. Our genetic data for B. vitiensis are the most extensive, which show that while virtually every island has at least one unique haplotype, the haplotypes are closely related, consistent with the recent isolation. However, this does not rule out inter-island rafting or human-mediated transportation as an important and possibly necessary mechanism to explain the occurrence of iguanas on many Fijian Islands that were not connected at glacial maxima, or in Tonga. Gibbons (1981) gave a detailed account of a storm (1980) that resulted in massive floating islands of vegetation that would be consistent with such dispersal events (sensu Censky et al. 1998).
(b) Origin of the Pacific iguanas
The distribution of true iguanas (Iguaninae) in the middle of the Pacific has long fascinated biogeographers because their distribution relative to their closest relatives is so difficult to explain. All extant members of the Iguaninae are in North, Central or South America—land masses over 8000 km from the Fijian Islands. Based on this unusual distribution, the Pacific iguanas are often given as an example of one of the longest known dispersal events by a large terrestrial vertebrate (Gibbons 1981). Long-distance rafting by either ancestral iguanas or eggs is the postulated mechanism (Cogger 1974; Gibbons 1981, 1985) and there is substantial distributional evidence that rafting has played an important role in the evolutionary and even recent history of the Iguaninae (Censky et al. 1998). Iguanas are diverse and endemic to many of the islands of the Caribbean in the western Atlantic and famously are found on the Galápagos islands in the eastern Pacific, which themselves are approximately 1000 km from the nearest mainland. Explaining the origin on the three species of Galápagos iguanas has been relatively uncontroversial in comparison because numerous morphological and molecular data demonstrate that the three species are nested within the true Iguaninae and are each other's closest relatives (Sites et al. 1996; Petren & Case 1997; Rassmann 1997; Wiens & Hollingsworth 2000). This implies both a single origin and an origin from the Americas via rafting since the Galápagos are volcanic in origin. While the precise timing of this rafting event is not well understood (Rassmann 1997), there are no other convincing theories to explain their presence in the Galápagos. The story is similar for the diverse Caribbean radiation of Cyclura, which had its origin in the Americas but arrived in and diversified through the Caribbean via rafting (Malone et al. 2000).
The distinction between the Galápagos and Caribbean iguanas on the one hand and the Pacific Fijian iguanas on the other hand is important because there are two viable alternative hypotheses to long-distance rafting which could explain the distribution of iguanas in the central Pacific. Strong cases have been made for both scenarios, but the weight of evidence favouring one over the other has not been evaluated. We review these hypotheses briefly in the light of the current evidence. In evaluating these hypotheses it is important to keep in mind that we are seeking explanations for a radiation of the Pacific iguanas that include the now extinct L. impensa and B. gibbonsi which were once present in both the Fijian Islands and Tonga, but apparently not other nearby island groups (Steadman 2006).
Long-distance rafting. Cogger (1974) and Gibbons (1981) each presented detailed physiological and climatic arguments to explain how the ancestors of the Pacific iguanas may have rafted from the Americas and this idea was supported by Zug (1991) as the most likely scenario. The well-studied South Equatorial surface currents pass by both the Galápagos and a number of other Pacific islands before reaching the islands in the southwest Pacific and turning south past New Zealand and back to South America. If such a current has existed in the Pacific over many millions of years, it is reasonable to suggest that opportunities for long-distance waif dispersal from the Americas would have occasionally been available to adults, juveniles or even eggs of ancestral iguanas. Brachylophus are unusual among iguanas in that they have an extremely long egg incubation (more than nine months) and Gibbons (1981) has suggested that dispersal could have been made by eggs. An extended long incubation in the ancestors of Brachylophus could have increased the chances of dispersal in this manner. A perceived difficulty with the rafting hypothesis is that iguanas are not present throughout the Pacific. Why just the archipelagos of Fiji and Tonga? This may seem a reasonable quibble because the South Equatorial currents pass by many hundreds of other Pacific islands, but neither extant nor extinct iguanas have been found (Steadman 2006). However, it is important to point out that the absence from some areas is a poor argument against dispersal. Gibbons presented compelling corroborating evidence from the distribution of the red mangrove, Rhizophora mangle, which disperses via floating seed pods on ocean currents. This plant is distributed around central West Africa, Central and South America, the Caribbean, Galápagos, Fiji and Tonga, but importantly, nowhere else in the Pacific. The red mangrove also forms a food source for Caribbean Cyclura and habitat for B. fasciatus (Gibbons 1981). Similarly, it has long been thought that the sweet potato (Ipomoea batatas) represented a human-mediated transfer to the Pacific from the Americas, but a recent genetic study has opened up the possibility of a more ancient natural dispersal to the region (Zhang et al. 2004).
Melanesian land bridge. Other authors have rejected the plausibility of such an extraordinary dispersal event and instead point to fossil, phylogenetic, distributional and geological evidence to suggest that the distribution of the Pacific iguanas can be explained by an ancient connection between some of the islands in Melanesia and Fiji and Tonga (Gorham 1965; McCarthy 2003, 2005). While the geological history of the Fijian and Tongan Islands is extremely complex, recent models have demonstrated that both island groups form the eastern extent of an ancient Melanesian arc that has been variously connected or in close proximity over the past 20–40 million years (Hall 2002; Schellart et al. 2006; Steadman 2006). There are numerous examples from the distribution of both plants and animals to support this connection. Three Fijian examples from amphibians and reptiles are: (i) the two species of endemic Platymantis frogs whose closest relatives are in New Guinea and the Solomon Islands, (ii) endemic Pacific boas (Candoia) whose closest relatives are in New Guinea and the Solomon Islands (Austin 2000; Noonan & Chippindale 2006) and (iii) the burrowing elapid snake Ogmodon vitiensis whose closest relatives are in New Guinea (Keogh 1998; Keogh et al. 1998). Of these only the Boidae represented by Candoia has a similar global pattern to the Iguaninae.
Several authors have suggested that recent fossil evidence from Asia support the Asian/Melanesian origin hypotheses for the Pacific iguanas (Worthy 2000; Burns et al. 2006). Indeed, there is now accumulated fossil evidence that ‘iguana’-like animals were both diverse and common in several parts of Asia (Kequin & Lianhai 1995; Evans et al. 2002; Evans 2003; Conrad & Norell 2007; Conrad et al. 2007). An Asian/Melanesian origin for the Pacific iguanas necessarily requires that ancestral stock of true Iguaninae was historically present in this region, but currently there are no extant members of the Iguaninae anywhere in the Old World, Asia, Melanesia or Australia. As far as we are able to interpret, none of the Asian fossils can be linked to the extant members of the Iguaninae, but instead are ‘iguanids’ in a broad sense (Nydam 2002; Conrad & Norell 2007; Conrad et al. 2007). These iguanids include a tremendous diversity of extinct and extant lizards (Schulte et al. 2003) and may represent basal clades within the Iguania. Indeed one of the oldest fossils that can clearly be allocated to the ‘Iguaninae’ is an Early Miocene fossil from the New World (Mexico) which, based on morphological data, appears to be most closely related to extant Dipsosaurus and together they form the sister group to other Iguaninae, including Brachylophus (Norell & De Queiroz 1991). While an Asian/Melanesian origin for the Pacific iguanas is tantalizing as an alternative to long-distance over-water dispersal, the evidence available thus far does not support it, as it would require not only conclusive proof of true Iguaninae ancestors in the region, but also complete extinction of all members of this group in the Old World.
(c) Conservation implications
The Pacific iguanas have suffered dramatically since human arrival, with both the gigantic L. impensa and B. gibbonsi eaten to extinction after human arrival in the Pacific some 2800 years ago (Steadman et al. 2002). The extant Brachylophus species are now under imminent threat due to habitat loss and modification, as well as the strong impact of feral cats, mongooses and goats, with B. vitiensis now extirpated on many islands that it formerly occupied. Brachylophus vitiensis is listed as Critically Endangered on the IUCN Red List with only a single secure population remaining on the island of Yadua Taba (Gibbons 1985; Harlow et al. 2007). The status of B. fasciatus in the wild is little known, but large populations still exist on the two uninhabited Aiwa islands in the Lau group (Harlow 2003).
Our study has two important implications for the conservation of extant Brachylophus. First, we have demonstrated that living Brachylophus comprise three distinct species rather than two, and this has immediate implications for conservation and land management so that each species is represented in secure long-term reserves. Second, one of the most important results from our study is that, with just one exception, every island for which we have samples has at least one unique haplotype and represents a distinct lineage in our phylogeny (figure 2). The only haplotype that is shared between two islands is found on Devilau and Monuriki. Protecting the genetic diversity in these iconic animals is paramount and our results stress the need for urgent conservation for all three Brachylophus species.
Acknowledgments
These samples were collected and exported under CITES permit EP 8/33/2 and through pre-CITES permits from the Republics of Vanuatu and Fiji.
This study would not have been possible without the cooperation and logistical help of the National Trust of Fiji Islands and Kula Eco Park over many years. In particular, we are indebted to Elizabeth Erasito and Pita Biciloa and family at the National Trust and Philip Felstead and Ramesh Chand at Kula Eco Park. Dr Niumaia Tabunakawai (principal veterinary officer) permitted the import of Vanuatu Brachylophus into Fiji and the export of these and the holotype from Fiji to the United States (California Academy of Sciences; CAS) for study. For help, advice and discussions, we thank Harold Cogger, Greg Pregill, Jay Savage and George Zug. We thank Suzanne Morrison for her unpublished data on snout–vent lengths and femoral pores, Craig Morley for facilitating tissue samples from Macuata, and Harold Cogger, Greg Pregill, Suzanne Morrison and Paddy Ryan for photographs. We thank the Margi Dykens and the Research Library of the San Diego Natural History Museum for the use of historic illustrations used in the electronic supplementary material, figure 2. For financial support, we thank the BP Conservation Programme, the International Iguana Foundation, NSF, CAS, and the Australian Research Council. For access to museum specimens, we thank the Australian Museum, the Museum of Comparative Zoology, Harvard, CAS, the American Museum of Natural History, the United States National Museum of Natural History, the Bernice Pauahi Bishop Museum, the Peabody Museum (BYU), the Field Museum of Natural History, the Academy of Natural Science, Philadelphia, and the British Museum of Natural History. The Zoological Society of San Diego (Donal Boyer, John Kinkaid) allowed access to live Brachylophus for measurements and colour assessments. We thank Paul Doughty, Conrad Hoskin, George Zug, Steve Trewick and an anonymous referee for their helpful comments on the manuscript. The use of trade, product or firm names in this publication is for descriptive purposes only and does not imply endorsement by the US Government.
Footnotes
One contribution of 15 to a Theme Issue ‘Evolution on Pacific islands: Darwin's legacy’.
Supplementary Material
Summary of locality information
Summary of uncorrected genetic distances
List of specimens examined
Individual gene trees
Historical iguana illustrations
References
- Alberts A.C. Phylogenetic and adaptive variation in lizard femoral gland secretions. Copiea. 1991;1991:69–79. doi:10.2307/1446249 [Google Scholar]
- Arévalo E, Davis S.K, Sites J. Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in Central Mexico. Syst. Biol. 1994;43:387–418. doi:10.2307/2413675 [Google Scholar]
- Austin C.C. Molecular phylogeny and historical biogeography of Pacific Island boas (Candoia) Copiea. 2000;2000:341–352. doi:10.1643/0045-8511(2000)000[0341:MPAHBO]2.0.CO;2 [Google Scholar]
- Avery D.F, Tanner W.W. Speciation in the Fijian and Tongan iguana (Sauria: Iguanidae) with the description of a new species. Great Basin Nat. 1970;30:166–172. [Google Scholar]
- Brongniart A. Imprimeur de I'Inst. Natural; Paris, Baudouin, France: 1805. Essai d'une classification naturelle des reptiles. p. 53. [Google Scholar]
- Burns E.L, Costello B.H, Houlden B.A. Three evolutionary significant units for conservation in the iguanid genus Brachylophus. Pacific Cons. Biol. 2006;12:65–77. [Google Scholar]
- Censky E.J, Hodge K, Dudley J. Over-water dispersal of lizards due to hurricanes. Nature. 1998;395:556. doi:10.1038/26886 [Google Scholar]
- Chase C.G. Tectonic history of the Fiji Plateau. Geol. Soc. Am. Bull. 1971;82:3087–3110. doi:10.1130/0016-7606(1971)82[3087:THOTFP]2.0.CO;2 [Google Scholar]
- Cogger H.G. Voyage of the banded iguana. Aust. Nat. Hist. 1974;18:144–149. [Google Scholar]
- Colgan D.J, Da Costa P. Genetic discrimination between the Iguanas Brachylophus vitiensis and Brachylophus fasciatus. J. Herpetol. 1997;31:589–591. doi:10.2307/1565617 [Google Scholar]
- Conrad J.L, Norell M.A. A complete Late Cretaceous iguanian (Squamata, Reptilia) from the Gobi and identification of a new iguanian clade. Am. Mus. Nov. 2007;3584:1–47. doi:10.1206/0003-0082(2007)3584[1:ACLCIS]2.0.CO;2 [Google Scholar]
- Conrad J.L, Rieppel O, Grande L. A Green River (Eocene) Polychrotid (Squamata: Reptilia) and a re-examination of iguanian systematics. J. Paleontol. 2007;81:1365–1373. doi:10.1666/06-005R.1 [Google Scholar]
- Daugherty C.H, Cree A, Hay J.M, Thompson M.B. Neglected taxonomy and continuing extinctions of tuatara (Sphenodon) Nature. 1990;347:177–179. doi:10.1038/347177a0 [Google Scholar]
- Etheridge R, De Queiroz K. A phylogeny of Iguanidae. In: Estes R, Pregill G, editors. Phylogenetic relationships of the lizard families, essays commemorating Charles L. Camp. Stanford University Press; Palo Alto, CA: 1988. pp. 283–368. [Google Scholar]
- Evans S.E. At the feet of the dinosaurs: the early history and radiation of lizards. Biol. Rev. 2003;78:513–551. doi: 10.1017/s1464793103006134. doi:10.1017/S1464793103006134 [DOI] [PubMed] [Google Scholar]
- Evans S.E, Prasad G.V.R, Manhas B.K. Fossil lizards from the Jurassic Kota Formation of India. J. Vert. Paleontol. 2002;22:299–312. doi:10.1671/0272-4634(2002)022[0299:FLFTJK]2.0.CO;2 [Google Scholar]
- Frost D.R, Etheridge R. A phylogenetic analysis and taxonomy of iguanian lizards (Reptilia: Squamata) Univ. Kansas Mus. Nat. Hist. Misc. Publ. 1989;81:1–65. [Google Scholar]
- Gibbons J.R.H. The biogeography of Brachylophus including the description of a new species, B. vitiensis, from Fiji. J. Hepetol. 1981;15:255–273. [Google Scholar]
- Gibbons J.R.H. The biogeography and evolution of Pacific Island reptiles and amphibians. In: Grigg G, Shine R, Ehmann H, editors. The biology of Australasian frogs and reptiles. Surrey Beatty and Sons; Sydney, Australia: 1985. pp. 125–142. [Google Scholar]
- Gibbons J.R.H, Clunie F. Sea level changes and Pacific prehistory: new insights into early human settlement of Oceania. J. Pacific. Hist. 1986;21:58–82. doi:10.1080/00223348608572529 [Google Scholar]
- Girard C. J. B. Lippincott & Co; Philadelphia, PA: 1858. United States exploring expedition during the years 1838, 1839, 1840, 1841, 1842. Under the command of Charles Wilkes, U.S.N. Herpetology. p. 496. [PMC free article] [PubMed] [Google Scholar]
- Gorham S.W. Fiji frogs with synopses of the genera Cornufer and Platymantis. Zool. Beitr. 1965;11:381–435. [Google Scholar]
- Günther A. Descriptions of new species of reptiles and fishes in the collection of the British Museum. Proc. Zool. Soc. Lond. 1862;1862(Pt. 2):187–194. [Google Scholar]
- Hall R. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer-based reconstructions, model and animations. J. Asian Earth Sci. 2002;2:353–431. doi:10.1016/S1367-9120(01)00069-4 [Google Scholar]
- Harlow P.S. Searching for banded iguanas in the Lau Islands, Eastern Fiji. Iguana. 2003;10:103–107. [Google Scholar]
- Harlow P.S, Biciloa P.N. Abundance of the Fijian crested iguana (Brachylophus vitiensis) on two islands. Biol. Cons. 2001;98:223–231. doi:10.1016/S0006-3207(00)00157-9 [Google Scholar]
- Harlow P.S, et al. The decline of the endemic Fijian crested iguana Brachylophus vitiensis in the Yasawa and Mamanuca archipelagos, western Fiji. Oryx. 2007;41:44–50. doi:10.1017/S0030605307001639 [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Keogh J.S. Molecular phylogeny of elapid snakes and a consideration of their biogeographic history. Biol. J. Linn. Soc. 1998;63:177–203. [Google Scholar]
- Keogh J.S, Shine R, Donnellan S.C. Phylogenetic relationships of terrestrial Australo-Papuan elapid snakes based on cytochrome b and 16S rRNA sequences. Mol. Phyl. Evol. 1998;10:67–81. doi: 10.1006/mpev.1997.0471. doi:10.1006/mpev.1997.0471 [DOI] [PubMed] [Google Scholar]
- Kequin G, Lianhai H. Iguanians from the Upper Cretaceous Djadochta Formation, Gobi Desert, China. J. Vert. Paleontol. 1995;15:57–78. [Google Scholar]
- Leviton A.E, Gibbs R.H, Jr, Heal E, Dawson C.E. Standards in herpetology and ichthyology: part I. Standard symbolic codes for institutional resource collections in herpetology and ichthyology. Copeia. 1985;1985:802–832. [Google Scholar]
- Malone C.L, Wheeler T, Taylor J.F, Davis S.K. Phylogeography of the Caribbean rock iguana (Cyclura): implications for conservation and insights on the biogeographic history of the West Indies. Mol. Phyl. Evol. 2000;17:269–279. doi: 10.1006/mpev.2000.0836. doi:10.1006/mpev.2000.0836 [DOI] [PubMed] [Google Scholar]
- McCarthy D. The trans-Pacific zipper effect: disjunct sister taxa and matching geological outlines that link the Pacific margins. J. Biogeogr. 2003;30:1545–1561. doi:10.1046/j.1365-2699.2003.00929.x [Google Scholar]
- McCarthy D. Biogeographical and geological evidence for a smaller, completely enclosed Pacific basin in the Late Cretaceous. J. Biogeogr. 2005;32:2161–2177. doi:10.1111/j.1365-2699.2005.01355.x [Google Scholar]
- Noonan B.P, Chippindale P.T. Dispersal and vicariance: the complex evolutionary history of boid snakes. Mol. Phyl. Evol. 2006;40:347–359. doi: 10.1016/j.ympev.2006.03.010. doi:10.1016/j.ympev.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Norell M.A, De Queiroz K. The earliest iguanine lizard (Reptilia: Squamata) and its bearing on iguanine phylogeny. Am. Mus. Nov. 1991;2997:1–16. [Google Scholar]
- Nydam R.L. Lizards of the Mussentuchit local fauna (Albian-Cenomanian boundry) and comments on the evolution of the Cretaceous lizard fauna of North America. J. Vert. Paleontol. 2002;22:645–660. doi:10.1671/0272-4634(2002)022[0645:LOTMLF]2.0.CO;2 [Google Scholar]
- Olson, D. et al In press. Priority forests for conservation in Fiji. Oryx
- Petren K, Case T.J. A phylogenetic analysis of body size evolution and biogeography in chuckwallas (Sauromalus) and other iguanines. Evolution. 1997;51:206–219. doi: 10.1111/j.1558-5646.1997.tb02402.x. doi:10.2307/2410974 [DOI] [PubMed] [Google Scholar]
- Pregill G.K, Dye T. Prehistoric extinction of giant iguanas in Tonga. Copeia. 1989;1989:505–508. doi:10.2307/1445455 [Google Scholar]
- Pregill G.K, Steadman D.W. South Pacific iguanas: human impacts and a new species. J. Herpetol. 2004;38:15–21. doi:10.1670/73-03A [Google Scholar]
- Pregill G.K, Worthy T.H. A new iguanid lizard (Squamata, Iguanidae) from the later Quaternary of Fiji, Southwest Pacific. Herpetologica. 2003;59:57–67. doi:10.1655/0018-0831(2003)059[0057:ANILSI]2.0.CO;2 [Google Scholar]
- Rassmann K. Evolutionary age of the Galápagos iguanas predates the age of the present Galápagos Islands. Mol. Phyl. Evol. 1997;7:158–172. doi: 10.1006/mpev.1996.0386. doi:10.1006/mpev.1996.0386 [DOI] [PubMed] [Google Scholar]
- Rodda P. Geology of Fiji. In: Stevenson A.J, Herzer R.H, Balance P.F, editors. Geology and submarine resources of the Tonga-Lau-Fiji region. South Pacific Applied Geosciences Commission (SOPAC) Technical Bulletin; Suva, Fiji: 1994. pp. 131–151. [Google Scholar]
- Schellart W.P, Lister G.S, Toy V.G. A Late Cretaceous ande Cenozoic reconstruction of the Southwest Pacific region: Tectonics controlled by subduction and slab rollback processes. Earth Sci. Rev. 2006;76:191–233. doi:10.1016/j.earscirev.2006.01.002 [Google Scholar]
- Schulte J.A, Valladares J.P, Larson A. Phylogenetic relationships within Iguanidae inferred using molecular and morphological data and a phylogenetic taxonomy of iguanian lizards. Herpetologica. 2003;59:399–419. doi:10.1655/02-48 [Google Scholar]
- Sites J.W, Davis S.K, Buerra T, Iverson J.B, Snell H.L. Character congruence and phylogenetic signal in molecular and morphological data sets: a case study in the living iguanas (Squamata, Iguanidae) Mol. Biol. Evol. 1996;13:1087–1105. doi: 10.1093/oxfordjournals.molbev.a025671. [DOI] [PubMed] [Google Scholar]
- Steadman D.W. University of Chicago Press; Chicago, IL: 2006. Extinction and biogeography of tropical Pacific birds. p. 594. [Google Scholar]
- Steadman D.W, Pregill G.K, Burley D.V. Rapid prehistoric extinction of iguanas and birds in Polynesia. Proc. Natl Acad. Sci. USA. 2002;99:3673–3677. doi: 10.1073/pnas.072079299. doi:10.1073/pnas.072079299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. Champaign, IL: Illinois Natural History Survey.
- Watling D. Rediscovery of a petrel and new faunal records on Gau Island. Oryx. 1986;20:31–34. [Google Scholar]
- Wiens J.J, Hollingsworth B.D. War of the iguanas: conflicting molecular and morphological phylogenies and long-branch attraction in iguanid lizards. Syst. Biol. 2000;49:143–159. doi: 10.1080/10635150050207447. doi:10.1080/10635150050207447 [DOI] [PubMed] [Google Scholar]
- Worthy T.H. The fossil megapode (Aves: Megapodiidae) of Fiji with description of a new genus and two new species. J. R. Soc. New Zealand. 2000;30:337–364. [Google Scholar]
- Zhang D, Rossel G, Kriegner A, Hijmans R. AFLP assessment of diversity in sweetpotato from Latin America and the Pacific region: its implications on the dispersal of the crop. Gen. Res. Crop Evol. 2004;51:115–120. doi:10.1023/B:GRES.0000020853.04508.a0 [Google Scholar]
- Zug G. Bishop Museum Press; Honolulu, HI: 1991. The lizards of Fiji: natural history and systematics. p. 136. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of locality information
Summary of uncorrected genetic distances
List of specimens examined
Individual gene trees
Historical iguana illustrations