Abstract
The human serum albumin (HSA) secondary structure modifications induced by the chrysotile surface have been investigated via computational molecular dynamics (MD) and experimental infrared spectroscopy (FTIR) on synthetic chrysotile nanocrystals coated with different amount of HSA. MD simulations, conducted by placing various albumin subdomains close to the fixed chrysotile surface, show an initial adsorption phase, accompanied by local rearrangements of the albumin motifs in contact with the chrysotile layer. Next, large-scale rearrangements follow with consequent secondary structure modifications.
Gaussian curve fitting of the FTIR spectra obtained for HSA-coated synthetic chrysotile nanocrystals has allowed the quantification of HSA structural modifications as a function of the amount of protein adsorbed. The experimental results support the atomistic computer simulations providing a realistic description of the adsorption of plasma proteins onto chrysotile and unravelling a key step in the understanding of asbestos toxicity.
Keywords: solid–liquid interface, molecular dynamics, Fourier transform infrared, human serum albumin
1. Introduction
A real understanding of life's processes requires an understanding of how proteins and other endogenous macromolecules perform their actions. Proteins can be viewed as copolymers consisting of different amino acids, connected through the so-called peptide bond, and can be studied at various levels of detail, from the quantum mechanical to the statistical mechanical, the choice depending on the type of investigation. The simplest amino acid characterization is in terms of their hydrophobic or hydrophilic nature, for example through the use of a number of different coding scales, such as the Kyte & Doolittle (1982), the Hopp & Woods (1983) and the Rose (Rose et al. 1985) hydropathy scales. However, it is clear that such description provides an oversimplified view of some important details of protein behaviour. The protein conformation determined by X-ray diffraction (solid state) and NMR (in solution) gives us a detailed description (or a ‘snapshot’) of the three-dimensional structure of a macromolecule, but other features are also important. Biological molecules are not rigid, and dynamic changes within a molecular structure may play a significant role in the physiological function of the molecule. For proteins to exploit their function, they must fold into the correct three-dimensional structure (incorrect folding is, for example, implicated in diseases such as Alzheimer's and BSE; Stanley 1997; Kostova & Wolf 2003). Large proteins called chaperonins can direct correct folding (Horwich et al. 2004; Horovitz & Willison 2005). The interaction of biological macromolecules, in particular proteins, with a foreign surface is a closely related field that play an important role in a wide range of biological processes (such as cell aggregation and interaction of ligands with membrane receptors) and is not easily amenable to theory owing to the structural details at the atomistic level and the denaturation process these molecules can undergo upon adsorption. In recent years, computer simulations have added significant new insights, complementing both theory and experiments (Noinville et al. 1995; Euston 2004; Ganazzoli & Raffaini 2005). The atomistic computer simulations are the only techniques that can deal with this task, providing a realistic description of the adsorption process, although the size and the complexity of the problem are often discouraging.
In this respect, the study of the adsorption mechanism of plasma proteins on the surface of a human health hazardous material is a key step in the possible understanding of its toxicity and mutagenic properties. Asbestos fibres are by far the most well-known environmentally dangerous agents: human health hazards associated with airborne asbestos fibres exposure are well documented in the medical literature (Schreier 1989; Mossman 1993).
Environmental and occupational exposure to asbestos fibres is frequently associated with pleural and parenchymal lung diseases, such as lung cancer and malignant mesothelioma (Churg & Warnock 1981; Churg & Green 1998).
Ninety-five per cent of manufactured asbestos is composed of chrysotile, Mg3Si2O5(OH)4 (Chissick 1987). This latter consists of sheets of tetrahedral (T) silica in a pseudo-hexagonal network joined to a brucite layer, in which Mg is in octahedral (O) coordination with the apical oxygen of the SiO4 layer and additional hydroxyl groups. The mismatch of the smaller lateral dimension of the SiO4 sheet with respect to the Mg(OH)2 layer is accommodated by the concentrically or spirally curled cylindrical chrysotile structure (Whittaker 1956, 1957; Yada 1967, 1971). The presence of impurities, ion substitutions and structural disorder in mineral chrysotile affects not only its morphology and chemical–physical properties, but also its interaction with biological systems (Whittaker & Wicks 1970; Stroink et al. 1985). Chrysotile fibres from different mines exhibit chemical–physical properties which change appreciably from sample to sample (Yada 1971).
Stoichiometric chrysotile nanotubes have been synthesized as a unique phase to be used as a reference sample with definite structure, morphology and chemical composition for investigation of the chemical and physical properties of chrysotile fibres, along with their interactions with biological systems (Falini et al. 2002, 2004; Bergamini et al. 2004; Gazzano et al. 2005, 2007; Roveri et al. 2006).
In the last decades, the adsorption of albumin onto mineral asbestos fibres has been investigated by employing X-ray photoelectron spectroscopy (XPS; Jaurand et al. 1983), FTIR and NMR spectroscopies (Dumitru-Stanescu et al. 1994), revealing the tendency of mineral chrysotile fibres to yield adducts with albumin.
Recently, the first morphological evidence of bovine serum albumin (BSA) adsorption onto synthetic stoichiometric chrysotile nanocrystals has been obtained, and BSA secondary structure modifications induced by the surface interaction have been quantified by FTIR spectroscopy on the BSA-coated chrysotile nanocrystals (Falini et al. 2006) and circular dichroism (CD) investigations on the protein solution exchanged from the chrysotile surface (Sabatino et al. in press).
In the present paper, the interaction of selected human serum albumin (HSA) subdomains with the chrysotile surface was investigated through molecular mechanics (MM) and molecular dynamics (MD) simulations. The chrysotile surface may form an idealized, zeroth-order approximation of asbestos surface and was used owing to its relative simplicity and rigidity, which allow to treat it as a fully rigid body for all purposes. It should be remembered, however, that serpentine asbestos is a more complex material, being formed by microcrystals with a size of the order of a few nanometres and randomly oriented. Different crystallographic planes may be exposed, producing additional edges and grooves able to modify protein adsorption. Such features do also affect to various degrees the interaction of blood proteins with asbestos, interactions largely due to the fast adsorption of albumin, the most abundant blood protein. The amount of this adsorption reflects the protein affinity towards the substrate and reaches a maximum value that corresponds probably to a monolayer. The non-specific adsorption of protein alone is unlikely to produce the thick layer found in the asbestos body, but all fibres reaching the alveoli are, within a short time, likely to be covered with a thin layer of protein that has been adsorbed from the blood (Churg & Green 1998).
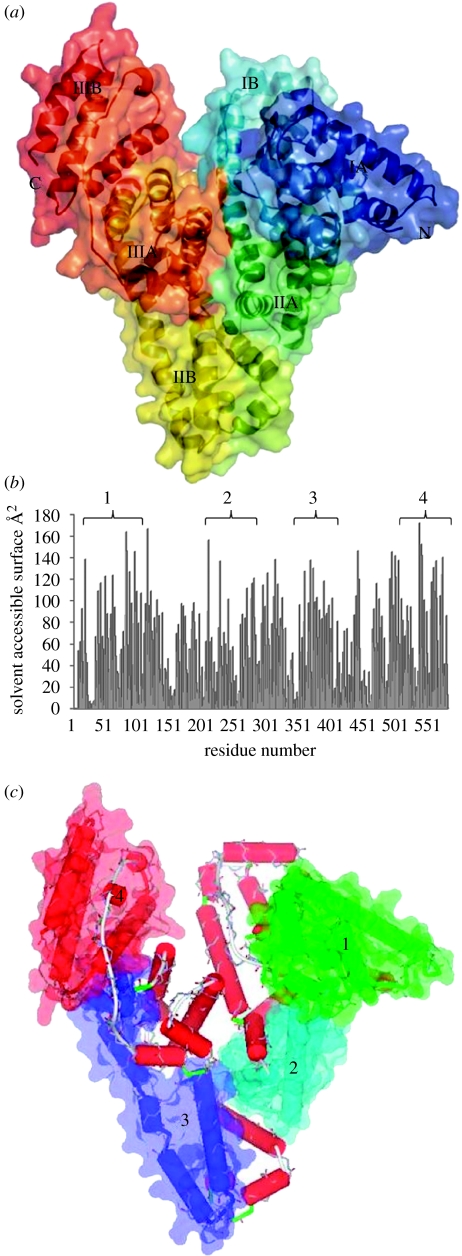
The first three-dimensional crystal structure determination of HSA at a resolution of 2.8 Å was obtained by He & Carter (1992) and shows that HSA has a heart-like asymmetric shape, 80 Å on the side, an average thickness of 30 Å and a volume of 88 249 Å3. These values are very close to those of human and bovine α-fetoprotein (Luft & Lorscheiser 1983). Its primary sequence was known before that derived from complementary DNA (cDNA) data (Minghetti et al. 1986) and shows that HSA is a single, 66 kDa monomeric polypeptide of 585 amino acid residues, stabilized by 17 disulphide bridges (Dugaiczyk et al. 1982; Sugio et al. 1999), whose locations were firstly derived from peptide studies by Brown (1974) and Saber et al. (1977). HSA shows a peculiar amino acid distribution, compared with that of a typical protein, with a low content of Met, Gly and Ile, while the ionic residues (Glu and Lys) are very abundant. This is reflected, at least partially, in the facility of interaction of albumin with anionic and cationic ligands (like drugs). The presence of only one Trp residue in the protein has been particularly advantageous in binding experiments using fluorescent probes (Peters 1996). HSA is characterized by a repeating pattern of three α-helical homologous domains derived from genic multiplication (Brown 1976) and numbered I, II and III, starting from the amino terminus, an architecture similar to other plasma proteins like transthyretin and transferrin. Each domain is divided into two subdomains, A and B (figure 1a), composed of six and four α-helices (Luft & Lorscheiser 1983; He & Carter 1992), respectively. Subdomain A is composed of a cluster of four α-helices (A-h1 to A-h4), flanked by two short α-helices (A-h5 and A-h6), while subdomain B is composed of one cluster of four α-helices (B-h7 to B-h10). These α-helices, hence also the subdomains A and B, are joined by an extended loop and show a comparable three-dimensional structure. They are related by a pseudo-binary axis between the two α-helices A-h2 and B-h8 (He & Carter 1992). In the HSA tertiary structure, some regions overlap as a consequence of the helical continuation, from the C-terminal of subdomains IB and IIB to the N-terminal of subdomains IIA and IIIA (Dockal et al. 1999).
Figure 1.
(a) The three-domain structure of HSA with the protein secondary structure succession shown in different colours (N- and C-termini are marked as N and C, respectively). Each domain is divided into two subdomains (A and B) composed, respectively, of a cluster of four α-helices flanked by two short α-helices (subdomain A) and one cluster of four α-helices (subdomain B). The α-helices and the subdomains A and B are joined by an extended loop and show a comparable three-dimensional structure. (b) Solvent accessible surface (SAS) versus residue number plot, obtained using a 1.4 Å probe resembling a water molecule, with the indication of the four simulated zones (1–4). (c) Position of the four studied zones (1–4) within the whole albumin molecule. The first zone (zone 1) is formed by five connected α-helices (ranging from h2 to h6) and short random sections derived from a single HSA domain (IA). The same behaviour is shown also by the second zone used in the simulation (zone 2), formed by four α-helices (h2–h5) belonging to domain IIA, and by zone 4 with four α-helices from the IIIB domain. Zone 3 is different because it consists of two α-helices from domain IIB (h3 and h4) and two α-helices from domain IIIA (h1 and h2). In all cases, the disulphide bridges are retained throughout the simulations.
However, in computer simulation methods, the very large size of albumin prevents accounting for the whole molecule, hence our choice to investigate the interaction and adsorption of four specific HSA subdomains onto the flat chrysotile surface. The simulations were carried out by using a two-step strategy, with direct energy minimizations of selected HSA subdomains close to the chrysotile surface, using an effective dielectric medium with a distance-dependent dielectric constant to account for the dipolar interactions (Raffaini & Ganazzoli 2003, 2004). Extensive MD runs of the previously minimized subdomains follow with subsequent energy minimization of the instantaneous snapshots. The first process can be viewed as a simulation of the initial adsorption step of HSA over the chrysotile plane, providing also the preferred conformations during the adsorption process at large surface coverage. Conversely, the second procedure yields the best overall conformation on a clean surface with the largest interaction energy, leading to the formation of a stable thin layer of amino acids, basically a monolayer.
The results obtained by this computational approach have been compared to the data collected through an experimental study by FTIR spectroscopy carried out on synthetic stoichiometric chrysotile nanocrystals coated with different amount of HSA, adsorbed in physiological conditions. This experimental approach allows us to quantitatively evaluate the HSA secondary structure modifications induced by the electrostatic interactions with the surface of the synthetic chrysotile nanocrystals.
In this work, we have used synthetic chrysotile nanocrystals that can be considered an ideal reference sample to overcome compositional and structural heterogeneity of mineral chrysotile. To the best of our knowledge, this is the first investigation on the in vitro interaction of chrysotile fibres with HSA after the pioneering work of Morgan (1974).
2. Material and methods
2.1 Simulation methods
The chrysotile planes were prepared starting from the experimental crystallographic structure (Whittaker 1956), adding the relevant hydrogen atoms at their calculated positions. The domains are made of two chrysotile parallel planes measuring approximately 100×100 Å. The optimal geometrical conformation in vacuo was found using a MM approach running the conjugate gradient algorithm up to a gradient better than 10−3 kcal mol−1 Å.
Starting coordinates of the non-hydrogen atoms of HSA used for the initial trial geometries were taken from the deposited crystallographic structure at 2.5 Å resolution (HSA, PDB ID code: 1AO6; He & Carter 1992; Curry et al. 1998) and consists of two independent and basically equivalent chains, denoted as A and B. Chain B, identical to chain A, was deleted and the structure was checked through Moleman2 (Kleywegt 2000; Kleywegt et al. 2001) and DeepView (Guex & Peitsch 1997) to guarantee the system conformity with the MD program (in particular, the names of the side chains that must be congruent with the used force field). Four specific HSA subdomains have been chosen, mainly on the basis of their ability to interact with a flat surface and, consequently, by their calculated solvent accessibility, to guarantee the selection of the highest contact area with the chrysotile surface. The solvent accessible surface plot is shown in figure 1b, while the location of the subdomains within the whole albumin molecule is shown in figure 1c. The first zone, denoted as zone 1 and ranging from Glu16 to His105, is formed by five connected α-helices (ranging from h2 to h6) and short random sections derived from a single HSA domain (IA). The same behaviour is shown also by the second zone used in the simulation (zone 2), formed by four α-helices (h2–h5) belonging to domain IIA, and by zone 4 with four α-helices from the IIIB domain. Zone 3, ranging from Ala510 to Ala582, is different because it consists of two α-helices from domain IIB (h3 and h4) and two α-helices from domain IIIA (h1 and h2). In all cases, the disulphide bridges are retained throughout the simulations.
The four selected subdomains (see later) were terminated with –COO− and –NH3+ groups in their zwitterionic form and the polar hydrogen atoms were added in their calculated positions. The protonation state for all the ionizable residues was set to the normal ionization state at pH 7.0, and both topology and connectivity of the molecule have been created. The isolated subdomains were then fully optimized up to an energy gradient lower than 10−3 kcal mol−1 Å.
The protein fragments were then placed close to the chrysotile surface and minimized with the assumption of an effective dielectric medium with a distance-dependent dielectric constant asymptotically yielding the correct water value (ϵ=78). The conformations obtained in the effective dielectric medium do essentially coincide with those found in the presence of water molecules, both being very similar to the experimental geometries. The hydration effects on the adsorption of the protein fragments on the chrysotile surface can affect the kinetics of adhesion to the surface and the time scale of the subsequent HSA rearrangement (spreading), a process that is mainly dictated by the interaction within the HSA fragment and the modified surface (Raffaini & Ganazzoli 2007). On the other hand, the final geometries obtained in the dielectric medium correspond in general to the thermodynamically preferred conformation, apart from some possible readjustments of the side groups in explicit water. Although some fragments may retain a globular shape, possibly with some residual secondary structure and one side of the fragments is not solvated, being in contact with the modified surface, the exposed sides can be efficiently hydrated forming a large number of hydrogen bonds with water. The hydration of the amino acids comprised within the α-helices and the β-sheets of the native secondary structure is somewhat enhanced, a feature that may largely compensate the net loss of hydration of the outer residues in contact with the surface. However, the energy minimizations carried out in the presence of a large number of water molecules cannot be used to calculate the relative stability of different geometries because, in the presence of a large number of solvent molecules, most of the atoms in the simulation belong to water which therefore dominates the total energy. Moreover, from a thermodynamic viewpoint, the energy minimizations are equivalent to freezing the system at 0 K, leading to the optimization of water molecules in some local glass-like configurations of the phase space. This procedure is however plagued by the problem of the huge number of local minima, typical of the glassy state. Owing to these results, all subsequent MD simulations were performed in a distance-dependent dielectric medium with periodic boundary conditions, with a pressure of 1 atm and a constant temperature of 300 K, controlled through a Berendsen thermostat. Integration of the dynamical equations was carried out through the Verlet algorithm with a constant time step of 1.0 fs. In all cases, the long MD runs in the dielectric medium lasted for 1.5 ns. The instantaneous coordinates (or frames) were periodically saved for further analysis or geometry optimization. Within these runs, the total and potential energy showed an initial decrease and then fluctuated around a constant value, indicating that an equilibrium state was finally reached. Each frame collected during the MD run was then minimized up to a gradient of less than 1×10−3 kcal mol−1.
All simulations steps, including data analysis, were performed with the NAMD1 package (Phillips et al. 2005), using a modified version of the AMBER force field (Cornell et al. 1995) on a dual-PIII workstation, working under LINUX RedHat v. 7.3 (SGI-SMP kernel v. 2.4.18-4SGI_XFS_1.1). All the information related to the trajectories (positions, energies and velocities) have been stored in a sequential series of files of appropriate dimensions (less than 700 Mb) for the back-up.
2.2 Chemical reagents
Reagents were from Sigma–Aldrich; 0.06 M phosphate buffer (pH 7.4) was from Riedel-de Haen Sigma–Aldrich.
2.3 Synthetic stoichiometric chrysotile nanocrystals
Stoichiometric chrysotile fibres were synthesized as a unique phase by means of hydrothermal reactions under controlled conditions (Falini et al. 2002, 2004). MCM41 (average pore size of 3.9 nm and specific area surface of 910 m2 g−1) was used as a silica source instead of reported silica gel, in order to satisfy the necessary required purity of the reactants in terms of metal ions. The reaction was carried out mixing MCM41 in 0.1 M MgCl2 aqueous solution; the Si/Mg molar ratio was 0.68. The pH was raised to 13.0 by adding 1 M NaOH solution, and then the hydrothermal treatment at 82 atm and 300°C for 24 h was performed. The above reported reaction conditions resulted in chrysotile nanocrystals as a unique stoichiometric phase with constant chemical composition, structure, crystallinity, size, morphology and surface area (Falini et al. 2004; Roveri et al. 2006).
2.4 Chrysotile fibres
A suspension of the synthetic chrysotile samples (50 mg per 30 ml) was ultrasonicated in order to disaggregate the nanocrystals (model ultrasonic UTA, Falc) for 2 min.
2.5 HSA-coated chrysotile nanocrystal preparation
Protein solutions were prepared by dissolution of HSA in 0.01 M phosphate buffer (pH 7.4) and used immediately. The adsorption experiments were performed in polypropylene centrifuge tubes containing the proper amount of synthetic chrysotile to achieve an adsorption area of 2.2 m2 and 10 ml of liquid. Different protein concentrations were prepared by adding phosphate buffer to the protein solution ranging from 0.3 to 2.5 mg ml−1. During incubation, the tubes were rotated for 12 h end-over-end at 37°C. The samples were then centrifuged and the protein concentration in the supernatant was measured by UV spectroscopy. The amount of adsorbed protein was calculated from the difference in the concentration between the initial and equilibrated solution. Adducts were then lyophilized from the buffer solution at −40°C until they reached a constant weight.
An HSA reference sample was prepared following the same procedure as the previous ones, in the absence of chrysotile nanocrystals.
2.6 Fourier transform infrared analysis
FTIR measurements were carried out on the samples lyophilized from the buffer solution. The infrared spectra were measured from 4000 to 400 cm−1 with 2 cm−1 resolution using a Bruker IFS 66v/S spectrometer. The sample compartment atmosphere had a total pressure of 2 mbar of air dried to an atmospheric dew point of −40°C by means of a Balston 76-01 Membrane Air Dryer. Other settings include an 8 mm aperture, 16 scans, velocity 10 kHz, DLATGS detector and a 3-term Blackman–Harris apodization function. KBr pellets were obtained under vacuum, using 2 mg of the powdered samples carefully mixed with 200 mg of infrared grade KBr.
Fourier self-deconvolution and second-derivative resolution enhancement were applied to narrow the widths of infrared bands and increase the separation of the overlapping components. The resolution enhancement resulting from self-deconvolution and the second derivative is such that the number and position of the component bands to be fitted are determined. The curve fitting was carried out employing Bruker opus peak software (v. 4.0). The number of bands was entered into the program along with their respective positions and half-heights. The program iterates the curve-fitting process to achieve the best Gaussian-shaped curves that fit the protein spectrum. A best fit is determined by the root mean square (r.m.s.) of differences between the original protein spectrum and the sum of all individual resolved bands. The assignment of component bands in amide I of HSA has been done according to the literature data. The percentages of each secondary structure were calculated from the integrated areas of the component bands in amide I.
3. Results and discussion
3.1 Initial adsorption by direct energy minimizations in the dielectric medium
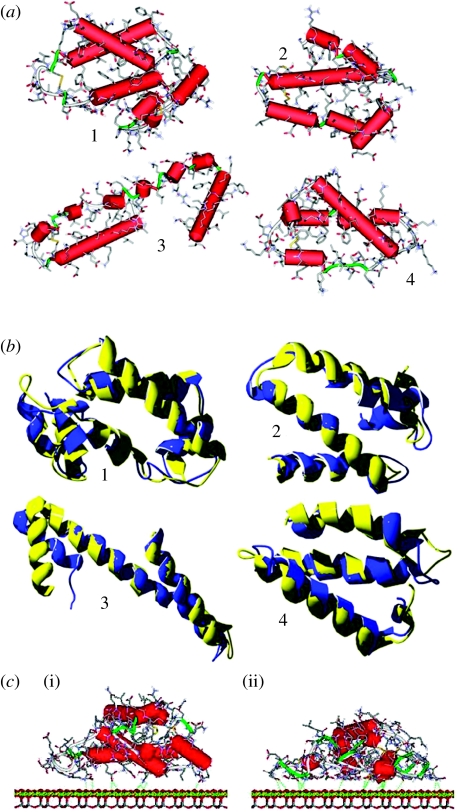
More details on the amino acids composition within the four simulated zones and their corresponding α-helices are reported in table 1, together with the Kyte & Doolittle (1982) and Rose (Rose et al. 1985) indexes. However, we note that some amino acids of contrasting hydropathy index are anyway present in all the α-helices, irrespective of the overall index, a situation that is particularly evident for zones 1 and 3. The structure of the selected zones is shown more clearly in figure 2a, where the secondary structure is highlighted by cylinders, denoting the α-helices, and ribbons, which indicate random strands and regular turns.
Table 1.
Amino acids composition within the four simulated zones and the corresponding α-helices, together with the Kyte–Doolittle and Rose indexes.
| zones | domain | α-helix | AA range | AA numbera | KD indexb | Rose index |
|---|---|---|---|---|---|---|
| 1 | E16–H105 | 90 | 29.1 | 66.14 | ||
| IA | h2 | E16–L31 | 16 | 11.9 | 12.66 | |
| h3 | P35–D56 | 22 | −2.7 | 16.04 | ||
| h4 | S65–T76 | 12 | 3.5 | 9.04 | ||
| h5 | A88–A92 | 5 | 5.1 | 3.92 | ||
| h6 | E95–H105 | 11 | −17.7 | 7.81 | ||
| 2 | E208–E280 | 73 | −29.5 | 52.59 | ||
| IIA | h2 | E208–F223 | 16 | −5.3 | 11.62 | |
| h3 | E227–H247 | 21 | −1.3 | 15.56 | ||
| h4 | L250–E266 | 17 | −0.9 | 12.49 | ||
| h5 | L275–E280 | 6 | −2.1 | 4.43 | ||
| 3 | W341–V415 | 75 | −18.2 | 54.30 | ||
| IIB | h3 | S342–A362 | 21 | 12.5 | 15.71 | |
| h4 | D365–E383 | 19 | −14.4 | 13.56 | ||
| IIIA | h1 | P384–L398 | 15 | −8.8 | 10.74 | |
| h2 | Y401–V415 | 15 | −5.9 | 10.71 | ||
| 4 | A510–A582 | 73 | −31.6 | 50.52 | ||
| IIIB | h1 | A510–T515 | 6 | 6.4 | 4.59 | |
| h2 | E518–K536 | 19 | −23.1 | 12.86 | ||
| h3 | E542–K560 | 19 | 0.9 | 13.88 | ||
| h4 | E565–A582 | 18 | −1.4 | 12.80 |
The number of amino acids in the subdomains exceeds the sum of those in the α-helices owing to the presence of regular turns and random strands.
Kyte–Doolittle index, calculated as the sum of the values for the individual amino acids. A positive hydropathy value indicates hydrophobicity and a negative one hydrophilicity.
Figure 2.
(a) Starting three-dimensional structures of the individual four subdomains. Hydrogen atoms have been omitted for clarity. Cylinders indicate α-helices (red), while ribbons denote random strands and regular turns (green). (b) Comparison between the starting secondary structures and the minimized ones for the four simulated subdomains, showing the little differences between the optimized (blue ribbon) and the experimental (yellow ribbon) geometries of the backbone. (c) Geometries of the (i) starting and (ii) initial adsorption states of the zone 1 on the chrysotile surface in the dielectric medium: the amino acids close to the surface locally optimize their interactions with the chrysotile planes by partially loosing their secondary α-helical structure. The retained helicoidal features suffer large distortions from their typical conformation, so that H-bonds between adjacent turns are absent. Consequently, these helicoidal strands cannot be classified as genuine α-helices. The detailed chemical structure is shown, omitting the hydrogen atoms for clarity.
The geometry of the isolated subdomains was optimized for the later determination of the interaction energy with the chrysotile surface. In the dielectric medium, the minimized energies amount to −5240.09, −4260.99, −4151.63 and −4595.16 kcal mol−1 Å for the four analysed zones, respectively. In all cases, there were little differences between the optimized and the experimental geometries of the backbone (figure 2b), but different orientations of the side chains were evident, mostly due to the lack of hydrogen bonds with the solvent.
Afterward, the geometries of the selected subdomains close to the chrysotile surface were optimized. In all cases, there was a significant initial adsorption on the chrysotile surface, usually accompanied by conformational rearrangements of the interacting strands, involving a partial loss of secondary structure in the vicinity of the surface. The results of these initial energy minimizations are reported in the first part of table 2. The lowest-energy minima, corresponding to the most stable states found by this procedure, are taken as zero values for either zones (Etot in table 2). The interaction energy was then defined as Eint=(Efree+Eplane)− Etot, where Efree is the energy of the free, isolated subdomain in the optimized geometries. According to this definition, Eint>0 is the energy released by the subdomains upon adsorption. In our simulations, we have that Eplane≡0 for the reason that the chrysotile planes are kept fixed, although all their atoms do correctly interact with the HSA subdomains. Estrain was defined as the difference between E0 and Efree, where E0 is the energy of the isolated subdomains in the geometry they adopt upon adsorption.
Table 2.
Results of the initial energy minimizations and of the long MD runs and subsequent optimizations in the dielectric medium. (nAA represents the number of AA in a 5 Å range from the chrysotile plane, Hintra the number of intramolecular H-bonds and AA the residues involved in the α-helix formation. Energies are in kcal mol−1.)
| CAa (Å2) | initial adsorption | final adsorption | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Etot | Eint | Estrain | nAA | Hintra | AA | Etot | Eint | Estrain | nAA | Hintra | AA | ||
| zone 1 | 1027 | −5804.13 | 564.04 | 34.64 | 24 | 19 | 36 | −6013.43 | 773.34 | 181.33 | 81 | 1 | 2 |
| zone 2 | 769 | −4768.90 | 507.91 | 28.27 | 16 | 18 | 32 | −5066.36 | 805.37 | 153.06 | 63 | 4 | 3 |
| zone 3 | 862 | −4407.42 | 455.79 | 20.80 | 15 | 26 | 49 | −4817.79 | 966.16 | 151.66 | 61 | 7 | 10 |
| zone 4 | 807 | −4971.14 | 575.97 | 85.26 | 20 | 11 | 20 | −6013.43 | 918.26 | 226.26 | 68 | 3 | 2 |
Initial contact area values.
In all the cases investigated, the amino acids close to the surface locally optimize their interactions with the chrysotile planes by partially loosing their secondary α-helical structure, as shown in figure 2c(i) for zone 1.
In particular, all the α-helices are significantly shortened when close to the surface, in order again to maximize the amino acid–surface interactions through a partial loss of the secondary structure. Some helicoidal features are retained, but with large distortions from the typical conformation of the α-helices, so that H-bonds between adjacent turns are absent. Therefore, these helicoidal strands cannot be classified as genuine α-helices.
The driving force for the α-helices unfolding consists here in their favourable electrostatic interactions with the chrysotile plane. As a general feature, upon adsorption the subdomains achieve a lower energy, hence a larger Eint, when more amino acids interact with the surface, irrespective of their overall hydropathy index, because of the apparently random distribution of hydrophobic and hydrophilic residues. However, it should be noted that, in this initial adsorption phase, Eint seems to be partially correlated with the hydropathy index values for the single α-helix (local effect). In fact, the highest Eint values are observed for those zones (like zones 4 and 3) containing hydrophilic helices (IIIB-h2 and IA-h6, respectively; table 1), while zone 3, containing the most hydrophobic α-helix (IIB-h3), shows the lowest Eint value. This latter finding is not unexpected due to the cooperative nature of the adsorption process of a complex copolymer (like a protein fragment) on a polar surface. The α-helices undergo a large strain in order to optimize their interactions with the surface. Therefore, we expect that a larger interaction should usually be accompanied by a greater strain energy of the adsorbed subdomains, hence a larger deformation, so as to allow more amino acids to be in contact with the surface. By extrapolation, we expect that the subdomains can also undergo much larger deformations than those reported in figure 2c owing to the increasingly stronger interactions with the chrysotile surface. The strain energy is mainly (though not exclusively) due to the energy cost of the H-bonds breaking between the amino acids belonging to the α-helices. Thus, Estrain is also roughly related with both the hydropathy index values for the single α-helix and the number of broken intrahelix H-bonds (table 2). Additionally, a larger Estrain is also accompanied by a smaller number of amino acids still structured in α-helices (table 2). Conversely, the higher energy states show only minor conformational changes of the adsorbed subdomains. Therefore, they show not only a smaller strain energy but also a smaller Eint, with fewer broken H-bonds and more amino acids retaining the original α-helical structure.
3.2 Adsorption by MD runs and energy minimizations in the dielectric medium
The geometries obtained in §3.1 were subjected to MD runs to search the best final adsorption geometry with the overall energy minimum. The MD runs show a relatively fast and steady energy decrease, lasting for approximately 100–150 ps for all the studied zones. At this time, an equilibrium state was quickly reached and the final part of the MD simulation, carried out for 1.5 ns, showed only relatively minor readjustments mainly involving some local features, with modest energy changes. Further energy minimization of selected frames provided no further significant changes, showing these final adsorbed states to be equivalent to better than ±15 kcal mol−1. Some details of these geometries are reported in the second part of table 2 and shall be discussed below. The large rearrangements undergone by the subdomains within the initial stage of the MD runs can be qualitatively described as a fast liquid-like spreading of the adsorbed subdomains on the surface, possibly followed by small later readjustments of minor importance. The whole process leads to a very large contact area between the subdomains and the chrysotile surface. For the albumin subdomains considered here, the whole process quickly leads to the formation of a monolayer of amino acids evenly coating the surface. Here, all the α-helices are fully denatured and nearly all the amino acids are in contact with the chrysotile plane (figure 2c(ii)). In these final states, the whole pattern of intramolecular H-bonds is disrupted. Only a few hydrogen bonds are found, mostly involving the side groups of topologically adjacent amino acids.
The large rearrangements observed in the final adsorption step greatly modify the size of the subdomains and, in particular, the contact surface area (the footprints of the subdomains) before and after the final adsorption stage. This contact area is most simply obtained through the difference between the area of the protein surface accessible to the solvent before and after adsorption. In turn, the accessible surface, or Connolly surface, is evaluated through a spherical probe of radius 1.4 Å, mimicking a water molecule, rolling on the exposed van der Waals surface. Upon full spreading on the chrysotile plane, the contact area of the four analysed zones increases by a factor of approximately 2.75, 2.98, 2.83 and 3.15, respectively (relative to the initial contact area values reported in table 2), comparable with the experimental values found for the initial spreading of albumin on a hydrophobic surface. The increase in contact area, together with the Estrain values, are now in agreement with the overall hydropathy index, suggesting the importance of the whole subdomains structure in the second (or final) adsorption phase. Note that we might expect the whole protein to show an even larger increase of its footprint upon surface denaturation owing to its overall size (figure 1a). On the other hand, it should be realized that, in addition to the intermolecular interactions still present between the protein and the chrysotile plane (mainly hydrogen bonds), the network of intramolecular disulphide bridges hinders the full albumin spreading on the surface, thus limiting somewhat the contact area.
3.3 FTIR analysis of HSA-coated chrysotile nanocrystals
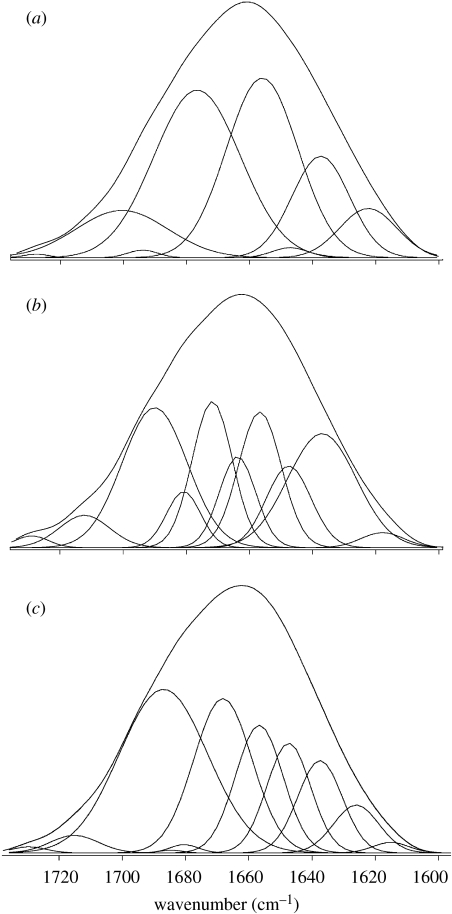
HSA structural modifications induced by interaction with the chrysotile surface have been investigated by FTIR spectroscopy. In fact, FTIR spectra allow an evaluation of the percentage content of each secondary structure by Gaussian curve fitting using the amide I spectral region according to the literature data (Jakobsen & Wasacz 1990; Fu et al. 1999; Pelton & McLean 2000; Servagent-Noinville et al. 2000; Carrasquillo et al. 2001a,b; Min et al. 2004). In the FTIR spectra, the band range 1610–1640 cm−1 is generally assigned to β-sheet, 1640–1650 cm−1 to random coil, 1650–1658 cm−1 to α-helix and 1660–1700 cm−1 to β-turn structures (Min et al. 2004). Figure 3a reports the FTIR spectra and their Gaussian curve fitting in the 1750–1600 cm−1 region (amide I bands) for the lyophilized HSA-coated chrysotile nanocrystals for low (0.5 mg ml−1) HSA starting concentration. The secondary structure elements, calculated in percentages from the integrated areas of the component bands, are reported in table 3, where they are compared with those obtained from native HAS-lyophilized powder from aqueous solution at pH 7.4, which are very close to the values previously reported (Griebenow & Klibanov 1995). The appreciable differences in protein conformation observed in the protein coating are due to the interaction of albumin with the inorganic surface. They are the experimental evidence of the results obtained by the modelling approach described above. Actually, the α-helix content appears drastically reduced in favour of an increase in β-sheet and random coils content, in agreement with the computational evidence of a partial loss of secondary structure during the adsorption first step.
Figure 3.
FTIR spectra and their Gaussian curve fitting from HSA-coated chrysotile nanocrystals at (a) low, (b) medium and (c) high surface coverage, where a shift at higher wavenumbers is clearly evident as a function of Γ.
Table 3.
HSA secondary structure (%). (The ‘±’ values are standard deviations calculated by analysing five individual spectra in each case.)
| HSA-coated chrysotile nanocrystals | ΓHSA (mg m−2) | α-helix | β-sheet | β-turn | random coil |
|---|---|---|---|---|---|
| 1.04 | 14±1 | 25±1 | 52±1 | 8±1 | |
| 1.55 | 15±2 | 18±2 | 57±2 | 10±2 | |
| 2.04 | 14±1 | 15±1 | 59±1 | 12±1 | |
| HAS lyophilized | 33±1 | 16±1 | 52±1 | 0 |
In addition, HSA-coated chrysotile nanocrystals have been investigated by FTIR spectroscopy as a function of the protein concentration which induces a different degree of surface coverage. Surface coating extent (Γ) represents the amount of protein adsorbed (mg m−2) at constant temperature on a given substrate and strongly depends on the surface features: a hydrophilic substrate, like chrysotile, may adsorb a much greater protein amount than a hydrophobic one (Nakanishi et al. 2001). It is important to take into account that at pH 7.4, HSA (isoelectric point 4.6) and chrysotile (isoelectric point 8–12, according to the different amount of magnesia) are predominantly negatively and positively charged, respectively; thus electrostatic forces play here an important role.
A given amount of chrysotile nanocrystals has been interacted with HSA solutions at different starting concentrations. The lyophilized solid samples obtained after centrifugation of the suspensions were examined by FTIR spectroscopy.
Figure 3b,c reports the FTIR spectra and their Gaussian curve fitting in the 1750–1600 cm−1 region (amide I bands) for the lyophilized HSA-coated chrysotile nanocrystals for medium (1.0 mg ml−1) and high (4 mg ml−1) HSA starting concentration, respectively. No appreciable modification in the α-helix bands range appears as the coating extent increases. At the same time, between 1640 and 1650 cm−1 and between 1660 and 1700 cm−1 an evident spectral modification can be appreciated, indicative of increased random and β-turn structure contents. In particular, the bands between 1660 and 1700 cm−1 (β-turns region) increase in intensity with increasing Γ, showing the strongest variation for the highest protein concentration. On the contrary, the β-sheet spectral evidence decreases in intensity for increasing Γ values, in agreement with a consistent reduction of their presence.
The HSA secondary structure elements, calculated in percentages from the integrated areas of the component bands, are reported in table 3 as a function of the surface coating extent (Γ), determined by the medium and high protein concentrations. On the whole, when comparing the three different surface coatings within themselves, the β-sheet and β-turn contents appreciably decrease and increase, respectively, with the increase in Γ value, while the α-helix amount appears to be quite independent of the surface coating extent. On the other hand, if compared to the native protein secondary structure, the β-sheet content, after the initial increase at low Γ value, tends to get lower up to the percentage observed for the protein not deposited on the inorganic surface.
These results support the protein structural changes first observed using bovine serum albumin on the same synthetic chrysotile surface for a very low surface coverage (Γ=0.18 mg m−2; Falini et al. 2006). Although for the highest surface coverage the major protein modification in terms of β-sheet, β-turn and random coil percentages can be fully observed, it must however be underlined that using a range of HSA initial concentrations (0.5–4 mg ml−1) allows one to appreciate that the greater differences in protein conformation appear on passing from the low to the medium surface coating extent. In fact, when the protein–surface interaction is prevailing on the protein–protein interactions, owing to the greater accessibility of the surface and the strong electrostatic effect that it can exert, the great protein flexibility can be fully displayed. The observed increase in β-turn structures, which are concentrated on the exterior of the protein, may be related to the protein unfolding in order to expose the interior regions. This change in secondary structure is probably driven by the formation of surface-mediated hydrogen bonds, with the polar side-chain groups serving as sites for molecular recognition.
Our future aim is to investigate structural perturbations of the adsorbed and hydrated protein by FTIR spectroscopy in order to better characterize protein–substrate interactions.
4. Conclusions
The HSA secondary structure modifications induced by the chrysotile surface have been studied via MD and experimental FTIR methods. The interaction of HSA subdomains selected on the basis of their solvent accessibility with the chrysotile surface was investigated through MM and MD simulations. HSA adsorption on the chrysotile surface, considered as a rigid body, is characterized by conformational rearrangements, involving a partial loss of secondary structure close to the surface. All the α-helices are significantly shortened while some helicoidal features are retained, but with large distortions, so that the H-bonds between adjacent turns are absent. The whole process leads to a very large contact area between the subdomains and the chrysotile surface, quickly leading to the formation of an amino acid monolayer. Contact area and energy values analysis suggest the importance (at least in the initial adsorption phase) of the α-helices local structure, while the final adsorption phase seems to be driven by the whole subdomains structure.
FTIR spectroscopy investigation carried out on HSA-coated chrysotile nanocrystals at different protein concentration allows one to experimentally quantify HSA secondary structure modifications due to interaction with the inorganic surface. Experimental data support the adhesion mechanism indicated by MD computational approach. In fact, for low Γ value corresponding to the initial adsorption step, the α-helix content appears drastically reduced when compared with the protein not deposited on the substrate, while an increase in β-sheet and random coils content is evident.
A second adsorption step may be singled out during which, with the increase in Γ value, the β-sheet and β-turn contents appreciably decrease and increase, respectively, while the α-helix amount does not change appreciably.
The present results give strong support to the atomistic computer simulations as a powerful tool to provide a realistic description of the mechanism of the adsorption process of plasma proteins on the chrysotile surface, which can be quantitatively investigated by FTIR spectroscopy, gaining insight into the mechanism of asbestos toxicity.
Acknowledgements
MIUR (Prin 2006032335), CIRCMSB, CNR and University of Bologna (funds for selected research topics) are acknowledged for financial support.
Footnotes
NAMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign.
References
- Bergamini C, Fato R, Biagini G, Pugnaloni A, Giantomassi F, Foresti E, Lesci G.I, Roveri N, Lenaz G. Mitochondrial changes induced by natural and synthetic asbestos fibers: studies on isolated mitochondria. Cell. Mol. Biol. 2004;50(Suppl.):691–700. (Sarreguemines, France, Print) [PubMed] [Google Scholar]
- Brown J.R. Structure of serum albumin: disulfide bridges. Fed. Proc. 1974;33:1389. [Google Scholar]
- Brown J.R. Structural origins of mammalian albumin. Fed. Proc. 1976;35:2141–2144. [PubMed] [Google Scholar]
- Carrasquillo K.G, Aponte-Carro J.C, Alejandro A, Diaz Toro D, Griebenow K. Reduction of structural perturbations in bovine serum albumin by non-aqueous microencapsulation. J. Pharm. Pharmacol. 2001a;53:115–120. doi: 10.1211/0022357011775091. [DOI] [PubMed] [Google Scholar]
- Carrasquillo K.G, Stanley A.M, Aponte-Carro J.C, De Jesus P, Constantino H.R, Bosques C.J, Griebenow K. Non-aqueous encapsulation of excipient-stabilized spray-freeze dried BSA into poly(lactide-co-glycolide) microspheres results in release of native protein. J. Control. Release. 2001b;76:199–208. doi: 10.1016/S0168-3659(01)00430-8. [DOI] [PubMed] [Google Scholar]
- Chissick S.S. In: Encyclopedia of physical science and technology. Mayers R.A, editor. vol. 2. Academic Press; Orlando, FL: 1987. pp. 79–108. [Google Scholar]
- Churg A, Green F.H.Y. 2nd edn. Williams and Wilkins; Baltimore, MD: 1998. Pathology of occupational lung disease. [Google Scholar]
- Churg A.M, Warnock M.L. Asbestos and other ferruginous bodies—their formation and clinical significance. Am. J. Pathol. 1981;102:447–456. [PMC free article] [PubMed] [Google Scholar]
- Cornell W.D, et al. A second-generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. doi: 10.1021/ja00124a002. [DOI] [Google Scholar]
- Curry S, Mandelkow H, Brick P, Franks N. Crystal structure of human serum albumin with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998;5:827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- Dockal M, Carter D.C, Rüker F.J. The three recombinant domains of human serum albumin. Structural characterization and ligand binding properties. Biol. Chem. 1999;274:29 303–29 310. doi: 10.1074/jbc.274.41.29303. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A, Law S.W, Dennison O.E. Nucleotide sequence and the encoded amino acids of human serum albumin mRNA. Proc. Natl Acad. Sci. USA. 1982;79:71–75. doi: 10.1073/pnas.79.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitru-Stanescu R, Mandravel C, Bercu C. Infrared and nuclear magnetic resonance studies of some surface properties of asbestos–albumin interaction. Analyst. 1994;119:689–691. doi: 10.1039/an9941900689. [DOI] [PubMed] [Google Scholar]
- Euston S.R. Computer simulation of proteins: adsorption, gelation and self-association. Curr. Opin. Colloid Interface Sci. 2004;9:321–327. doi: 10.1016/j.cocis.2004.09.005. [DOI] [Google Scholar]
- Falini G, Foresti E, Lesci I.G, Roveri N. Structural and morphological characterization of synthetic chrysotile single crystals. Chem. Commun. 2002;14:1512–1513. doi: 10.1039/b203430a. [DOI] [PubMed] [Google Scholar]
- Falini G, Foresti E, Gazzano M, Gualtieri A.F, Leoni M, Lesci I.G, Roveri N. Tubular-shaped stoichiometric chrysotile nanocrystals. Chem. Eur. J. 2004;10:3043–3049. doi: 10.1002/chem.200305685. [DOI] [PubMed] [Google Scholar]
- Falini G, Foresti E, Lesci I.G, Lunelli B, Roveri N, Sabatino P. Bovine serum albumin interaction with chrysotile: spectroscopic and morphological studies. Chem. Eur. J. 2006;12:1968–1974. doi: 10.1002/chem.200500709. [DOI] [PubMed] [Google Scholar]
- Fu K, Griebenow K, Hsieh L, Klibanov A.M, Langer R. FTIR characterization of the secondary structure of proteins encapsulated within PLGA microspheres. J. Control. Release. 1999;58:357–366. doi: 10.1016/S0168-3659(98)00192-8. [DOI] [PubMed] [Google Scholar]
- Ganazzoli F, Raffaini G. Computer simulation of polypeptide adsorption on model biomaterials. Phys. Chem. Chem. Phys. 2005;7:3651–3663. doi: 10.1039/b506813d. [DOI] [PubMed] [Google Scholar]
- Gazzano E, Foresti E, Lesci I.G, Tomatis M, Riganti C, Fubini B, Roveri N, Ghigo D. Different cellular responses evoked by natural and stoichiometric synthetic chrysotile asbestos. Toxicol. Appl. Pharmacol. 2005;206:356–364. doi: 10.1016/j.taap.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Gazzano E, et al. Iron-loaded synthetic chrysotile: a new model solid for studying the role of iron in asbestos toxicity. Chem. Res. Toxicol. 2007;20:380–387. doi: 10.1021/tx600354f. [DOI] [PubMed] [Google Scholar]
- Griebenow K, Klibanov A.M. Lyophilization-induced reversible changes in the secondary structure of proteins. Proc. Natl Acad. Sci. USA. 1995;92:10 969–10 976. doi: 10.1073/pnas.92.24.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- He X.M, Carter D.C. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- Hopp T.P, Woods K.R. A computer program for predicting protein antigenic determinants. Mol. Immunol. 1983;20:483–489. doi: 10.1016/0161-5890(83)90029-9. [DOI] [PubMed] [Google Scholar]
- Horovitz A, Willison K.R. Allosteric regulation of chaperonins. Curr. Opin. in Struct. Biol. 2005;15:646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Horwich A.L, Fenton W.A, Farr G.W. Chaperonins. Encycl. Biol. Chem. 2004;1:393–398. [Google Scholar]
- Kleywegt G.J. Validation of protein crystal structures. Acta Crystallogr. 2000;D56:249–265. doi: 10.1107/s0907444999016364. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J, Zou J.Y, Kjeldgaard M, Jones T.A, Around O. In: International tables for crystallography. Crystallography of biological macromolecules. Rossmann M.G, Arnold E, editors. vol. F. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2001. pp. 353–367. [Google Scholar]
- Jakobsen R.J, Wasacz F.M. Infrared spectra-structure correlations and adsorption behavior for helix proteins. Appl. Spectr. 1990;44:1478–1489. doi: 10.1366/0003702904417724. [DOI] [Google Scholar]
- Jaurand M.C, Baillif P, Thomassin J.H, Magne L, Touray J.C. X-ray photoelectron and chemical study of the adsorption of biological molecules on the chrysotile asbestos surface. J. Colloid Interface Sci. 1983;95:1–9. doi: 10.1016/0021-9797(83)90065-6. [DOI] [Google Scholar]
- Kostova Z, Wolf H.D. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Luft A.J, Lorscheiser F.L. Structural analysis of human and bovine alpha-fetoprotein by electron microscopy, image processing, and circular dichroism. Biochemistry. 1983;22:5978–5980. doi: 10.1021/bi00294a043. [DOI] [PubMed] [Google Scholar]
- Min J, Meng-Xia X, Dong Z, Yuan L, Xiao-Yu L, Xing C. Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin. J. Mol. struct. 2004;692:71–80. doi: 10.1016/j.molstruc.2004.01.003. [DOI] [Google Scholar]
- Minghetti P.P, Ruffner D.E, Kuang W.J, Dennison O.E, Hawkins J.W, Beattie W.G, Dugaiczyk A. Molecular structure of the human albumin gene is revealed by nucleotide sequence within q11-22 of chromosome 4. J. Biol. Chem. 1986;261:6747–6757. [PubMed] [Google Scholar]
- Morgan A. Adsorption of human serum albumin by asbestiform minerals and its application to the measurement of surface areas of dispersed samples of chrysotile. Environ. Res. 1974;7:330–341. doi: 10.1016/0013-9351(74)90034-6. [DOI] [Google Scholar]
- Mossman B.T. Cellular and molecular mechanisms of disease. In: Guthrie G.D, Mossman B.T, editors. Health effects of mineral dust. Reviews in Mineralogy. vol. 28. Mineralogical Society of America; Washington, DC: 1993. pp. 513–521. [Google Scholar]
- Nakanishi K, Sakiyama T, Imamura K. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J. Biosci. Bioeng. 2001;91:233–244. doi: 10.1263/jbb.91.233. [DOI] [PubMed] [Google Scholar]
- Noinville V, Vidal-Madjar C, Sébille B. Modeling of protein adsorption on polymer surfaces. Computation and adsorption potential. J. Phys. Chem. 1995;99:1516–1522. doi: 10.1021/j100005a023. [DOI] [Google Scholar]
- Pelton J.T, McLean L.R. Spectroscopic methods for analysis of protein secondary structure. Anal. Biochem. 2000;277:167–176. doi: 10.1006/abio.1999.4320. [DOI] [PubMed] [Google Scholar]
- Peters T., Jr . Academic Press; San Diego, CA: 1996. All about albumin: biochemistry, genetics, and medical applications. [Google Scholar]
- Phillips J.C, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaini G, Ganazzoli F. Simulation study of the interaction of some albumin subdomains with a flat graphite surface. Langmuir. 2003;19:3403–3412. doi: 10.1021/la026853h. [DOI] [Google Scholar]
- Raffaini G, Ganazzoli F. Molecular dynamics simulation of the adsorption of a fibronectin module on a graphite surface. Langmuir. 2004;20:3371–3378. doi: 10.1021/la0357716. [DOI] [PubMed] [Google Scholar]
- Raffaini G, Ganazzoli F. Sequential adsorption of proteins and the surface modification of biomaterials: a molecular dynamics study. J. Mater. Sci. Mater. Med. 2007;18:309–316. doi: 10.1007/s10856-006-0694-5. [DOI] [PubMed] [Google Scholar]
- Rose G.D, Geselowitz A.R, Lesser G.J, Lee R.H, Zehfus M.H. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Roveri N, Falini G, Foresti E, Fracasso G, Lesci I.G, Sabatino P. Geo-mimetic synthetic chrysotile nanocrystals. J. Mater. Res. 2006;21:2711–2725. doi: 10.1557/jmr.2006.0359. [DOI] [Google Scholar]
- Sabatino, P., Casella, L., Granata, A., Iafisco, M., Lesci, G.I., Monzani, E., & Roveri, N. In press. Synthetic chrysothile nanocrystals as a reference standard to investigate surface-induced serum albumin structural modifications. J. Coll. Int. Sci. [DOI] [PubMed]
- Saber M.A, Stöckbauer P, Morávek L, Meloun B. Disulfide bonds in human serum albumin. Collect Czech. Chem. Commun. 1977;42:564–579. [Google Scholar]
- Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 angstrom resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- Schreier H. Elsevier; New York, NY: 1989. Asbestos in the natural environment. [Google Scholar]
- Servagent-Noinville S, Revault M, Quiquampoix H, Baron M.H. Conformational changes of bovine serum albumin induced by adsorption on different clay surfaces: FTIR analysis. J. Coll. Int. Sci. 2000;221:273–283. doi: 10.1006/jcis.1999.6576. [DOI] [PubMed] [Google Scholar]
- Stanley P. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- Stroink G, Hutt D, Lim D, Dunlap R.A. The magnetic properties of chrysotile asbestos. IEEE Trans. Magnetics. 1985;21:2074–2076. doi: 10.1109/TMAG.1985.1064007. [DOI] [Google Scholar]
- Whittaker E.J.W. Structure of chrysotile. II. Clinochrysotile. Acta Crystallogr. 1956;9:862–864. doi: 10.1107/S0365110X56002461. [DOI] [Google Scholar]
- Whittaker E.J.W. The structure of chrysotile. V. Diffuse reflections and fiber texture. Acta Crystallogr. 1957;10:149–156. doi: 10.1107/S0365110X57000511. [DOI] [Google Scholar]
- Whittaker E.J.W, Wicks F.J. Chemical differences among the serpentine “polymorphs”. Am. Mineral. 1970;55:1025–1047. [Google Scholar]
- Yada K. Study of chrysotile asbestos by a high resolution electron microscope. Acta Crystallogr. 1967;23:704–707. doi: 10.1107/S0365110X67003524. [DOI] [Google Scholar]
- Yada K. Study of microstructure of chrysotile asbestos by high resolution electron microscopy. Acta Crystallogr. A. 1971;27:659–664. doi: 10.1107/S0567739471001402. [DOI] [Google Scholar]