Abstract
We introduce another view of sequence evolution. Contrary to other approaches, we model the substitution process in two steps. First we assume (arbitrary) scaled branch lengths on a given phylogenetic tree. Second we allocate a Poisson distributed number of substitutions on the branches. The probability to place a mutation on a branch is proportional to its relative branch length. More importantly, the action of a single mutation on an alignment column is described by a doubly stochastic matrix, the so-called one-step mutation matrix. This matrix leads to analytical formulae for the posterior probability distribution of the number of substitutions for an alignment column.
Keywords: maximum likelihood, maximum parsimony, substitution model, posterior probability, tree reconstruction
1. Introduction
Tree reconstruction is nowadays a matter of routinely applying the available programs, comparing the resulting trees and then concluding what might be the best tree (e.g. Hillis et al. 1996; Felsenstein 2004; Schmidt & von Haeseler 2008). With the advent of Baysian approaches, it is possible to model increasingly complex evolutionary scenarios (Huelsenbeck & Ronquist 2001). However, a detailed understanding about what can and cannot be inferred (Baake 1998; Mossel & Steel 2005) from a sequence alignment is still missing. In recent years, some theoretical insights were obtained by studying very simple models of sequence evolutions on binary sequence data (e.g. Erdös et al. 1999a,b). Although these models are not realistic in a biological sense, they have provided some profound insights in the reconstruction process per se.
Here, we will introduce another description of the evolutionary process on trees. More precisely, given a phylogenetic tree and an alignment that evolved along the tree, we now ask the following question: how does the alignment change if an additional substitution on an arbitrary branch of the tree takes place? This rather abstract question is motivated by the following biological problem. Consider a collection of morphological traits that are either in an ancestral (0) or derived (1) state. Each derived character state characterizes a monophyletic group and represents a cluster in the tree. For such a data matrix (or alignment), the tree reconstruction problem is easy. However, stochastic effects that act somewhere on the branches of the tree may disturb this signal. This noise is modelled by the assumption of throwing an arbitrary number of changes on the tree and measuring their impact on the otherwise perfect data matrix. To this end, we construct a one-step mutation (OSM) matrix. The matrix description allows a linear algebra view of evolution and comprises distance methods, maximum parsimony as well as maximum likelihood. Moreover, the description reveals a surprising connection to Hadamard matrices that were employed for phylogenetic questions (Hendy & Penny 1989; Steel et al. 1998). An immediate application of OSM matrices is the analytical computation of posterior probabilities that count the number of evolutionary changes on a tree. So far, these posterior probabilities have been estimated using Bayesian simulation (Nielsen 2002; Huelsenbeck et al. 2003) or by applying the theory of counting processes (Minin & Suchard 2008).
In the following we refrain from most technicalities but rather outline the general ideas by discussing illustrative examples. The technical details and proofs will appear elsewhere.
2. The Binary model on an n-taxon tree
(a) Notation
We consider a set of n taxa S={1, …, n}. With S comes some information about the common properties and differences of the taxa, typically displayed in an alignment. In the following a (sequence) alignment is an n×ℓ-array with entries either 0 or 1, where ℓ is the length of the alignment. Each of the ℓ columns (sites) aj of the alignment represents a pattern of n homologous characters, where aij∈{0,1} is the state of character j in taxon i. For binary character states, 2n patterns are possible.
We are interested in the evolution of such patterns along a (rooted) tree T=(V, E) with node set V and branch set E⊂V×V (Semple & Steel 2003). The node set V contains the taxon set S that forms the leaf set. Avoiding the technical details, each branch is uniquely encoded by the subsets X of S that originates from the branch. Such a set X specified by a branch will be called a cluster. A leaf is a trivial cluster.
Finally, we introduce a function λ:E→+, such that λ(e)>0 represents the length of a branch e∈E. The tree-length ΛT is the sum of the branch lengths. The relative branch length
| (2.1) |
denotes the probability that a substitution hits branch e of the rooted tree T.
(b) The effect of substitutions on an alignment
We now describe how a single mutation on the tree changes the current character states at the leaves. Obviously the outcome will depend on the branch where the substitution occurred. Moreover, each of the 2n possible patterns will be affected differently by such a substitution. Therefore we introduce a 2n×2n matrix that describes the action of a substitution on the patterns for a specific branch.
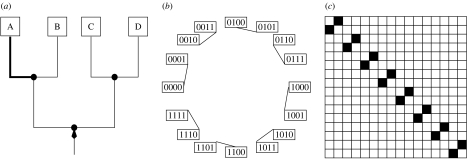
Figure 1 describes the model on an example tree T4 with four taxa. For instance, a substitution at the branch defined by cluster {A} changes the pattern 1011 to the pattern 1010 because only the character of taxon A is affected. Please note that order of taxa is (D, C, B, A) for each pattern. All possible changes between the patterns identified by a substitution on branch eA are depicted in the substitution graph (figure 1b). The corresponding adjacency matrix σA is displayed in figure 1c, where a black square stands for one (the patterns are connected by an edge in the substitution graph) and a white square represents zero. The structure of matrix σA constitutes an example of the so-called permutation matrices with entries equal to one if the substitution converts one pattern into another (Bona 2004, p. 75).
Figure 1.
(a) The rooted phylogenetic tree T4. The branch defined by cluster {A} is highlighted. A substitution on this branch gives rise to the unique change in patterns depicted in (b) the graph or (c) its corresponding adjacency matrix.
For each branch we easily construct the corresponding permutation matrix. Without proof we state that each matrix is fix point free (a substitution changes every pattern) and has 2n−1 transpositions (applying a substitution twice returns the original pattern). Then it follows that this type of permutation matrix is self-inverse with respect to matrix multiplication and that they form an Abelian group if the identity matrix is included.
We point out that the permutation matrix for a non-trivial cluster is the product of the permutation matrices of its elements. In other words the action of one mutation on a branch e can be replaced by any partition of the cluster associated with e, such that each set of the partition is represented by a branch in the tree. For tree T4 we obtain six permutation matrices σA, σB, σC, σD and σAB=σA·σB, σCD=σC·σD.
Because we study symmetric permutation matrices, it is well known that the eigenvalues are either 1 or −1. Further, we observe that all those permutation matrices are diagonalized by the 2n×2n-dimensional Hadamard matrix (Hendy & Penny 1989; Hendy 2005). The 2×2 Hadamard matrix is defined as
The 2n×2n Hadamard matrix is then
where the symbol ⊗ represents the Kronecker product. Hence, for the cluster (e) associated with branch e,
is the diagonal matrix of eigenvalues of .
To take the relative contribution of the branch lengths into account, we weight each permutation matrix with pe as described in (2.1). Such matrices are a special case of the generalized permutation matrices. Then the so-called OSM matrix of the tree T4 is simply the following convex sum:
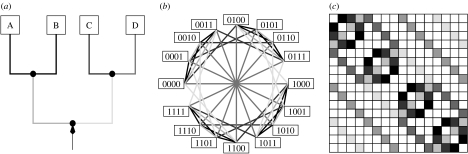
Figure 2c shows the result of this computation. The substitution graph in figure 2b displays the effect of a substitution on all the branches of the tree on the patterns. Two patterns are connected by an edge if a substitution switches between the two patterns.
Figure 2.
(a) Substitutions on different branches of T4 give rise to branches of (b) the graph, and (c) the corresponding entries in the OSM matrix. Corresponding components are identified by common shades of grey.
For an arbitrary phylogenetic tree T on n taxa, the OSM matrix is obtained by
| (2.2) |
where (e) is the cluster identified by branch e∈E. Owing to its construction, MT is also diagonalized by the Hadamard matrix.
The entry MT(i, j) is positive if the tree T contains a cluster where a substitution on the corresponding branch implies that pattern i is changed to j. Hence, each row and each column has 2n−2 non-zero entries, one entry for each branch in the tree. Thus, the OSM matrix belongs to the class of doubly stochastic Markov transition matrices, where the relative branch lengths are represented exactly once in each row and each column. Consequently, the k-th power provides the probabilities to move from one pattern to another in k substitutions. Thus, the repeated application of MT describes a random walk on the state space of the 2n patterns. This random walk is very different from the standard random walk on the hypercube (Eigen et al. 1989). If k is large, then will lose the phylogenetic information of the original alignment and will approximate the uniform distribution, that is each pattern occurs at the same frequency.
(c) Poisson weights
Our setting does not yet assume a probability distribution for the number of substitutions on the tree. In molecular evolution, one typically assumes that the number of substitutions is Poisson distributed with parameter ΛT (Uzzell & Corbin 1971). Under this assumption we can compute the average OSM matrix by
| (2.3) |
which is equivalent to
The exponential of the matrix ΛTMT is easy to compute, because MT is a sum of generalized permutation matrices (2.2), which commute with respect to matrix multiplication. Thus, we obtain
| (2.4) |
3. Relation to tree reconstruction
The OSM matrix leads to a very general description of character-based phylogenetic inference techniques. Moreover, the explicit model assumptions in maximum likelihood and the implicit assumptions in maximum parsimony are directly comparable.
The OSM matrix and its powers describe the substitution process between arbitrary patterns. However, in classical phylogeny the starting point of a substitution process is ancestral states on trees. In particular, one assumes a stationary distribution π=(π0, π1) of character states at the root, and the characters evolve along the tree according to a Markov transition matrix (Tavaré 1986). In our framework, this is equivalent to starting in the constant patterns 0=(0, …, 0) or 1=(1, …, 1) and letting it evolve according to the OSM matrix. This process has a non-stationary pattern distribution which starts at , i.e. with zero substitutions only constant patterns exist, and in each step the pattern distribution is given by . If the number of substitutions is not weighted as in equation (2.3), then will approach the uniform distribution as k goes to infinity. To overcome the loss of phylogenetic signal, we assume in the following that the number of substitutions on a tree is Poisson distributed with parameter ΛT. Moreover, we assume that the substitution process is described by the symmetric Cavender–Farris–Neyman mutation model (CFN, Neyman 1971; Farris 1973; Cavender 1978). Under these assumptions, the probability of observing pattern a when starting in a constant pattern is then calculated employing (2.3)
| (3.1) |
where π0 and π1 are taken from the stationary distribution of character states. The resulting probability distribution for all possible patterns is then identical to the standard way of computing the probabilities of pattern (Felsenstein 2004).
(a) Distance approaches
Now, we briefly illustrate how to derive distance corrections from the OSM matrix. To this end, we consider the rooted tree with two leaves A, B and branch lengths λA and λB. Then the corresponding OSM matrix M2 has the following structure:
where pA and pB are computed according to (2.1). From (2.4) it is straightforward to compute using (*).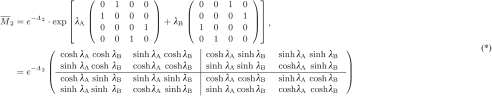
The first and the last row of yield the probabilities to observe one of the four patterns. If evolution starts with character state 0 or 1 at the root of the tree and the character states are in equilibrium (π0=π1), then we quickly compute
as the probability to observe a constant pattern in an alignment. Similarly we compute the probability to observe different character states between taxa A and B. From this it is straightforward to get the distance correction of the CFN model.
(b) Maximum likelihood
The maximum-likelihood principle for an alignment and a tree T is easily formulated in terms of the OSM matrix. We introduce as parameter vector θ the branch lengths of T. Then the probability of is given by
| (3.2) |
where the factors on the right-hand side are defined by equation (3.1). The parameter vector θ enters the equation via the OSM and in the obvious way. As usual, we want to find parameter assignments such that (3.2) is maximized.
(c) Maximum parsimony
We associate the adjacency matrix AOSM (e.g. Cormen et al. 2001, §22.1), or simply A, with the OSM matrix. A is obtained as the unweighted sum of the permutation matrices . Hence, an entry Aij is equal to one when there is a branch in the tree which changes pattern i into pattern j, and is zero otherwise. Finally, we note that Ak(i, j) describes the number of paths of length k between pattern i and j. Each path specifies a series of branches in the tree where a substitution occurred.
Now, fix a column ai in alignment , and a tree T. We ask for the minimal number kmin such that or is greater than zero. In other words, for an alignment column ai, the minimal number of mutations on T equals
Thus the minimal number of mutations for an alignment equals
| (3.3) |
This is another description of the maximum-parsimony principle.
4. Mapping substitutions
From the computation of the powers of the OSM matrix, it is possible to derive the (posterior) probability distribution ppdf(k|x) of the number of mutations that generated an observed pattern x, when the process started in pattern 0 or 1. The posterior probabilities have been estimated before, employing Bayesian simulation methods (Nielsen 2002; Huelsenbeck et al. 2003; Minin & Suchard 2008), but an analytic approach has not previously been attempted.
In general, the posterior probabilities ppdf(k|a) for a pattern a are calculated in the following way using (3.1):
i.e. we compute for pattern a the proportion of its occurrence after k substitutions.
For the two-taxon case, we observe that
Thus, applying hyperbolic identities, we end up with
Taylor expansion of cosh(x) around x=0 then leads to the posterior probability
For the remaining patterns, we obtain the following ppdf:
Thus, on a two-taxon tree, an even number of substitutions leads to a constant pattern when starting in a constant pattern; whereas an odd number of substitutions will be reflected by the non-constant pattern 01 or 10.
Now, it is an easy calculation to obtain closed formulae for the posterior mean number μ(a) of substitutions for pattern a by the following calculations:
Only if Λ is large, then the posterior mean number of substitutions will approach the expected number of substitutions per site Λ. For a constant pattern the posterior mean is always smaller than Λ, and for non-constant patterns the posterior mean is larger than Λ.
Similarly we extend the calculations to a four-taxon tree. For instance, consider the four-taxon tree T4 (figure 1a). This tree has two non-trivial clusters {A, B} and {C, D}. We want to compute the posterior probability of the number of substitutions if the constant pattern 0000 is observed. Let us assume that the two character states occur with uniform probability; then we can compute:
where is the sum of the lengths of the interior branches. Now Taylor expansion leads to the desired posterior probability distribution.
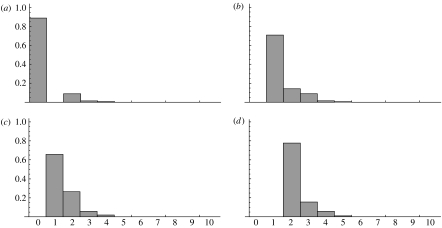
Figure 3 shows the resulting posterior probability distributions for the 16 possible patterns, assuming branch lengths and . The symmetries in the CFN model are reflected in the symmetries of the posterior distributions. Complementary patterns (i.e. 0000 and 1111) show the same distribution. Because the tree is clock-like, the parsimonious uninformative patterns (0001, 0010, 0100, 1000) and their complements show identical distributions, as do the patterns that need at least two substitutions (0101, 0110, 1010, 1001) on T4. Posterior probabilities may be used to compute, for instance, the number of unvaried sites (Fitch & Ayala 1994), which is exactly the proportion of the constant patterns with zero substitutions. In our example we expect approximately 42 per cent constant patterns of which approximately 90 per cent are unvaried. This is only one application for posterior probabilities of the number of substitutions.
Figure 3.
Posterior probabilities for representative patterns of the four-taxon tree T4 with branch lengths and , and character distribution π0=π1=1/2. The selected patterns (a) 0000, (b) 0001, (c) 0011 and (d) 0101 represent constant, parsimonious uninformative, compatible and incompatible patterns, respectively.
As in the two taxon case, we compute the posterior mean of substitutions for pattern a as
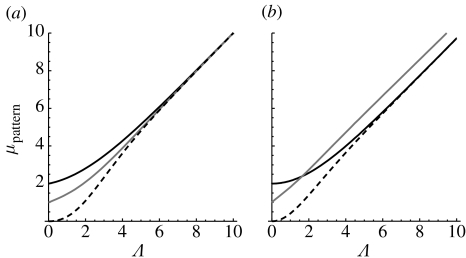
Figure 4a shows the posterior mean number of substitutions for the topology of T4 with branch probabilities and for a constant pattern (0000), a pattern compatible with an interior branch (0011) and a pattern incompatible with the tree (0110). The difference between posterior mean and tree lengths is smaller than 0.01 if the tree lengths exceed 10 substitutions per site.
Figure 4.
Posterior mean number of substitutions as a function of the tree length Λ for the tree topology T4. The posterior means for patterns 0000 (dashed line), 0011 (grey solid line) and 0110 (black solid line) are shown. (a) The posterior means on the tree with relative branch lengths and . (b) The result for relative branch lengths , and .
Figure 4b displays the posterior means for a tree with branch probabilities , and pAB=pCD=0.01. The proportion of pA and pD is so large that the incompatible pattern 0110 will be observed more often than the pattern 0011, which is compatible with a branch of the tree. Thus, this tree is an instance where maximum parsimony will reconstruct the wrong tree (Felsenstein 1978). The figure also shows that the compatible pattern 0011 has a lower posterior mean number of substitutions than 0110 for short tree lengths. However, if the tree length exceeds 1.64 substitutions per site, then the situation is reversed. The posterior mean of the incompatible pattern quickly approaches the tree length, whereas the mean posterior substitutions of the compatible pattern are only close to the tree length if Λ≥54 substitutions per site. In other words, if we observe a compatible pattern, then this pattern has typically experienced more substitutions than the incompatible pattern.
5. Summary and discussion
Here, we have presented an alternative description of how to model sequence evolution on a tree. Our approach lifts the commonly used stochastic models of sequence evolution that act on nucleotides to the set of all possible patterns for n taxa.
We have shown that available tree reconstruction principles are included in our description of the process. Moreover, the definition of the OSM matrix leads to analytical formulae to compute the posterior probability distribution of the number of substitutions for each pattern. From this distribution it is then straightforward to compute the posterior mean of the substitutions. The formulation of the substitution process as an OSM matrix leads to the introduction of the Hadamard matrix that allows an easy computation of matrix powers. Recently, Bryant (submitted) has presented a continuous version of the OSM approach and showed its connection to the Hadamard matrix (Hendy 2005).
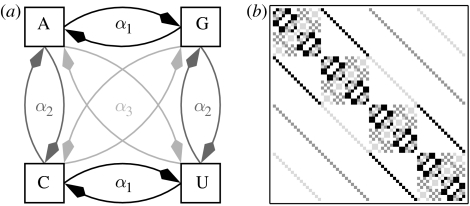
While we have outlined only the simplest model of sequence evolution, several extensions are easily possible. The OSM approach can be augmented to the Kimura 3st model (Kimura 1981); see figure 5a for an illustration. In this framework every substitution class (transition, transversion 1 and transversion 2) uniquely generates a fix-point free 4n×4n-dimensional permutation matrix for each branch in a tree. Let α1+α2+α3=1 denote the probabilities for the three substitution classes, then the OSM matrix for the Kimura 3st model is defined as
| (5.1) |
i.e. we look at the sum of generalized permutation matrices. Figure 5a shows an OSM Kimura matrix on a rooted triplet tree. Each row and each column contain 12=(number of branches)×(number of substitution classes) non-zero entries, where each entry is the product of a mutation class parameter αi and a branch probability pe (equation (5.1)). All the results for binary character state models can be expanded to the Kimura 3st model. If one wants to abandon the assumption that evolution proceeds along a tree, then this is also possible within the OSM framework. Consider a set of rooted trees that give rise to a collection of possibly conflicting clusters. The associated OSM matrix is then given by
where is the normalized sum of branch lengths of those trees in which the branch depicting is existent. Here issues such as the meaning of the overall length of the cluster set or the meaning of a root in such sets need to be discussed. This extension bears some similarity to a maximum-likelihood reconstruction of networks (von Haeseler & Churchill 1993).
Figure 5.
(a,b) The transition scheme and an example of an OSM matrix under the Kimura 3st model of a rooted triplet tree. All four branch lengths are taken to be equal. Hence, black squares indicate a transition, dark grey squares an α2 transversion and light grey squares an α3 transversion.
Another question concerns the Poisson weights for the number substitutions. Generally, the argument is that the process of distributing substitutions along a tree is memoryless and therefore the number of substitutions is Poisson distributed. Our framework permits a different probability distribution to be assigned to the substitution process. One possible weighting scheme could be a contagious distribution, which has been used to evaluate accident data (Kemp 1967). This approach might provide an alternative description of the evolutionary history of an alignment.
In summary, the OSM description offers a variety of potential applications in molecular systematics, which will be explored in the near future.
Acknowledgements
We are grateful to a number of colleagues who helped to increase our understanding of the model by asking the small questions necessary to get that kind of understanding. Helpful comments by two anonymous reviewers are gratefully acknowledged. Support by the Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF) is kindly acknowledged. T.G. and A.v.H. are also supported by the Austrian Ministry for Science and Research (GEN-AU Bioinformatics Integration Network II).
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Statistical and computational challenges in molecular phylogenetics and evolution’.
References
- Baake E. What can and what cannot be inferred from pairwise sequence comparisons? Math. Biosci. 1998;154:1–21. doi: 10.1016/s0025-5564(98)10044-5. doi:10.1016/S0025-5564(98)10044-5 [DOI] [PubMed] [Google Scholar]
- Bona M. Chapman and Hall–CRC; Boca Raton, FL: 2004. Combinatorics of permutations. [Google Scholar]
- Bryant, D. Submitted. Hadamard phylogenetic methods and the n-taxon process. (http://arXiv.org/abs/0806.1378) [DOI] [PubMed]
- Cavender J.A. Taxonomy with confidence. Math. Biosci. 1978;40:271–280. doi:10.1016/0025-5564(78)90089-5 [Google Scholar]
- Cormen T.H, Leiserson C.E, Rivest R.L, Stein C. 2nd edn. MIT Press and McGraw-Hill; Cambridge, MA: 2001. Introduction to algorithms. [Google Scholar]
- Eigen M, Lindemann B.F, Tietze M, Winkler-Oswatitsch R, Dress A, von Haeseler A. How old is the genetic code? Statistical geometry of tRNA provides an answer. Science. 1989;244:673–679. doi: 10.1126/science.2497522. doi:10.1126/science.2497522 [DOI] [PubMed] [Google Scholar]
- Erdös P, Steel M, Székely L, Warnow T. A few logs suffice to build (almost) all trees (part 1) Random Struct. Algor. 1999a;14:153–184. doi:10.1002/(SICI)1098-2418(199903)14:2<153::AID-RSA3>3.0.CO;2-R [Google Scholar]
- Erdös P, Steel M, Székely L, Warnow T. A few logs suffice to build (almost) all trees (part 2) Theor. Comput. Sci. 1999b;221:77–118. doi:10.1016/S0304-3975(99)00028-6 [Google Scholar]
- Farris J. A probability model for inferring evolutionary trees. Syst. Zool. 1973;22:250–256. doi:10.2307/2412305 [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. doi:10.2307/2412923 [Google Scholar]
- Felsenstein J. Sinauer Associates; Sunderland, MA: 2004. Inferring phylogenies. [Google Scholar]
- Fitch W.M, Ayala F.J. The superoxide dismutase molecular clock revisited. Proc. Natl Acad. Sci. USA. 1994;91:6802–6807. doi: 10.1073/pnas.91.15.6802. doi:10.1073/pnas.91.15.6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy M.D. Hadamard conjugation: an analytic tool for phylogenetics. In: Gascuel O, editor. Mathematics of evolution and phylogeny. Oxford University Press; Oxford, UK: 2005. pp. 143–177. [Google Scholar]
- Hendy M.D, Penny D. A framework for the quantitative study of evolutionary trees. Syst. Zool. 1989;38:297–309. doi:10.2307/2992396 [Google Scholar]
- Hillis D.M, Moritz G, Mable B.K, editors. Molecular systematics. 2nd edn. Sinauer Associates; Sunderland, MA: 1996. [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Nielsen R, Bollback J.P. Stochastic mapping of morphological characters. Syst. Biol. 2003;52:131–158. doi: 10.1080/10635150390192780. doi:10.1080/10635150390192780 [DOI] [PubMed] [Google Scholar]
- Kemp C.D. On a contagious distribution suggested for accident data. Biometrics. 1967;23:241–255. doi:10.2307/2528159 [PubMed] [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl Acad. Sci. USA. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. doi:10.1073/pnas.78.1.454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V.N, Suchard M.A. Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 2008;56:391–412. doi: 10.1007/s00285-007-0120-8. doi:10.1007/s00285-007-0120-8 [DOI] [PubMed] [Google Scholar]
- Mossel E, Steel M. How much can evolved characters tell us about the tree that generated them? In: Gascuel O, editor. Mathematics of evolution and phylogeny. Oxford University Press; Oxford, UK: 2005. pp. 384–412. [Google Scholar]
- Neyman J. Molecular studies of evolution: a source of novel statistical problems. In: Gupta J.Y.S.S, editor. Statistical decision theory and related topics. Academic Press; New York, NY: 1971. pp. 1–27. [Google Scholar]
- Nielsen R. Mapping mutations on phylogenies. Syst. Biol. 2002;51:729–739. doi: 10.1080/10635150290102393. doi:10.1080/10635150290102393 [DOI] [PubMed] [Google Scholar]
- Schmidt H.A, von Haeseler A. Phylogenetic inference using maximum likelihood methods. In: Lemey P, Salemi M, Vandamme A, editors. The phylogenetic handbook. 2nd edn. Cambridge University Press; Cambridge, UK: 2008. [Google Scholar]
- Semple C, Steel M. Oxford Lectures Series in Mathematics and its Applications. Oxford University Press; Oxford, UK: 2003. Phylogenetics. [Google Scholar]
- Steel M, Hendy M.D, Penny D. Reconstructing phylogenies from nucleotide pattern probabilities: a survey and some new results. Discret. Appl. Math. 1998;88:367–396. doi:10.1016/S0166-218X(98)00080-8 [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lec. Math. Life Sci. 1986;17:57–86. [Google Scholar]
- Uzzell T, Corbin K.W. Fitting discrete probability distributions to evolutionary events. Science. 1971;172:1089–1096. doi: 10.1126/science.172.3988.1089. doi:10.1126/science.172.3988.1089 [DOI] [PubMed] [Google Scholar]
- von Haeseler A, Churchill G.A. Network models for sequence evolution. J. Mol. Evol. 1993;37:77–85. doi: 10.1007/BF00170465. doi:10.1007/BF00170465 [DOI] [PubMed] [Google Scholar]