Abstract
The continuous accumulation of sequence data, for example, due to novel wet-laboratory techniques such as pyrosequencing, coupled with the increasing popularity of multi-gene phylogenies and emerging multi-core processor architectures that face problems of cache congestion, poses new challenges with respect to the efficient computation of the phylogenetic maximum-likelihood (ML) function. Here, we propose two approaches that can significantly speed up likelihood computations that typically represent over 95 per cent of the computational effort conducted by current ML or Bayesian inference programs. Initially, we present a method and an appropriate data structure to efficiently compute the likelihood score on ‘gappy’ multi-gene alignments. By ‘gappy’ we denote sampling-induced gaps owing to missing sequences in individual genes (partitions), i.e. not real alignment gaps. A first proof-of-concept implementation in RAxML indicates that this approach can accelerate inferences on large and gappy alignments by approximately one order of magnitude. Moreover, we present insights and initial performance results on multi-core architectures obtained during the transition from an OpenMP-based to a Pthreads-based fine-grained parallelization of the ML function.
Keywords: phylogenetic inference, maximum likelihood, RAxML, multi-gene phylogenies, multi-core architectures, OpenMP
1. Introduction
The accumulation of molecular sequence data coupled with recent advances in computer architectures, in particular the availability of multi-core processors on every new desktop or laptop, poses new challenges for the efficient computation and parallelization of the broadly accepted and widely used phylogenetic maximum-likelihood (ML) function (Felsenstein 1981). Putting aside the important problem of multi-gene alignment assembly (Delsuc et al. 2005), the need for computationally more efficient likelihood calculations on huge, ‘gappy’, as well as extremely memory-intensive multi-gene alignments has become apparent. Current analyses of such datasets require months of inference time (McMahon & Sanderson 2006; Dunn 2008) or dedicated supercomputer architectures such as the IBM Blue Gene/L (Ott et al. 2007).
In this paper, we initially present a straightforward adaptation of the ML function to gappy multi-gene alignments, which has the potential to accelerate inferences on such large datasets by one order of magnitude. At the same time, though not yet implemented, this adaptation can reduce the memory footprint, which is mainly due to the space required for the partial likelihood arrays (also called likelihood vectors) and proportional to the gappyness of the respective alignment. With gappyness we refer to the percentage of gaps that have been inserted into such an alignment to represent missing per-gene sequences. It is important to note that this potential reduction in memory footprint will further accelerate the proposed method due to an increase in cache efficiency. For instance, the memory footprint of a large DNA multi-gene alignment of mammals (Bininda-Emonds 2007) containing approximately 2200 sequences with a length of approximately 50 000 bp and a gappyness exceeding 90 per cent has a memory footprint of approximately 9 GB under the standard GTR+Γ model. With our new approach, this can be reduced to approximately 1 GB or even significantly less if the GTR+CAT approximation of rate heterogeneity is used (Stamatakis 2006a). Thus, a full analysis of such a dataset including a sufficient number of bootstrap (Felsenstein 1985) and ML searches will not require supercomputers any more (Ott et al. 2007). In combination with the general parallelization scheme for ML on multi-core architectures using the low-level Pthreads programming library, which we present in the second part of this paper, problems of comparable size can be solved on medium-sized systems with 8–16 cores within reasonable times.
It is important to emphasize that the concepts presented in this paper are not RAxML specific. They can be applied to every Bayesian or ML-based phylogenetic inference algorithm, and partially also apply to the maximum parsimony criterion.
The remainder of this paper is organized as follows: in §2 we describe the data structures and method to efficiently compute the likelihood score on gappy multi-gene alignments. In §3 we discuss the lessons learned during the Pthreads-based parallelization of RAxML. In §4, we provide performance results for the new multi-gene likelihood function implementation and for the Pthreads-based parallelization of RAxML. We conclude our paper with a brief outline of current and future work.
2. Mapping ML to multi-gene alignments
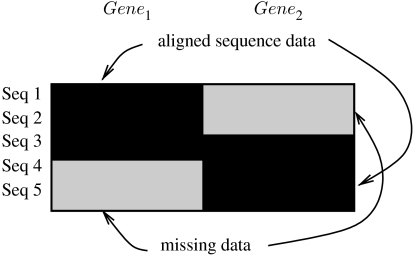
We will initially outline the basic idea by the example of a two-gene alignment with five sequences and a gappyness of 40 per cent as shown in figure 1. The black areas represent the sequences for which data are available, i.e. there are data for three sequences available from gene 1 (Sequence 1, Sequence 2, Sequence 3) and for three sequences from gene 2 (Sequence 3, Sequence 4, Sequence 5). The shaded grey regions represent the areas of gappyness, i.e. those parts of the multi-gene alignment that have been filled with gaps to account for the fact that there is simply no sequence data available for that particular taxon/gene pair. Since for the sake of the example, we assume that the two genes, gene 1 and gene 2, have the same length, and two out of five sequences are missing in each gene, this alignment has a gappyness of 40 per cent. Note that, the ‘true’ alignment gaps that lie in the black areas of figure 1 are not counted.
Figure 1.
Example of a gappy multi-gene alignment.
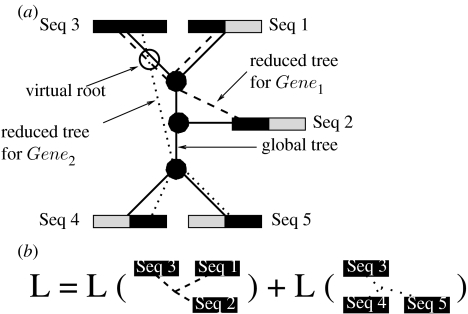
We can now consider an assignment of these gappy sequences to a fixed tree topology as outlined in figure 2a. The global tree topology that represents the relationships of all five sequences for both genes is represented as a black line. In order to compute the likelihood for this tree, one might, for instance, place a virtual root in the branch of the global tree that leads to Sequence 3. In order to appropriately adapt the computation of the likelihood function, one can now use distinct sets of branch lengths for every individual gene, as commonly done in partitioned models for such multi-gene analyses (e.g. McGuire et al. 2007). In order to account for the missing data, one can use a reduced set of branch lengths that only connects those sequences of genes 1 and 2 (as outlined by the dotted lines in figure 2a) for which sequence data are available. This means that for each gene or partition, we reduce the global tree topology T to a per-gene topology, T|Gene1 (read as T restricted to Gene1) and T|Gene2 by successively removing all the branches that lead to leaves for which there is no sequence data available for the respective genes or partitions. In our example, and as outlined in figure 2b, the likelihood of the tree can then simply be computed as . Note that this approach can significantly reduce the computational cost since we only have to compute the likelihood score for two three-taxon trees, instead of two five-taxon trees. In addition, this also reduces memory requirements for the inner likelihood vectors (also called conditional or partial likelihood arrays) since instead of—depending on the data structures used—at least three full-length likelihood arrays over the entire alignment length, we only require one such vector, since only one inner node is required for each gene tree. Essentially, this way of computing the likelihood on a global tree for gappy multi-gene alignments accounts for the fact that one does not need to conduct useless computations for data that are not present. It is important to note that, provided that per-partition optimization of individual branch lengths is carried out and disregarding slight numerical deviations, the procedure to compute the likelihood introduced here yields exactly the same likelihood scores as the standard method. This is due to the fact that missing data as well as gaps are modelled as undetermined characters in all current ML implementations (PHYML: Guindon & Gascuel 2003; IQPNNI: Minh et al. 2005; GARLI: Zwickl 2006), i.e. the probabilities for observing nucleotides A, C, G, T at the tips are all set to 1.0 (analogously for protein data).
Figure 2.
(a,b) Data structures and likelihood computation for gappy multi-gene alignments.
While the proposed concept is straightforward, the actual implementation is more complicated, especially with respect to an efficient mechanism to compute the topology reduction T|Gi for a specific gene Gi on the global topology T. In addition, tree searches, e.g. using the commonly used subtree pruning regrafting (SPR) technique for ML-based optimization of the tree, need to be appropriately adapted to determine on the fly which partial likelihood vectors for which genes need to be updated, i.e. those which are affected by a certain SPR move. In general, a specific SPR that is conducted within a subtree, which might for instance only contain sequences of one single gene, does not require any re-computation of likelihood values or update of likelihood vectors for the remaining genes. To this end, we have currently implemented only two basic operations on a fixed tree topology in a proof-of-concept implementation in RAxML (Stamatakis 2006b) that demonstrates the significant computational advantages of the method we propose: a full tree traversal to compute the likelihood score on a fixed tree topology with fixed per-gene branch lengths and a full branch length optimization on a fixed tree topology. In the following, we will outline the procedures to compute these two basic functions as well as the data structures used in more detail.
(a) A data structure for the multi-gene likelihood function
To describe the implementation of the multi-gene likelihood function method in RAxML, we will initially review the memory and data-structure organization for the single-gene case. The amount of memory space required by current ML implementations is largely dominated by the length and number of likelihood vectors. Thus, the memory requirements are of order O(n×m), where n is the number of sequences and m essentially the alignment length or, to be more precise, the number of distinct column patterns. An unrooted phylogenetic tree for an alignment of dimensions n×m has n tips or leaves and n−2 inner nodes, such that n−2 internal likelihood vectors of length m are required to compute the likelihood bottom up towards a given virtual root vr. Note that, the computation of the vectors at the tips of the tree (leaf vectors) is significantly less expensive and requires less memory than the computation of inner vectors (for details see Bader et al. 2006).
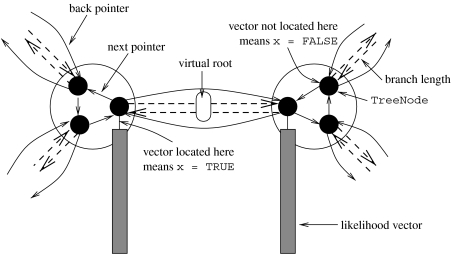
In RAxML (fastDNAml (Olsen et al. 2001) and GARLI (Zwickl 2006) use similar techniques) only one inner likelihood vector per internal node is allocated, as opposed to, e.g. IQPNNI (Minh et al. 2005), PAML (Yang 2007), or PHYML (Guindon & Gascuel 2003). This vector is relocated to one of the three outgoing branches of an internal node pointer-to-TreeNode next (see data structure below) of the inner node, which points towards the current virtual root. If the likelihood vector is already located at the correct branch, i.e. the value of x=TRUE, it must not be recomputed. The infrastructure to move likelihood vectors is implemented via a cyclic list of three data structures of type TreeNode (one per outgoing branch pointer-to-TreeNode back, see figure 3), which represents one internal node of the tree. At all times, two of the entries for x in the cyclic list representing an inner node are set to x≔FALSE; whereas the remaining one is set to x≔TRUE. The actual likelihood array data are then accessed via the node number. Finally, the vector z[NUM_BRANCHES] contains the branch length (branch lengths for analyses with per-partition branch length optimization) towards the node that is addressed via the respective back pointer. Note that, this type of data structure requires each branch to be stored twice, i.e. for two TreeNode data structures q and p for which q.back=p and p.back=q: q.z[0]=p.z[0].
Figure 3.
Likelihood vector organization.
From the perspective of the likelihood vectors that are oriented towards the virtual root, based on their location in the cyclic list of TreeNode, the tree is always rooted. In addition, at each movement of the virtual root, e.g. in order to optimize a branch, a certain amount of vectors must be recomputed. The same holds for changes in tree topology. However, there is a trade-off between additional computations and reduced memory consumption for inner likelihood vectors.
In the following, we will describe how the data-structure TreeNode can be extended to accommodate the global overall tree topology as well as the reduced per-partition tree topologies (see data structure below). We extend the data structure by an array of back pointers backs that point to the neighbouring nodes for the reduced trees of each individual partition. In general, the address of the back pointer of partition 0, for instance, might be located further away, i.e. backs[0]!=back. In addition, the array xs[NUM_BRANCHES] provides analogous information as x, but for each gene separately. If a certain inner node represented by a linked cyclic list of three TreeNode structures does not form part of a reduced tree for gene i T|Gi, all respective entries are set to NULL-POINTER and FALSE, respectively: backs[i]:=NULL-POINTER;, xs[i]:=TRUE;. If they do form part of the reduced tree, all three entries of backs[i] !=NULL-POINTER and one of the xs[i] must be set to TRUE.
(b) Computing a tree traversal
Given a starting tree, we initially place virtual roots for each gene/partition. If there exists one taxon for which there are sequence data available in every partition, the virtual roots are all placed in the branch that leads to this sequence. Otherwise, we assign virtual roots to the branches leading to the tip/leave node of the first taxon in the alignment that has data for a specific partition. Given the virtual root, we then conduct a full tree traversal to set up the backs[] data structures by starting at the branch between inner node reference and tip node p where we placed the virtual root for the respective partition. To set up the entire data structure, we loop over all partitions and call the function outlined below.
Once the above function has been executed for all partitions, the data structures are set up as outlined for the five-taxon case in figure 2a. The dotted lines in figure 2a indicate the reduced tree data structures given by the backs[] arrays, while the straight line represents the overall tree topology as provided by the single back pointers. Note that, in the current proof-of-concept implementation, we do not yet exploit the potential memory footprint reduction, which can be achieved by allocating memory space at inner nodes only for those partitions for which backs[partition]!=NULL-POINTER, i.e. the memory allocation scheme is essentially the same as outlined in figure 3. The implementation of an appropriately adapted data structure for inner likelihood vectors requires a significant amount of re-engineering.
Given the initial determination of the per-partition pointer meshwork, we can compute the overall likelihood score for a given tree and fixed branch lengths by summing over the per-gene log likelihood scores. The log likelihood for each individual partition is computed by conducting a full tree traversal based on the topology induced by the backs[] arrays, i.e. for example we use backs[0] to navigate to the internal nodes of the global tree topology that also form part of the topology for gene 0.
(c) Optimizing branch lengths
The general branch length optimization procedure in RAxML conducts several iterations (multiple optimization traversals) over all branches of the tree until a convergence criterion is reached. Each branch is individually optimized via a Newton–Raphson procedure. The adapted procedure for the multi-gene data structure works in an analogous way. The tree is traversed individually for each partition via the backs[] pointer structure. In addition, since branch-length optimization requires constant virtual re-rooting of the tree at each branch and re-computation of partial likelihood arrays due to constant changes in branch lengths, the xs[] vectors are updated accordingly to determine which partial likelihood arrays need to be re-computed. For each partition, only the corresponding portion of the likelihood array for that specific partition will be updated. As outlined by the results in §4a, the overhead induced by setting up and maintaining additional data structures as well as conducting multiple tree traversals, one for each partition, is insignificant compared with the computational advantages of the method.
3. Mapping ML to multi-core architectures
A large part of our recent work has focused on orchestrating and mapping the parallelism inherent to the phylogenetic ML function to a broad variety of emerging parallel architectures ranging from graphics processing units (GPUs; Charalambous et al. 2005), over the IBM CELL (Blagojevic et al. 2007a–c; Stamatakis et al. 2007) and typical single-core shared-memory machines (Stamatakis et al. 2005) up to the SGI ALTIX as well as IBM Blue Gene/L supercomputers (Ott et al. 2007, 2008).
Here we focus on software engineering aspects and programming paradigms for exploitation of fine-grained loop-level parallelism on emerging multi-core architectures such as the AMD Barcelona system. While OpenMP (http://www.openmp.org) provides a generic approach with low programming overhead to parallelize the ML function (Stamatakis et al. 2005), we have recently decided to replace it by the significantly less generic low-level POSIX Threads Programming (Pthreads) library (available as of RAxML v. 7.0.0, current release: 7.0.4). One main reason for this transition is that the usage of Pthreads allows for a more fine-grained and more complete control over the computer architecture, which we deem essential for multi-core architectures, in particular with respect to cache congestion (see Parkhurst et al. (2006) for on overview of challenges) and direct enforcement of memory locality in non-uniform memory access (NUMA) architectures. Moreover, the explicit implementation of memory locality and appropriate synchronization mechanisms facilitates the integration of the message passing interface (MPI)-based parallelization of the ML function for distributed and massively parallel machines (see Ott et al. 2007) into one single piece of software. Apart from increased efficiency and control due to usage of Pthreads, OpenMP exhibits some sources of potential non-determinism. For example, the parallel computation of the likelihood score, i.e. the sum over all per-column likelihood values, requires a global reduction operation. Given an alignment of length 1000 and four threads, t0, …, t3 the likelihood will be computed as follows: each thread ti will initially compute its local likelihood score l(ti) by summing over 250 per-column likelihoods. Then, the overall likelihood will be computed by summing over the partial sums, i.e. . However, OpenMP does not guarantee that the l(ti) will always be added in the same order, i.e. an exactly identical numerical operation with exactly equal results in the sequential version of the code can yield slightly distinct results for the final likelihood score (of the order of 10−5–10−6) among different invocations in the parallel OpenMP version due to small rounding errors induced by this non-determinism. Thus, when using OpenMP without enforcing a deterministic addition order for reduction operations, two successive tree traversals on exactly the same topology, set of branch lengths, and with the same virtual root, can yield slightly different scores. This phenomenon has in fact been observed with the OpenMP-based version of RAxML on a four-way AMD Opteron processor for an analysis of a long multi-gene alignment of mammals. Another important issue is that Pthreads allows for use of more sophisticated parallelization concepts than OpenMP which is based on the fork-join concept that is centred around the parallelization of for loops. For instance, the more flexible Pthreads approach allows synchronization points to be reduced in a similar way as outlined in Ott et al. (2007) by conducting the descent into a subtree and the update of several partial likelihood arrays simultaneously with one single synchronization at the end of such a partial tree traversal.
The main disadvantages of Pthreads are the significantly higher programming overhead (four weeks) and the missing implementation of efficient basic synchronization mechanisms such as a barrier construct, which have to be implemented using a busy-wait strategy.
To assess the effects of memory locality on NUMA architectures, we developed two versions of the Pthreads-based parallelization, one that uses a single global data structure for the likelihood arrays and tip data vectors, and the other that uses local data structures allocated by the individual threads on their local core or CPU.
One important lesson learned in terms of software engineering is that alignment columns and likelihood array data structures should not be distributed in single contiguous chunks to the threads, i.e. given an alignment with 1000 columns, t0 conducts computations on columns 0–499, and t1 on columns 500–999. Instead, a modulo distribution should be used, i.e. t0 computes columns 0, 2, 4, …, and t1 columns 1, 3, 5, …, etc. The rationale for this approach is that such a modulo-based distribution of columns will guarantee a better load balance in the case of partitioned analyses with per-partition branch length optimization, since every thread will be able to work on an approximately equally large portion of sites per partition. This is also important for concatenated analyses of DNA and amino acid alignments, since the computation of the likelihood score for a protein site is significantly more compute intensive than for a DNA site.
Finally, Pthreads-based applications are easier to compile under Linux/Unix than OpenMP-based programs, which require a dedicated compiler. Despite the fact that OpenMP has become a standard feature of the widely used GCC compiler since v. 4.2, this has led to a significantly higher use of the parallel RAxML version since the release of the Pthreads-based version in January 2008.
Moreover, initial tests with the OpenMP implementation in GCC show that, in contrast to commercially available OpenMP compilers, the fork-join thread synchronization mechanism is highly inefficient and thus GCC is currently not suited for the parallelization of the phylogenetic likelihood function. Because of the aforementioned inefficiency of the barrier mechanism we even observed parallel slowdowns with GCC in some cases.
4. Experimental setup and results
(a) Multi-gene alignment method
We tested our approach on three gappy real-world multi-gene DNA alignments with 59 sequences (d59_8, 6951 bp, 8 partitions), 404 sequences (d404_11, 13 158 bp, 11 partitions) and 2177 sequences (d2177_68, 51 089 bp, 68 partitions) with increasing degree of sampling-induced gappyness due to missing sequences in individual genes (partitions) of 28, 73 and 91 per cent, respectively.
We implemented the adapted method to compute the likelihood on such alignments under the GTR+Γ model for DNA data in RAxML. The current proof-of-concept implementation, including the test datasets, partition files and trees, is available at http://icwww.epfl.ch/∼stamatak/MULTI-GENE.tar.bz2 (use make -f Makefile.MULTIGENE, raxmlHPC -m GTRGAMMA -M -q partitionFile -s alignmentFile -t tree -n runID to execute). The program reads the input tree specified via -t and initially conducts 50 complete tree traversals (to average over the comparatively small traversal times on dataset d59_8) and a full branch length optimization under the standard method. Thereafter, the program will conduct 50 full tree traversals and a complete branch length optimization using the improved method presented here.
Computational experiments were conducted on an unloaded 2.4 GHz 8-way AMD Opteron processor of the CyberInfrastructure for Phylogenetic RESEARCH (CIPRES, www.phylo.org) project cluster located at the San Diego Supercomputer Center. As already mentioned, the log-likelihood scores after branch-length optimization returned by the standard and fast method were almost identical: d59_8 -53509.78614 (slow), -53509.78607 (fast); d404_11 -158659.88 (slow), -158659.85 (fast); d2177_68 -2379892.76 (slow) -2379894.17 (fast). The insignificant deviations in log-likelihood scores are due to the significantly lower amount of floating point operations (and thereby reduced numerical error propagation) conducted to compute the per-partition likelihood on the respective reduced gene trees that are partially up to 10 times smaller, in terms of number of nodes and branch lengths, than the global tree. Those small deviations are in the same order of magnitude as observed for branch length and model parameter optimizations on fixed tree topologies among different programs. Those deviations are, for example, due to differences in the way the tree is traversed to optimize branch lengths, the method deployed to optimize individual branch lengths as well as the convergence criterion or threshold. Finally, log likelihood scores also vary among binaries generated with different compilers from the same source code when typical high compiler-based optimization levels are used.
Table 1 provides the sequential execution times in seconds for the standard and fast tree traversals (columns S-Travers and F-Travers) as well as the standard and fast branch-length optimization procedures (columns S-Opt and F-Opt). Column gaps provides the percentage of sampling-induced gappyness, i.e. ‘real’ alignment gaps are not counted. As can be derived from table 1, the speedup for tree traversals amounts to 1.57 (d59_8), 4.58 (d404_68), 10.98 (d2177_68) and to 4.67 (d59_8), 33.69 (d404_68), 45.11 (d2177_68) for branch-length optimization.
Table 1.
Execution times on an AMD Opteron for standard and fast likelihood computations on gappy multi-gene alignments.
| dataset | S-Travers | F-Travers | S-Opt | F-Opt | gaps (%) |
|---|---|---|---|---|---|
| d59_8 | 1.87 | 1.19 | 13.32 | 2.85 | 28 |
| d404_11 | 37.04 | 8.08 | 303.18 | 9.00 | 73 |
| d2177_68 | 756.96 | 68.69 | 7483.53 | 165.87 | 91 |
(b) Multi-core platforms
In order to test scalability of the Pthreads-based parallelization of RAxML, we extracted a DNA dataset containing 50 taxa and 50 000 bp (loop length: 23 385 unique patterns) from the 2177 taxon 68 gene mammalian dataset mentioned in §4a. To measure the speedup on various multi-core architectures, we started RAxML tree searches under the GTR+Γ model on a fixed maximum parsimony starting tree. As test platforms we used a two-way quad-core AMD Barcelona system (eight cores), a two-way quad-core Intel Clovertown system (eight cores), and an eight-way dual core Sun x4600 system (16 cores) that is based on AMD processors. We measured execution times for sequential execution, 2, 4, 8 and 16 (applies only to x4600) cores. In addition, we only report the best speedup values for every number of cores used with respect to the thread to CPU assignment/mapping. Note that due to architectural issues, an execution on two cores that are located on a single socket can be much slower than an execution with two cores, located on two distinct sockets (see Ott et al. (2008) for a more detailed study of thread-to-core mapping effects on performance). For instance, we observed execution time differences of approximately 40 per cent on the Intel Clovertown system for different assignments of two threads to the eight cores of the system and over 50 per cent for distinct mappings of four threads.
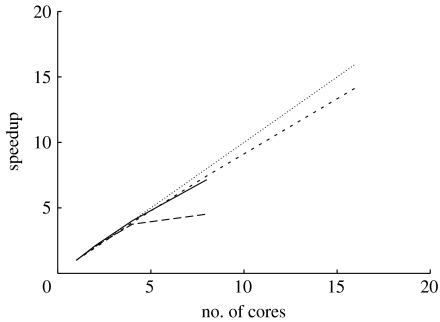
Figure 4 provides the speedup values on the three architectures for the respective optimal thread-to-core mapping on the three systems analysed. The graph clearly illustrates the performance differences induced by the distinct memory access architectures in the AMD (Barcelona, x4600) and Intel (Clovertown) systems. While both the Barcelona as well as the x4600 scale well up to the full number of cores due to the AMD HyperTransport protocol, the Intel system clearly suffers from the shared bus, which represents the major bottleneck for memory-intensive applications such as ML-based phylogenetic inference. Thus, the performance improvement by doubling the number of cores only amounts to 20 per cent. However, Intel is expected to release a NUMA-based memory access architecture in autumn 2008 (with the introduction of the QuickPath interconnect), which will help to solve this problem. Note that the above experiments have all been conducted with the Pthreads version that allocates one single global data structure for likelihood arrays and tip vectors. Our initial experiments with the Pthreads version that uses local memory assignment did not show any major performance improvement or degradation (execution times typically vary around ±1%) most probably due to a sufficiently accurate system-level mechanism that moves cache lines to the cores that conduct the first write operation on them.
Figure 4.
Speedup of Pthreads-based parallel RAxML version on different multi-core architectures. Solid line, Barcelona; long-dashed line, Clovertown; short-dashed line, x4600; linear speedup, dotted line.
Finally, in table 2 we provide execution times in seconds for Pthreads-based, OpenMP-based and sequential RAxML tree searches on DNA data under GTR+Γ on fixed starting trees. Experiments were conducted on a four-way SMP (four single cores) AMD Opteron processor. We used three real-world single-gene datasets of 150, 500 and 714 sequences, and one multi-gene dataset of 125 sequences with approximately 30 000 bp and approximately 20 000 distinct column patterns (the actual length of the compute-intensive for-loops). The table underlines the performance improvements achieved by using Pthreads instead of OpenMP, which range between 2 and 23 per cent. Note the significant super-linear speedup of 4.5 (owing to increased cache efficiency) for the Pthreads-based version on dataset 125. The remaining speedup values are sub-linear, because the computation to communication ratio is less favourable in the single-gene case, i.e. loop lengths are significantly shorter. In particular on long multi-gene datasets, super-linear speedups due to increased cache efficiency can be frequently observed on a broad variety of distributed as well as shared memory systems. A recent paper (Stamatakis & Ott 2008) comparing MPI, Pthreads and OpenMP to exploit loop-level parallelism in the phylogenetic ML function provides a more detailed performance study.
Table 2.
Execution times in seconds on a four-way AMD Opteron for the Pthreads, OpenMP and sequential RAxML versions.
| dataset | Pthreads | OpenMP | sequential | loop length |
|---|---|---|---|---|
| d125 | 3682.30 | 4722.05 | 16 604.25 | 19 436 |
| d150 | 214.25 | 216.22 | 782.47 | 1130 |
| d500 | 1282.22 | 1393.85 | 4749.67 | 1193 |
| d714 | 1675.64 | 1694.64 | 6207.23 | 1231 |
5. Conclusions and future work
We have presented an efficient data structure and a proof-of-concept implementation that has the potential to accelerate the computation of the ML function on large and gappy multi-gene alignments by approximately one order of magnitude. In addition, we have addressed technical issues and lessons learned regarding the exploitation of loop-level parallelism in the phylogenetic ML function on multi-core architectures. We have also conducted an initial parallel performance study on current multi-core architectures.
Future work will cover the development of appropriate rules for conducting SPR-based tree searches on gappy multi-gene alignments, based on the data structures introduced here.
Acknowledgments
We are grateful to Nicolas Salamin, Olaf Bininda-Emonds, Markus Göker and Nikos Poulakakis for providing us their alignments for performance tests. The Exelixis Lab (A.S.) is funded under the auspices of the Emmy–Noether programme by the German Science Foundation (DFG).
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Statistical and computational challenges in molecular phylogenetics and evolution’.
References
- Bader D, Roshan U, Stamatakis A. Computational grand challenges in assembling the tree of life: problems & solutions. Elsevier; Amsterdam, The Netherlands: 2006. Advances in computers. [Google Scholar]
- Bininda-Emonds O, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. doi:10.1038/nature05634 [DOI] [PubMed] [Google Scholar]
- Blagojevic, F., Nikolopoulos, D. S., Stamatakis, A. & Antonopoulos, C. D. 2007a Dynamic multigrain parallelization on the cell broadband engine. In Proc. 12th ACM SIGPLAN Symp. on Principles and Practice of Parallel Programming, San Jose, CA, March 2007, pp. 90–100. New York, NY: Association for Computing Machinery.
- Blagojevic F, Nikolopoulos D.S, Stamatakis A, Antonopoulos C.D, Curtis-Maury M. Runtime scheduling of dynamic parallelism on accelerator-based multi-core systems. Parallel Comput. 2007b;33:700–719. [Google Scholar]
- Blagojevic, F., Stamatakis, A., Antonopoulos, C. D. & Nikolopoulos, D. S. 2007cRAxML-Cell: parallel phylogenetic tree inference on the cell broadband engine. In Proc. 21st IEEE Int. Parallel and Distributed Processing Symposium, Long Beach, CA, March 2007 Los Alamitos, CA: IEEE.
- Charalambous, M., Trancoso, P. & Stamatakis, A. 2005 Initial experiences porting a bioinformatics application to a graphics processor Springer Lecture Notes in Computer Science, no. 3746, pp. 415–425. Berlin, Germany: Springer.
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005;6:361–375. doi: 10.1038/nrg1603. doi:10.1038/nrg1603 [DOI] [PubMed] [Google Scholar]
- Dunn C.W, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;10:745–749. doi: 10.1038/nature06614. doi:10.1038/nature06614 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. doi:10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. doi:10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- McGuire J.A, Witt C.C, Altshuler D.L, Remsen J.V., Jr Phylogenetic systematics and biogeography of hummingbirds: Bayesian and maximum likelihood analyses of partitioned data and selection of an appropriate partitioning strategy. Syst. Biol. 2007;56:837–856. doi: 10.1080/10635150701656360. doi:10.1080/10635150701656360 [DOI] [PubMed] [Google Scholar]
- McMahon M.M, Sanderson M.J. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst. Biol. 2006;55:818–836. doi: 10.1080/10635150600999150. doi:10.1080/10635150600999150 [DOI] [PubMed] [Google Scholar]
- Minh B.Q, Vinh L.S, von Haeseler A, Schmidt H.A. pIQPNNI: parallel reconstruction of large maximum likelihood phylogenies. Bioinformatics. 2005;21:3794–3796. doi: 10.1093/bioinformatics/bti594. doi:10.1093/bioinformatics/bti594 [DOI] [PubMed] [Google Scholar]
- Olsen G, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Bioinformatics. 2001;10:41–48. doi: 10.1093/bioinformatics/10.1.41. doi:10.1093/bioinformatics/10.1.41 [DOI] [PubMed] [Google Scholar]
- Ott, M., Zola, J., Aluru, S. & Stamatakis, A. 2007 Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. In Proc. IEEE/ACM Supercomputing Conf. 2007 (http://sc07.supercomputing.org/schedule/pdf/pap271.pdf)
- Ott, M., Klug, T., Weidendorfer, J. & Trinitis, C. 2008 autopin—automated optimization of thread-to-core pinning on multicore Systems. In Proc. 1st Workshop on Programmability Issues for Multi-Core Computers (MULTIPROG), Göteborg, Sweden, January 2008
- Ott M, Zola J, Aluru S, Johnson A.D, Janies D, Stamatakis A. Large-scale phylogenetic analysis on current HPC architectures. Sci. Program. 2008;16:255–270. [Google Scholar]
- Parkhurst, J., Darringer, J. & Grundmann, B. 2006 From single core to multi-core: preparing for a new exponential. In Proc. 2006 IEEE/ACM Int. Conf. on Computer-Aided Design, San Jose, CA, November 2006, pp. 67–72. Los Alamitos, CA: IEEE.
- Stamatakis, A. 2006a Phylogenetic models of rate heterogeneity: a high performance computing perspective. In Proc. 20th IEEE/ACM Int. Parallel and Distributed Processing Symposium (IPDPS2006), Rhodos, Greece, April 2006 Los Alamitos, CA: IEEE.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006b;22:2688–2690. doi: 10.1093/bioinformatics/btl446. doi:10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. & Ott, M. 2008 Exploiting fine-grained parallelism in the phylogenetic likelihood function with MPI, Pthreads, and OpenMP: a performance study. In Proc. Third IAPR Int. Conf. on Pattern Recognition in Bioinformatics (PRIB 2008). Springer Lecture Notes in Bioinformatics, no. 5265, pp. 424–436. Berlin, Germany: Springer.
- Stamatakis, A., Ott, M. & Ludwig, T. 2005 RAxML-OMP: an efficient program for phylogenetic inference on SMPs Springer Lecture Notes in Computer Science, no. 3606, pp. 288–302. Berlin, Germany: Springer.
- Stamatakis A, Blagojevic F, Antonopoulos C.D, Nikolopoulos D.S. Exploring new search algorithms and hardware for phylogenetics: RAxML meets the IBM Cell. J. VLSI Signal Process. Syst. 2007;48:271–286. doi:10.1007/s11265-007-0067-4 [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586. doi: 10.1093/molbev/msm088. doi:10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zwickl, D. 2006 Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas at Austin.