Abstract
Models of molecular evolution tend to be overly simplistic caricatures of biology that are prone to assigning high probabilities to biologically implausible DNA or protein sequences. Here, we explore how to construct time-reversible evolutionary models that yield stationary distributions of sequences that match given target distributions. By adopting comparatively realistic target distributions, evolutionary models can be improved. Instead of focusing on estimating parameters, we concentrate on the population genetic implications of these models. Specifically, we obtain estimates of the product of effective population size and relative fitness difference of alleles. The approach is illustrated with two applications to protein-coding DNA. In the first, a codon-based evolutionary model yields a stationary distribution of sequences, which, when the sequences are translated, matches a variable-length Markov model trained on human proteins. In the second, we introduce an insertion–deletion model that describes selectively neutral evolutionary changes to DNA. We then show how to modify the neutral model so that its stationary distribution at the amino acid level can match a profile hidden Markov model, such as the one associated with the Pfam database.
Keywords: variable-length Markov model, profile hidden Markov model, insertion–deletion model, scaled selection coefficient, fitness, Pfam
1. Introduction
The broad biological theme that has benefitted from the most experimental interrogation is likely to be the relationship between genotype and phenotype. This relationship is not solely a focus of experimental biology. Diverse computational schemes have been developed for leveraging experimental results by predicting aspects of phenotype from genotype. These in silico prediction systems can be harnessed to quantify the influence of phenotype on the evolution of genotype (Parisi & Echave 2001; Robinson et al. 2003; Rodrigue et al. 2005; Yu & Thorne 2006; Choi et al. 2007).
Most phenotypic features cannot yet be accurately predicted via computational approaches. This greatly hampers efforts to assess the evolutionary impact of phenotype. When a genotype–phenotype link has not been established, it is still desirable to determine what patterns of evolution could yield observed patterns of sequence conservation. Implicit information about genotype–fitness mapping is often available and would ideally be captured in evolutionary models. For example, it is widely appreciated that conserved sequence motifs are likely to be functionally important. Evolutionary models could be improved by assigning high stationary probabilities to the sequences that most consistently match empirical evidence.
Many models of sequence change have been proposed and gainfully employed, but few have been carefully parametrized to assign higher probabilities to the most biologically plausible sequences. A recent and notable exception (Lartillot & Philippe 2004) has learned from data to account for the variation of preferred amino acid residues among protein sequence positions. By contrast, many widely used models of nucleotide substitution or amino acid replacement allow nucleotide or amino acid types to have different probabilities, but force these probabilities to be shared among sequence positions. As a result, the most likely sequences will typically be homopolymers that consist of solely the nucleotide or amino acid type with the highest probability.
Here, we follow Lartillot and Philippe by considering evolutionary models that assign higher probabilities to the sequences that are likely to be more biologically plausible. However, we do not assume that sequence positions or codons change independently. Although the assumption of independence has strong computational advantages, it is less attractive from the standpoint of studying natural selection. With independence, local maxima on the fitness landscape are also global maxima. A better understanding of how the evolutionary process traverses the fitness landscape might be achieved with the more realistic fitness landscapes that are possible with dependence.
A strategy pioneered by Jensen and Pedersen (Jensen & Pedersen 2000; Pedersen & Jensen 2001) exists for performing likelihood-based evolutionary inference when dependence among changes at different positions makes conventional approaches intractable. The Jensen–Pedersen and other inference techniques have been applied when dependence stems from context-dependent mutation (Hwang & Green 2004; Siepel & Haussler 2004; Christensen et al. 2005) and natural selection (Robinson et al. 2003; Rodrigue et al. 2005; Yu & Thorne 2006). Here, we focus on dependence due to natural selection but not on parameter inference. Instead, we consider ways to construct time-reversible evolutionary models so that the induced stationary distribution matches a desired target distribution. A desirable target distribution might assign the highest probabilities to those sequences predicted to be the most biologically plausible.
We describe two alternative strategies for constructing evolutionary models with a desired stationary distribution of sequences. The parametric form of the first strategy has been employed in earlier studies to assess the evolutionary impact of phenotype on sequence change (Robinson et al. 2003; Rodrigue et al. 2005; Yu & Thorne 2006), but here we stress how this strategy can match desired stationary distributions. Although both strategies can be interpreted with respect to population genetics, the connection to population genetics is more direct in the second one.
We illustrate our modelling strategies with two applications. In the first application, sequence lengths are invariant because insertions and deletions are not permitted. Rates of codon substitution are parametrized to produce an evolutionary model with a stationary distribution of amino acid sequences that matches the distribution specified by a variable-length Markov model (VLMM) trained on human protein sequences. As a consequence, the rate of a particular codon substitution at a particular sequence position will be affected by the amino acids specified at codons that are nearby in the sequence. In the second application, the stationary distribution of amino acid sequences follows a profile hidden Markov model (HMM) trained on a protein family of interest (see Durbin et al. 1998). This is appealing because variation in sequence lengths is possible with a profile HMM. Therefore, the evolutionary model permits insertions and deletions as well as codon substitutions.
We contrast the VLMM and profile HMM evolutionary models to those that would result if all sequence changes were neutral. The contrasts permit evolutionary rates in the VLMM and profile HMM models to be decomposed into the product of factors that represent the mutation rate and the probability that the mutation is fixed. Following the pioneering approach of Halpern & Bruno (1998), this decomposition results in a crude estimate of the product of effective population size and the difference in relative fitnesses of the two sequences involved in a change.
(a) Neutral model for codon substitution
Differences between a target distribution and a stationary distribution for a neutral model of sequence change can be attributed to natural selection. We first describe our neutral model of codon substitution. In subsequent sections, we explain how its comparison with other evolutionary models can help quantify natural selection.
The Hasegawa–Kishino–Yano (HKY) substitution model has sequence positions evolved independently and identically with the rate to a nucleotide type h () being proportional to πh (, ) for transversions and κπh for transitions (Hasegawa et al. 1985). The stationary distribution of a DNA sequence i of length L for the HKY model is
| (1.1) |
where ik is the nucleotide at position k of sequence i and π collectively represents the parameters πA, πC, πG and πT. If the HKY model is coupled to the assumption that all mutations are selectively neutral, then the mutation rate must be proportional to κπh for transitions and πh for transversions. This perspective of the HKY model can easily be converted to a model for protein-coding DNA evolution where all point mutations that introduce stop codons are lethal and all other point mutations are selectively neutral. This resulting codon-based model resembles those of Goldman & Yang (1994) and Muse & Gaut (1994), but it does not differentiate between synonymous and non-synonymous substitutions. The stationary probability of a protein-coding DNA sequence i with L(i) codons (and 3L(i) nucleotides) for this simple model is
| (1.2) |
where ik1, ik2 and ik3, respectively, refer to the nucleotides at the first, second and third positions of the kth codon. The (1/Y)L(i) term arises because sequences containing premature stop codons are assumed not to survive. For the universal genetic code, the stop codons are TAA, TAG and TGA, and this means that .
At the protein level, the stationary probability of amino acid sequence I for this neutral model will be the sum of over all DNA sequences i that yield I when translated. Throughout, lowercase letters denote the nucleotide level and uppercase letters denote the corresponding amino acid information. The notation T(i) represents the amino acids that result from translating the DNA sequence i. Henceforth, we will use ik to refer to the kth codon of DNA sequence i rather than to the kth position of i. With this notation,
| (1.3) |
(b) Indirect matching of target and stationary distributions
In this section, we illustrate how a previously developed strategy for constructing evolutionary models can be employed to match a stationary distribution to a desired target distribution. We (Robinson et al. 2003; Yu & Thorne 2006) have been investigating the models of DNA sequence change that have evolutionary rates
| (1.4) |
when sequences i and j differ at exactly one site, where j has nucleotide type h. Rates of other changes are set to 0. The values of E(i) and E(j) can represent scores of phenotypes encoded by i and j, whereas the parameter f can convert the phenotypic effects induced by a change from i to j into an effect on evolutionary rate. These models are time reversible and yield a stationary distribution for a protein-coding DNA sequence i that we write where
| (1.5) |
Because the models that we have been investigating do not permit insertions and deletions, the sum in the denominator is over all sequences k with the same length as i. The stationary distribution for the neutral case of equation (1.2) results when fE(k) is identical among all DNA sequences k.
To represent the probability of amino acid sequence I for the desired target distribution, we write P(I). The stationary distribution of equation (1.5) matches P(I) when
| (1.6) |
By incorporating equation (1.6) and treating synonymous changes as selectively neutral, equation (1.4) can be rewritten as
where
| (1.7) |
Let the relative fitness of allele (sequence) i be wi. When convenient, we use allele i as a reference allele and set wi=1 so that the relative fitness of any other allele k can be written wk=1+sk. We assume multiplicative fitnesses so that the fitness of a genotype is the product of the fitnesses of the alleles that constitute it. Following Halpern & Bruno (1998) and others (Nielsen & Yang 2003; Berg et al. 2004; Knudsen & Miyamoto 2005; Sella & Hirsh 2005), we showed for a low mutation rate and a constant effective population size of diploid organisms that an approximation of is (Thorne et al. 2007). This means that the departure between the target distribution and the neutral model can be used to assess natural selection,
| (1.8) |
A weakness of this approximation is that it is justified only when 2Nsj is relatively close to 0. In §1c, we describe a modelling strategy with a closer connection to population genetics, but that is more difficult to employ for evolutionary inference. Interestingly, the strategies presented in both this and the next sections have the stationary distribution form of equation (1.5) and the 2Nsj approximation of equation (1.8).
(c) More direct matching of target and stationary distributions
In §1b, existing modelling strategies were retrofitted to population genetic interpretations. To more directly relate models of sequence change and population genetics, we modify our neutral model of codon substitution to reflect the insights of Halpern & Bruno (1998) that an interspecific rate should be proportional to the product of mutation rate and fixation probability. We have
| (1.9) |
where Zij is the event that a new mutant allele j eventually gets fixed in a population that otherwise consists of 2N−1 alleles of type i. The fixation probability approximation of Sella & Hirsh (2005),
| (1.10) |
then yields a stationary distribution (Thorne et al. 2007)
| (1.11) |
We can force to match our target distribution P(J) for all J by choosing suitable selection coefficients, as described by the following equation:
| (1.12) |
If the selection coefficients are chosen in this way, the fixation probability becomes
| (1.13) |
Substituting this formula into equation (1.9), the resulting evolutionary rates depend on N as well as the target distribution P(J). This dependence could complicate inference, but can be neglected when is well approximated by 2Nsj. With this approximation, we again have the result of equation (1.8) that , and we reproduce in different notation the Halpern & Bruno (1998) approximation (see also Yang & Nielsen 2008) that .
2. Variable-length Markov models
In this section, we use equation (1.8) to obtain 2Nsj estimates from an evolutionary model with a stationary distribution of sequences that matches a VLMM trained on human proteins. Let L(i) and L(I), respectively, represent the number of codons in protein-coding sequence i and the number of amino acids in protein sequence I. The kth amino acid will be denoted Ik whereas the subsequence consisting of the first k residues will be Ik. Taking liberty with conventional probabilistic notation by not distinguishing between random variables and their values, a zeroth-order discrete-state discrete-time Markov model for protein sequence organization has , whereas a Markov model of order r≥1 has . Well-developed statistical techniques are available for the analysis of data generated according to discrete-state discrete-time Markov chains of fixed order (e.g. see Guttorp 1995).
The VLMMs offer a parametrization advantage over models of fixed order, and their value for association mapping has been demonstrated (Browning 2006). Consider the transition probability that is associated with a Markov model of fixed order r and with a subsequence matching . If this subsequence is rare in the training data, estimates of are apt to be unreliable. In such a situation, one may instead desire to find the biggest integer l satisfying l<r for which can be well estimated for all Ik. Likewise, if subsequences matching are abundant in the training data, then one may want to find the largest integer l>r for which can be well estimated for all Ik. The key insights of VLMMs are to realize that and that can be approximated by a transition probability , where the value of l will vary among possible subsequences that immediately precede Ik. Choosing a value of l for each subsequence is a somewhat subjective endeavour but a sensible software implementation for training VLMMs from protein sequence data is available (Bejerano & Yona 2001; Bejerano 2004). Our goal here is not to provide alternative techniques for training VLMMs from protein sequence data. Instead, we are motivated by the potentially realistic descriptions of protein sequences by trained VLMMs. We focus on constructing evolutionary models of protein-coding DNA so that they yield a stationary distribution of protein sequences that matches a trained VLMM. We can then apply equation (1.8) to approximate 2Nsj for a change from sequence i to j.
We downloaded all annotated human protein sequences from NCBI human genome build 36.1 (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/). We used these sequences to train a VLMM with software that incorporates probabilistic suffix trees (Bejerano & Yona 2001; see also Bejerano 2004). Using the notation employed by this software, we elected to train with the settings of L=9, Pmin=0.00005, r=1.5, γmin=0.0000001 and α=0.
We also needed estimates of the nucleotide composition parameters π. If DNA experiences solely neutral evolution according to the HKY model, then stationary distributions of sequences obey equation (1.1) and π can be estimated by the proportions of nucleotide types found in the DNA. The vast majority of the human genome consists of DNA with no known biological function (International Human Genome Sequencing Consortium 2001) and, for the sake of this analysis, we assume that nucleotide frequencies in the human genome can produce reasonable estimates of π. Approximately 41 per cent of the genome consists of GC base pairs and the remaining 59 per cent consists of AT base pairs (International Human Genome Sequencing Consortium 2001). We therefore set π to and . Treatment of π could be improved by allowing values to vary among genes so as to incorporate regional and strand differences in mutation patterns, but this is not pursued here.
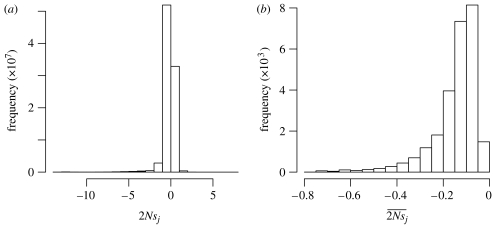
By combining the VLMM trained from human data with these π estimates, we can calculate τIJ of equation (1.7) because the VLMM specifies P(I) and P(J) and the π values determine the neutral model probabilities and . With the resulting value for τIJ, we can apply equation (1.8) to estimate the value of 2Nsj for a non-synonymous change from a sequence i that is in the human genome to a possible sequence j. From the NCBI human genome build 36.1, we downloaded 25 925 human messenger RNA sequences. For each possible non-synonymous change to these sequences, we estimated 2Nsj (figure 1a). The mean and standard deviation of these 2Nsj estimates are −0.211 and 0.796, respectively. Among the 89 486 730 possible non-synonymous mutations, 33 317 376 (37.2%) yielded a positive 2Nsj value. We also found the average 2Nsj estimate for each of the 25 925 protein-coding genes and investigated how these vary (figure 1b). The mean and standard deviation of these average 2Nsj values are −0.148 and 0.102, respectively, and none of the averages were positive.
Figure 1.
Estimates of 2Nsj from the VLMM trained by human protein sequences. (a) Distribution of 2Nsj estimates among possible non-synonymous changes in the human genome. (b) Distribution of the mean 2Nsj estimate per gene among human genes.
3. Profile hidden Markov models
Evolutionary inference with proper handling of insertion and deletion events can be challenging, but progress is being made (Fleissner et al. 2005; Lunter et al. 2005; Redelings & Suchard 2005, 2007). In this section, we first describe a model with codon substitutions and insertions and deletions that represent a neutral process of sequence change. We then explain how the neutral process can be modified to yield a stationary distribution of protein sequences that matches the probability distribution specified by a profile HMM. As with the VLMM case, we interpret departures from the neutral stationary distribution as being attributable to natural selection.
(a) Neutral model for insertions and deletions
To represent neutral sequence changes due to insertions and deletions, we consider a modification of the TKF92 insertion–deletion model (Thorne et al. 1992). This model was not originally framed at the codon level, but we present it at the codon level here. We make the convenient but restrictive assumption that insertions and deletions can only insert or delete entire codons and that insertions can occur only between codon boundaries. This prevents stop codons and frameshift mutations. More general and realistic insertion–deletion processes could also be considered but we reserve this for the future.
The TKF92 model has the insertion–deletion process operating independently of the substitution process. It describes the birth and death of entities termed ‘links’ and considers a type of link termed ‘immortal’ and another termed ‘normal’. Each normal link is associated with one or more consecutive codons. The normal link and its associated codons are referred to as a fragment. The death rate per normal link is μ. When a normal link dies, the entire fragment with which it is associated is deleted from the sequence. At the extreme 5′-end of each DNA sequence is the immortal link. The immortal link is not associated with any codons and is not subject to death. Both kinds of links can become parents of normal links and each experiences births at rate λ. A newborn link and its associated fragment are assumed to be inserted directly to the right (i.e. 3′-end) of the parental fragment. Conditional upon the length of a newly inserted subsequence, the inserted residues are sampled from the stationary distribution of the substitution process (equation (1.2)).
With this model, the stationary distribution of the number of links in a sequence is geometric (Thorne et al. 1991) and the stationary distribution for the number of codons per sequence depends on the probability distribution of the number of codons per fragment. It is computationally convenient to have the number of codons per fragment to be geometrically distributed so that the probability that a fragment has k codons is (1−r)rk−1, where k is greater than or equal to 1. The resulting stationary distribution for the number of codons in a sequence i is (Thorne et al. 1992)
| (3.1) |
A flaw of the TKF92 model is that fragment boundaries cannot change over time. If two consecutive codons are inserted together, then neither can later be deleted unless both are deleted. Here, we consider a variant that rectifies the flaw of unchanging fragment boundaries. Our variant differs from the TKF92 model because it does not consider fixed fragment boundaries. A more general improvement upon the TKF92 model has been outlined by Miklós et al. (2004).
Let 0(i, c, q) be the rate at which sequence i experiences a deletion that begins at the cth codon and removes from i the subsequence q that has a total of L(q) codons (i.e. the deletion eliminates the codons in positions ). Obviously, 0(i, c, q)=0 if or if the subsequence beginning at codon c and having length L(q) codons does not actually match subsequence q. The TKF92 deletion rate corresponding to 0(i, c, q) is 0 unless there is a fragment that begins at codon c and ends at codon . When fragment boundaries permit a particular deletion, the TKF92 model has the rate of that deletion as μ.
Let 0(i, c, q) be the rate at which i experiences an insertion between the codons at positions c and c+1 (if 0≤c<L(i)) or, in the case of c=L(i), that begins at the 3′-end of the codon at position L(i). The TKF92 insertion rate corresponding to 0(i, c, q) is 0 unless the fragment boundaries of i are such that an insertion is possible. If an insertion is possible, the TKF92 model has the insertion rate .
Owing to the multiple ways by which sequences can be fragmented, different sequences of the same length that all evolve according to the TKF92 model may experience different overall insertion and deletion rates and also different insertion and deletion rates at individual sequence locations. Our variant is much the same as the TKF92 model, except that all sequences of a particular length experience the same insertion and deletion rates as each other. Specifically, the insertion rates 0(i, c, q) and deletion rates 0(i, c, q) for this variant model are obtained by averaging the TKF92 rates over all possible fragmentations of a sequence of length L(i). We note that the probability of observing a fragment boundary between any two codons is . Some algebra shows that the rates are
| (3.2) |
and, assuming that a subsequence q begins at codon position c in sequence i,
| (3.3) |
These rates specify a time-reversible insertion–deletion model with a stationary distribution of sequence lengths that is identical to equation (3.1). A nice feature of the TKF92 model is the availability of explicit transition probabilities for transforming one sequence into another. Our modification does not share these transition probabilities, but our purposes here do not require explicit transition probabilities.
(b) The profile HMM rate matrix
If codon substitutions and insertion and deletion events are exclusively neutral, then the stationary probability of i would be . We want to consider departures due to natural selection from these neutral probabilities. For a change from sequence i to j, the rate Ri,j with natural selection will again be assumed to be multiplied by the neutral rate. This means that the rates of point mutations are given by equation (1.4), and the insertion and deletion rates are, respectively,
| (3.4) |
and
| (3.5) |
With insertions and deletions, sequence lengths are not fixed and equation (1.5) needs to be modified,
| (3.6) |
where the sum is now over all possible sequences of all possible lengths.
In order to fit our model to a profile HMM, we match equation (3.6) to the probability PHMM(I) of amino acid sequence I according to a profile HMM. To do this, we set
| (3.7) |
If we again assume that the relative fitnesses of i and j are 1 and 1+sj, we obtain the result of equation (1.8) rewritten to emphasize the relevance of the lengths of I and J,
| (3.8) |
In the above equation, the terms and depend on λ and μ. Instead of estimating λ and μ, we reason that protein sequences tend to be long relative to insertion and deletion lengths. This means the ratio of and should be close to 1 if the lengths of I and J are not too different. The approximation to 2Nsj then becomes
| (3.9) |
Although the insertion and deletion rates in equations (3.4) and (3.5) parallel the substitution rate form of equation (1.4), an alternative form of the insertion and deletion rates parallels the substitution rates defined by equation (1.9) and the fixation probability of equation (1.13). This alternative also yields the stationary distribution of equation (3.6) and the 2Nsj estimate of equation (3.9).
(c) Profile HMM example
We used a profile HMM from the Pfam database (Sonnhammer et al. 1997) that was trained from members of the tumour protein p53 family. We obtained a protein-coding DNA sequence from humans (GenBank accession NM_000546) that belongs to this family. By applying equation (3.9) with the nucleotide frequencies used for the VLMM analyses (i.e. πA=πT=0.295, πC=πG=0.205), we estimated 2Nsj value for possible mutations to the human p53 gene.
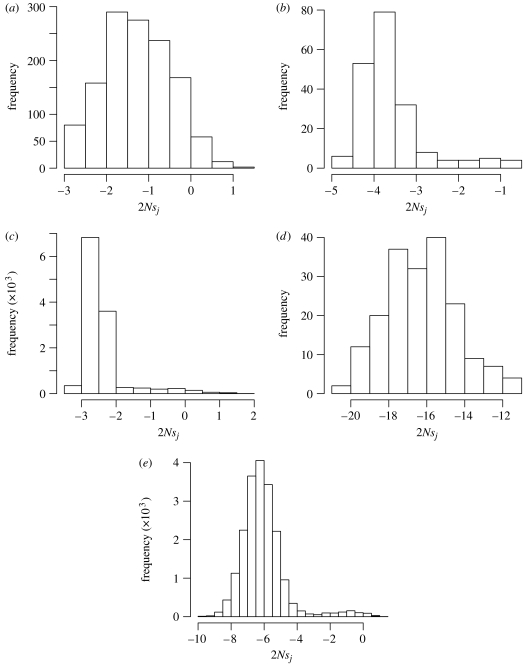
The mean and standard deviation of the 2Nsj for the 1280 possible non-synonymous point mutations were, respectively, −1.27 and 0.79 with only 72 (approx. 5.6%) yielding a positive estimate of 2Nsj. For the 195 possible single-codon deletions to this human p53-coding sequence, the mean and standard deviation of the 2Nsj estimates were −3.60 and 0.78, respectively, and none of the estimates were positive. For the (195+1)×61=11 956 possible single-codon insertions that could affect the p53 gene according to our insertion–deletion process, the mean 2Nsj estimate was −2.43 with a standard deviation of 0.68. There were 247 single-codon insertions (approx. 2.1%) with a positive 2Nsj value. We also examined the 186 possible deletions of 10 consecutive codons that could affect the human p53 sequence. The mean and standard deviation for these deletions were −16.30 and 1.90, respectively, with none exceeding 0. Finally, we randomly generated 100 insertions of 10 consecutive codons for each of the 196 possible insertion locations. The inserted subsequences were simulated in a simplistic fashion. For each of the 30 positions in the 10-codon insertions, the four nucleotides were equally likely to occupy the position except that subsequences containing a stop codon were discarded. The mean and standard deviation for the 19 600 insertions of 10 codons were, respectively, −6.074 and 1.37 with 121 (approx. 0.6%) exceeding 0. Histograms representing the 2Nsj estimates for the human p53 sequence are shown in figure 2.
Figure 2.
Estimates of 2Nsj for possible mutations to the human p53 gene. (a) Non-synonymous mutations, (b) single-codon deletions, (c) single-codon insertions, (d) deletions of 10 consecutive codons and (e) insertions of 10 consecutive codons.
4. Discussion
Our 2Nsj estimates are undeniably flawed. For example, some non-synonymous changes are lethal or extremely deleterious and should yield 2Nsj estimates that are much farther below 0 than any estimates that we obtained. This shortcoming is a reflection of the inadequacies of the VLMM and profile HMM target distributions and our assumption that these probability distributions fully reflect the relationship between sequence and fitness. For the profile HMM, improved handling of phylogenetic correlations among sequences in the training data might yield improved target distributions. The emission and transition probabilities of the profile HMMs can be viewed as being inferred in a Bayesian framework (e.g. see Durbin et al. 1998). Our 2Nsj estimates are apt to be sensitive to the prior distributions that are employed in training the Pfam models.
Our current approach also has other departures from rigorous statistical technique. For example, we treated the VLMM and profile HMM target distributions as if they were known rather than estimated, thus ignoring uncertainty in the estimates. Also, the sequences that yielded the 2Nsj estimates were not independent of those from which the target distributions were inferred.
Weaknesses of our population genetic interpretations include the assumption of low mutation rate and constant population size. These limitations need to be addressed, but we believe making them explicit is a good start. If population size varies during evolution, then the balance between mutation and selection will also vary. A consequence would be non-homogeneity and loss of stationarity in the process of sequence change. Although there are exceptions (e.g. Galtier & Gouy 1998; Blanquart & Lartillot 2006, 2008), the models of sequence change that are widely used in phylogenetics assume stationarity. These models can be viewed as implicitly relying upon the assumption that effective population sizes are constant.
An advantage of explicitly connecting population genetics to interspecific evolutionary models is that biological criteria can be more easily brought to bear on model selection. The general time-reversible model of nucleotide substitution (Tavaré 1986) has the four nucleotide types as possible states and many special cases exist. A general time-reversible model that uses each possible DNA sequence as a possible state has many more special cases. Even when the stationary distribution of such a model is constrained to match a desired target distribution, the number of ways to design the model could be enormous. Employing solely statistical criteria for selecting among the possible modelling strategies would be daunting and potentially ill-advised. A superior approach might favour the modelling strategies with clear population genetic interpretations.
A shortcoming of most models of sequence evolution is that they can inappropriately assign much probability to selectively deleterious sequences. Computational biologists have devised a variety of useful probabilistic descriptions of molecular sequences for the purpose of organizing and classifying molecular sequence data. They have had substantial motivation to create descriptions that assign high probability to the sequences that seem most biologically plausible. VLMMs and profile HMMs are prominent examples of such descriptions. Natural selection and mutation shape molecular sequences; however, the connections between these forces and the computational biology techniques for analysing sequence data are often weak (but see Berg et al. 2004). Our goal here has been to help lay the groundwork for establishing such connections. Reconciliation of the VLMM and profile HMM descriptions of sequence data with their population genetic implications seems to us to be a desirable endeavour. Other sorts of probabilistic descriptions of sequences (e.g. Bussemaker et al. 2000) could be matched to evolutionary models in a similar fashion.
The 2Nsj values that are depicted in figures 1 and 2 represent predicted distributions of fitness effects of new mutations. These distributions are central to evolutionary theory and diverse attempts to characterize them have been made (e.g. see Eyre-Walker & Keightley 2007). Although our approach for inferring 2Nsj values has the aforementioned flaws, worthwhile improvements seem feasible.
In the future, inference techniques that use these or other target distributions could be developed for analysing sets of homologous DNA sequences. Inference with the VLMM model could be performed via relatively straightforward modifications of previously published statistical techniques (e.g. see Robinson et al. 2003; Rodrigue et al. 2005). Inference with the profile HMM model would be more difficult due to the possibility of insertion and deletion events and this is a challenge that we are currently considering.
Acknowledgments
We thank P. Higgs for suggestions that facilitated some of this work. We thank R. Cartwright, A. Griffing, K. Lamm, Z. Yang and an anonymous reviewer for their comments. S.C.C., B.D.R. and J.L.T. were supported by NSF grant DEB-0445180 and NIH grant GM070806.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Statistical and computational challenges in molecular phylogenetics and evolution’.
References
- Bejerano G. Algorithms for variable length Markov chain modeling. Bioinformatics. 2004;20:788–789. doi: 10.1093/bioinformatics/btg489. doi:10.1093/bioinformatics/btg489 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Yona G. Variations on probabilistic suffix trees: statistical modeling and prediction of protein families. Bioinformatics. 2001;17:23–43. doi: 10.1093/bioinformatics/17.1.23. doi:10.1093/bioinformatics/17.1.23 [DOI] [PubMed] [Google Scholar]
- Berg J, Willmann S, Lässig M. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 2004;4:42. doi: 10.1186/1471-2148-4-42. doi:10.1186/1471-2148-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol. Biol. Evol. 2006;23:2058–2071. doi: 10.1093/molbev/msl091. doi:10.1093/molbev/msl091 [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A site- and time-heterogeneous model of amino acid replacement. Mol. Biol. Evol. 2008;25:842–858. doi: 10.1093/molbev/msn018. doi:10.1093/molbev/msn018 [DOI] [PubMed] [Google Scholar]
- Browning S.R. Multilocus association mapping using variable-length Markov chains. Am. J. Hum. Genet. 2006;78:903–913. doi: 10.1086/503876. doi:10.1086/503876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker H.J, Li H, Siggia E.D. Building a dictionary for genomes: identification of presumptive regulatory sites by statistical analysis. Proc. Natl Acad. Sci. USA. 2000;97:10 096–10 100. doi: 10.1073/pnas.180265397. doi:10.1073/pnas.180265397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.C, Hobolth A, Robinson D.M, Kishino H, Thorne J.L. Quantifying the impact of protein tertiary structure on molecular evolution. Mol. Biol. Evol. 2007;24:1769–1782. doi: 10.1093/molbev/msm097. doi:10.1093/molbev/msm097 [DOI] [PubMed] [Google Scholar]
- Christensen O.F, Hobolth A, Jensen J.L. Pseudo-likelihood analysis of codon substitution models with neighbor-dependent rates. J. Comput. Biol. 2005;12:1166–1182. doi: 10.1089/cmb.2005.12.1166. doi:10.1089/cmb.2005.12.1166 [DOI] [PubMed] [Google Scholar]
- Durbin R, Eddy S, Krogh A, Mitchison G. Cambridge University Press; Cambridge, UK: 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. ch. 5, pp. 100–133. [Google Scholar]
- Eyre-Walker A, Keightley P.D. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 2007;8:610–618. doi: 10.1038/nrg2146. doi:10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Fleissner R, Metzler D, von Haeseler A. Simultaneous statistical multiple alignment and phylogeny reconstruction. Syst. Biol. 2005;54:548–561. doi: 10.1080/10635150590950371. doi:10.1080/10635150590950371 [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol. Biol. Evol. 1998;15:871–879. doi: 10.1093/oxfordjournals.molbev.a025991. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Guttorp, P. 1995 Stochastic modeling of scientific data London, UK: Chapman and Hall; Boca Raton, FL: CRC.
- Halpern A.L, Bruno W.J. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol. Biol. Evol. 1998;15:910–917. doi: 10.1093/oxfordjournals.molbev.a025995. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. doi:10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- Hwang D.G, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl Acad. Sci. USA. 2004;101:13 994–14 001. doi: 10.1073/pnas.0404142101. doi:10.1073/pnas.0404142101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Jensen J.L, Pedersen A.M.K. Probabilistic models of DNA sequence evolution with context dependent rates of substitution. Adv. Appl. Prob. 2000;32:499–517. doi:10.1239/aap/1013540176 [Google Scholar]
- Knudsen B, Miyamoto M.M. Using equilibrium frequencies in models of sequence evolution. BMC Evol. Biol. 2005;5:21. doi: 10.1186/1471-2148-5-21. doi:10.1186/1471-2148-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. doi:10.1093/molbev/msh112 [DOI] [PubMed] [Google Scholar]
- Lunter G, Miklós I, Drummond A, Jensen J.L, Hein J. Bayesian coestimation of phylogeny and sequence alignment. BMC Bioinform. 2005;6:83. doi: 10.1186/1471-2105-6-83. doi:10.1186/1471-2105-6-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklós I, Lunter G.A, Holmes I. A “Long Indel” model for evolutionary sequence alignment. Mol. Biol. Evol. 2004;21:529–540. doi: 10.1093/molbev/msh043. doi:10.1093/molbev/msh043 [DOI] [PubMed] [Google Scholar]
- Muse S.V, Gaut B.S. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 2003;20:1231–1239. doi: 10.1093/molbev/msg147. doi:10.1093/molbev/msg147 [DOI] [PubMed] [Google Scholar]
- Parisi G, Echave J. Structural constraints and emergence of sequence patterns in protein evolution. Mol. Biol. Evol. 2001;18:750–756. doi: 10.1093/oxfordjournals.molbev.a003857. [DOI] [PubMed] [Google Scholar]
- Pedersen A.M.K, Jensen J.L. A dependent-rates model and an MCMC-based methodology for the maximum-likelihood analysis of sequences with overlapping reading frames. Mol. Biol. Evol. 2001;18:763–776. doi: 10.1093/oxfordjournals.molbev.a003859. [DOI] [PubMed] [Google Scholar]
- Redelings B.D, Suchard M.A. Joint Bayesian estimation of alignment and phylogeny. Syst. Biol. 2005;54:401–418. doi: 10.1080/10635150590947041. doi:10.1080/10635150590947041 [DOI] [PubMed] [Google Scholar]
- Redelings B.D, Suchard M.A. Incorporating indel information into phylogeny estimation for rapidly emerging pathogens. BMC Evol. Biol. 2007;7:40. doi: 10.1186/1471-2148-7-40. doi:10.1186/1471-2148-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.M, Jones D.T, Kishino H, Goldman N, Thorne J.L. Protein evolution with dependence among codons due to tertiary structure. Mol. Biol. Evol. 2003;20:1692–1704. doi: 10.1093/molbev/msg184. doi:10.1093/molbev/msg184 [DOI] [PubMed] [Google Scholar]
- Rodrigue N, Lartillot N, Bryant D, Philippe H. Site interdependence attributed to tertiary structure in amino acid sequence evolution. Gene. 2005;347:207–217. doi: 10.1016/j.gene.2004.12.011. doi:10.1016/j.gene.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Sella G, Hirsh A.E. The application of statistical physics to evolutionary biology. Proc. Natl Acad. Sci. USA. 2005;102:9541–9546. doi: 10.1073/pnas.0501865102. doi:10.1073/pnas.0501865102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 2004;21:468–488. doi: 10.1093/molbev/msh039. doi:10.1093/molbev/msh039 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L, Eddy S.R, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. doi:10.1002/(SICI)1097-0134(199707)28:3<405::AID-PROT10>3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Tavaré, S. 1986 Some probabilistic and statistical problems in the analysis of DNA sequences. In Lectures in mathematics in the life sciences, vol. 17 (ed. R. Miura), pp. 56–86. Providence, RI: American Mathematical Society.
- Thorne J.L, Kishino H, Felsenstein J. An evolutionary model for maximum likelihood alignment of DNA sequences. J. Mol. Evol. 1991;33:114–124. doi: 10.1007/BF02193625. doi:10.1007/BF02193625 [DOI] [PubMed] [Google Scholar]
- Thorne J.L, Kishino H, Felsenstein J. Inching toward reality: an improved likelihood model of sequence evolution. J. Mol. Evol. 1992;34:3–16. doi: 10.1007/BF00163848. doi:10.1007/BF00163848 [DOI] [PubMed] [Google Scholar]
- Thorne J.L, Choi S.C, Yu J, Higgs P.G, Kishino H. Population genetics without intraspecific data. Mol. Biol. Evol. 2007;24:1667–1677. doi: 10.1093/molbev/msm085. doi:10.1093/molbev/msm085 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 2008;25:568–579. doi: 10.1093/molbev/msm284. doi:10.1093/molbev/msm284 [DOI] [PubMed] [Google Scholar]
- Yu J, Thorne J.L. Dependence among sites in RNA evolution. Mol. Biol. Evol. 2006;23:1525–1537. doi: 10.1093/molbev/msl015. doi:10.1093/molbev/msl015 [DOI] [PubMed] [Google Scholar]