Abstract
The rate at which a given site in a gene sequence alignment evolves over time may vary. This phenomenon—known as heterotachy—can bias or distort phylogenetic trees inferred from models of sequence evolution that assume rates of evolution are constant. Here, we describe a phylogenetic mixture model designed to accommodate heterotachy. The method sums the likelihood of the data at each site over more than one set of branch lengths on the same tree topology. A branch-length set that is best for one site may differ from the branch-length set that is best for some other site, thereby allowing different sites to have different rates of change throughout the tree. Because rate variation may not be present in all branches, we use a reversible-jump Markov chain Monte Carlo algorithm to identify those branches in which reliable amounts of heterotachy occur. We implement the method in combination with our ‘pattern-heterogeneity’ mixture model, applying it to simulated data and five published datasets. We find that complex evolutionary signals of heterotachy are routinely present over and above variation in the rate or pattern of evolution across sites, that the reversible-jump method requires far fewer parameters than conventional mixture models to describe it, and serves to identify the regions of the tree in which heterotachy is most pronounced. The reversible-jump procedure also removes the need for a posteriori tests of ‘significance’ such as the Akaike or Bayesian information criterion tests, or Bayes factors. Heterotachy has important consequences for the correct reconstruction of phylogenies as well as for tests of hypotheses that rely on accurate branch-length information. These include molecular clocks, analyses of tempo and mode of evolution, comparative studies and ancestral state reconstruction. The model is available from the authors' website, and can be used for the analysis of both nucleotide and morphological data.
Keywords: heterotachy, covarion, Markov chain Monte Carlo, mixture model, phylogeny
1. Introduction
These are times of plenty for molecular phylogenetics. By the spring of 2008 GenBank (www.ncbi.nlm.nih.gov) could boast over 80 million distinct gene sequences in its database. After a slow start in the early 1980s GenBank's growth was catalysed by the discovery of the polymerase chain reaction and has been growing at an exponential or nearly exponential pace ever since, currently with a period doubling time of approximately 2–4 years. Many of these sequences are used in establishing the phylogenetic relationships among species and this is reflected in the growing use of phylogenies in biological research. The Web of Science database catalogued by the end of 2007 over 25 000 articles using or describing molecular phylogenies and this number is currently growing quadratically, increasing by over 3000 published articles per annum (figure 1; cf. Pagel 1999). Another growth phenomenon has, over this same time period, increasingly enabled computational biologists to exploit gene sequences on acceptable time scales. What has come to be known as Moore's Law was the playful but shrewd suggestion by one of the co-founders of the Intel Corporation that computational power would raise exponentially throughout the decades of 1970s, 1980s, 1990s and now into the twenty-first century. Moore was proved right and the trend continues with no end currently in sight.
Figure 1.
Growth in the use of molecular phylogenies in scientific research. Source: Web of Science, April, 2008. Search terms molecular AND phylogen*. Line of best fit is a quadratic spline. Dashed lines indicate that period doubling time is increasing.
The confluence of these three trends means that investigators can, as never before, attempt to infer phylogenetic relationships among ever-greater numbers of species and using ever-greater numbers of genes. The increasing size and taxonomic range of gene-sequence alignments also means that the models of sequence evolution used to characterize these data must be up to the task of identifying a varied and potentially complex range of signals. Different sites may show different patterns and rates of evolution and these patterns and rates may vary among genes, regions of genes, between ribosomal and protein coding genes, spacers and introns or even repetitive DNA. One source of variation in gene-sequence alignments that has received relatively little attention in phylogenetic models is the phenomenon of heterotachy, despite being pointed out by Walter Fitch and colleagues (e.g. Fitch & Markowitz 1970; Fitch 1971) close to 40 years ago. Heterotachy refers to a site in a gene-sequence alignment having a different rate of evolution in different parts of the tree. Fitch supposed that heterotachy could arise if evolutionary changes to one region of a gene made it more likely that other regions of the gene became less constrained. He called this the covarion hypothesis for ‘concomitantly variable codons’. Heterotachy, whether of the specific sort that Fitch envisioned or arising from other evolutionary considerations such as varying selective pressures, appears to be common in gene-sequence alignments (Lopez & Philippe 2002; Ane et al. 2005; Philippe et al. 2005; Taylor et al. 2006).
Tuffley & Steel (1998) proposed a simple and elegant model of covarion-like behaviour applicable to gene-sequence data. Their model supposes that a given site stochastically switches between being ‘on’ or ‘off’ throughout the tree. When ‘on’ a conventional Markov substitution process among nucleotides describes its evolution. When ‘off’ the site's rate of evolution goes to zero. Switches are also described by a Markovian process and therefore can occur anywhere in the tree, including within a branch, giving rise to a variety of possible rates of evolution throughout the phylogeny. The model achieves this complexity with just two extra parameters over that of a conventional model of sequence evolution, those describing the rate at which sites switch from on to off and back again. Modifications to the original Tuffley and Steel model include allowing more than one category of on rates (Galtier 2001), variable on rates and an off state (Wang et al. 2007), a parametric distribution of rates in the on state (Huelsenbeck 2002), and allowing the covarion model itself to vary throughout the tree (Penny et al. 2001).
An alternative approach to accounting for heterotachy exchanges the simple parametric elegance of the covarion model for a model requiring more parameters, but not linked to parametric assumptions about the degree or distribution of heterotachy throughout the tree. Consider two sites, one of which evolves at a more or less constant rate throughout the tree and another whose rate of evolution accelerates sporadically in one or more regions. Given a common underlying model of sequence evolution, the differing behaviour of these two sites can be captured by assigning them different branch lengths on a shared topology. Generalizing this approach across all sites, a model emerges in which some number of extra branch-length sets is sufficient to describe the heterotachy in the data. It is unlikely that the information about which branch-length sets correspond to which site or sites would be known in advance, but an approach to statistical inference known as mixture models removes the need to know this.
Mixture models, as applied to phylogenetic inference, sum the likelihood at each site of the alignment over more than one model of evolution (e.g. Koshi & Goldstein 1998; Huelsenbeck & Nielsen 1999; Lartillot & Philippe 2004; Pagel & Meade 2004; Blackburne et al. 2008). For example, we (Pagel & Meade 2004, 2005) reported a mixture model for characterizing what we called ‘pattern heterogeneity’ in the evolution of nucleotide sequences. This model fits more than one model of gene-sequence evolution to an alignment, without specifying in advance how many models there will be or to which sites they best correspond. Instead, the method finds the optimal number of models and sums the likelihood at each site over all of them, weighted by their prior probabilities. At the same time, Lartillot & Philippe (2004) reported a similar mixture model for protein sequence data. Both models routinely return large improvements in likelihood, and reduce long-branch attraction. They also avoid the problem of assigning sites to partitions and have been shown to improve on partitioned likelihoods, despite requiring no advance information from the user, as well as reduce the so-called ‘node-density’ artefacts (Venditti et al. 2008).
Applied to the problem of heterotachy, a branch-length set mixture model will seek to find some optimal number of extra branch-length sets, summing the likelihood at each site over all of them. Kolaczkowski & Thornton (2004) presented a model with two branch-length sets but calculated the likelihood incorrectly (Spencer et al. 2005) and their model was only applicable to data in which it was known in advance which branch-length set best characterized a given site. In more recent papers, these same authors, others and we report more fully developed branch-length sets mixture models for heterotachy applicable to phylogenetic inference (Zhou et al. 2007; Meade & Pagel 2008; Kolaczkowski & Thornton 2008).
An unwanted feature of the multiple branch-length set approaches is the potentially large number of extra parameters required to describe the heterotachy. Each extra branch-length set requires 2s−3 additional parameters (for an unrooted tree) where s is the number of species or taxa in the alignment, plus an empirical estimated weighting component. Even for a modest tree of 40 species this means estimating 77+1 additional parameters just to accommodate one extra branch-length set. Zhou et al. (2007) suggested that, given the burden of these extra parameters, the mixture model may, despite its better absolute likelihood, not be favoured over the simpler covarion model. These authors report that the relatively lenient Akaike information criterion (AIC) may prefer the branch lengths mixture model to the covarion (Akaike 1974; Felsenstein 2004) but that the more stringent Bayesian information criterion (BIC; Schwarz 1978; Felsenstein 2004) prefers the covarion. We have found similar results in five published nucleotide alignments—the mixture model with extra branch-length sets is often favoured under the AIC but is far less likely to be supported using the BIC (Meade & Pagel 2008).
Our goal here is to describe and evaluate a mixture-model approach to accounting for heterotachy that potentially reduces the number of extra parameters required to explain the data. Our approach is motivated by the belief that heterotachy is a common source of variation in gene-sequence data, but that it may be confined to only a small number of sites or to small regions of the tree. If either of these situations is true, then many of the branches in the tree will not require an additional branch length. Our approach considers an extra branch-length set but then tests pairs of branches corresponding to the same edge of the phylogenetic tree to see whether they can be collapsed into a single branch, or if two additional branch-length sets are considered, to see whether three branches can be collapsed to two, and so on. At the same time, the model continually proposes adding a branch length if one is found inadequate to explain the data, and performs these actions while exploring different tree topologies.
We implement our model using Markov chain Monte Carlo (MCMC; Gilks et al. 1996; Gelman 2003) methods. MCMC methods employ a Markov chain to explore the potential universe of models of evolution that might describe the data. Most implementations of MCMC methods explore models of a fixed number of dimensions, such as the parameters of a model of gene-sequence evolution. However, a variant of MCMC known as reversible-jump MCMC (RJMCMC; Green 1995) allows the Markov chain to move among models of different dimensionality. Boys & Henderson (2001) and Huelsenbeck et al. (2004) used the reversible-jump approach to explore models of sequence evolution, Suchard et al. (2001) used it to choose among alternative phylogenetic topologies, Pagel & Meade (2006) used it in a model of correlated evolution of binary traits, and we also use the RJMCMC approach in our model of pattern heterogeneity to determine the number of extra models of gene-sequence evolution (available in BayesPhylogenies package, www.evolution.reading.ac.uk). Here, we adopt the reversible-jump approach in a multiple branch-length sets mixture model for heterotachy to identify how many extra parameters—corresponding to distinct additional branch lengths—are needed to explain the data. Our hope is that this RJMCMC algorithm can return substantial improvements to the likelihood but often with far fewer parameters than simply force-fitting an entire extra branch-length set. We implement this reversible-jump heterotachy model in combination with our earlier model of pattern heterogeneity (Pagel & Meade 2004). This means that the heterotachy that our model identifies is that which exists over and above any tendency for different sites to adopt different patterns or rates of evolution.
In what follows we describe the model in more detail, then apply it to simulated and real datasets. It is not our goal to provide a general evaluation of branch-length mixture models for heterotachy as we and others have already reported results from simulated and real data. Our primary interests are to evaluate the reversible-jump procedure as a way of overcoming the burden of extra parameters, and as a method for identifying where in the tree and for which sites or genes in the alignment rates of evolution have been accelerated or slowed.
2. Reversible-jump branch-length sets mixture model
We define the likelihood of a model of gene-sequence evolution as an amount proportional to the probability of the data given the model of sequence evolution and a phylogenetic tree
| (2.1) |
where D will normally be an aligned set of sequence data; Q is the familiar substitution rate matrix that defines the model of evolution; and T is the phylogenetic tree. In the case of nucleotide data, Q is a 4×4 matrix of transition rates among A, C, G and T (Swofford et al. 1996). For protein data Q is a 20×20 matrix representing the transition rates among all pairs of amino acids.
Given an aligned set of gene-sequence or other character-state data, the probability of the data in D is found as the product over all of the sites of the individual probabilities of each site.
| (2.2) |
A mixture model for additional branch-length sets modifies this basic framework by including more than one set of branches for any given tree topology T. The branching structures (topology) of the trees are fixed while independent branch lengths, represented as a vector t, are free to vary. The probability of the data is now calculated by summing the likelihood at each site over all of the different t for a given tree. Thus, defining the branch-length sets as t1, t2, …, tJ, the probability of the data under the mixture model is
| (2.3) |
where the summation over j now specifies that the likelihood of the data at each site is summed over J separate branch-length sets, the summation being weighted by the wi where w1+w2+⋯+wJ =1.0 and represents our prior beliefs about the suitability of a set of branches for a given site. The number of branch-length sets, J, can be determined either by prior knowledge of how many different patterns are expected in the data, or they can be empirically estimated, along with the prior weights, from the data. Equation (2.3) describes the model that Zhou et al. (2007), Meade & Pagel (2008) and Kolaczkowski & Thornton (2008) use.
Although our primary interest is in heterotachy we need to digress from equation (2.3) for a moment to describe the wider context of the heterotachy mixture model. Nothing in equation (2.3) describes the variation that one might expect among sites in the alignment, such as the well-known gamma rate heterogeneity (Yang 1994) or the pattern heterogeneity that we describe elsewhere (Pagel & Meade 2004). But there is every reason to expect that these across-site sources of variation will exist alongside the within-site variation in rates that models of heterotachy are designed to describe, and so a complete model should account for both. Failure to do this will not just mis-characterize the data, it could conflate one form (such as pattern heterogeneity) with another (e.g. heterotachy) if their effects were somehow correlated.
It is straightforward to rewrite equation (2.3) to accommodate both pattern heterogeneity and heterotachy. We now consider a model in which we sum the likelihood at each site over a series of k models of sequence evolution, denoted by Qk, and over more than one branch-length set. The summation is nested as shown in equation (2.4), and simultaneously accounts for pattern heterogeneity and heterotachy:
| (2.4) |
A simple addition to this model can accommodate gamma-distributed rate variation across sites (see Pagel & Meade 2004 for a discussion of this with respect to pattern heterogeneity on its own). Although equation (2.4) describes the more complete model we will, for purposes of discussion, refer to the model as described in equation (2.3). Nevertheless, in all of the analyses we report below the heterotachy that emerges is above and beyond that which can be attributed to pattern heterogeneity and to gamma-distributed rate heterogeneity. None of the previous models of heterotachy simultaneously account for pattern heterogeneity, although Huelsenbeck (2002) and Zhou et al. (2007) allowed rate heterogeneity.
Elsewhere, we have described (Meade & Pagel 2008) the mixture model of equation (2.3) and illustrated its application to simulated and real data. We show that it can accurately estimate multiple sets of branch lengths for a given alignment and tree topology, as well as correctly identify the weights that should be assigned to those extra branch-length sets (see also Zhou et al. 2007). Here, we wish to define and investigate a variant of the model in equation (2.3) that responds to the realization that fitting two or more complete additional branch-length sets may introduce many redundant parameters, paired branch lengths in different branch-length sets that are in fact not different from each other.
This model can be written identically to that of equation (2.3) but now we wish to determine for how many of the 2s−3 edges in an unrooted phylogenetic tree topology are two or more distinct lengths needed to describe the data, and for how many a single length suffices. This is the job of the reversible-jump algorithm.
We can write the probability model to explain the data as where the parameters are as defined previously. Then the likelihood of the data given M can be written as
where p(Q), p(T) and p(t) are the prior probabilities of these terms, and for simplicity we represent the J vectors of branch lengths by a single matrix t. In practice, this integral is difficult to evaluate but MCMC methods can be used to estimate the posterior distribution of , and MCMC methods are now widely used in phylogenetic inference (e.g. Gascuel 2005).
In a phylogenetic context, we construct a Markov chain that jumps among possible models of sequence evolution, phylogenetic trees and vectors and elements of the branch-length space. At each iteration of the chain some new version of M is proposed either by altering the parameters in Q, or by changing the tree topology or its branch lengths. Changes to the topology move all of the lengths associated with a given branch to its new position. Successive steps of the chain are sampled using the Metropolis–Hastings algorithm (Metropolis et al. 1953; Hastings 1970). A newly proposed model that improves on the previous model in the chain is always sampled or accepted; otherwise it is accepted with probability proportional to the ratio of its likelihood to that of the previous model.
Formally, the acceptance probability rule computes the ratio of these likelihoods, the ratio of the prior probabilities, and two additional quantities called the proposal ratio and the Jacobian such that a new model is accepted according to
The product of the prior ratio and likelihood ratio is written as
where the terms are as defined above and the primes refer to the proposed model. This ratio keeps track of the relative performance of the current and proposed models, and assigns a cost, in the form of the prior ratio, to models with more parameters. The proposal ratio compares the probability of moving towards some new model to the probability of moving back to the original state. Its role is to ensure that the Markov chain searches the parameter space in an unbiased way. The Jacobian measures the volume of the two state-spaces defined by the current and proposed models. A Markov chain following this acceptance rule will in principle eventually converge to a state in which successive values of the chain sample what is known as the stationary distribution of states of the model. At stationarity, the chain samples the posterior distributions of the model's parameters.
For most applications of MCMC, the dimensionality of the current and proposed models is the same and thus the Jacobian can be ignored, as it takes the value of 1. An unusual feature of the approach we describe here, however, is that we wish to explore models with differing numbers of parameters, corresponding to some branches of the tree topology being assigned more than one length. We define ‘split’ and ‘merge’ moves as proposals to either add a new branch length to the tree by splitting an existing branch length into two distinct lengths, or to combine two such lengths into one. Whenever a split or merge move is proposed the resulting change to the likelihood is either accepted or rejected following the acceptance rule described above. Over large numbers of iterations these moves will settle on a stationary distribution of the number of extra branch lengths that are required to explain the data.
To incorporate the split and merge moves into the RJMCMC algorithm requires carefully constructed proposal mechanisms and calculation of the Jacobian term. We present details of these mechanisms, the proposal ratio and the Jacobian terms in the electronic supplementary materials.
3. Priors and running the Markov chain
The Markov chain requires specification of the prior distributions of the parameters of the model. We used uniform priors on the interval 0–100 for all parameters of the models of sequence evolution, uniform (0–1) priors on the empirical weights of the mixture models, and exponential (10) priors on the branch lengths for changes that are proposed independently of splitting or merging. Trees are given a uniform prior. We normally ran at least four independent chains for each dataset to check that they converged to the same region of the parameter space. Chains were allowed to reach an apparent steady state (this being operationally defined as a chain whose mean likelihood remains unchanged over many iterations) and then they were run at least 10 million further iterations. We sampled from ‘converged’ chains every 10 000 iterations retrieving at least 1000 trees for purposes of drawing inferences. Log-likelihoods reported below are the logarithms of the harmonic means of the posterior distribution of likelihoods.
4. Application to simulated data
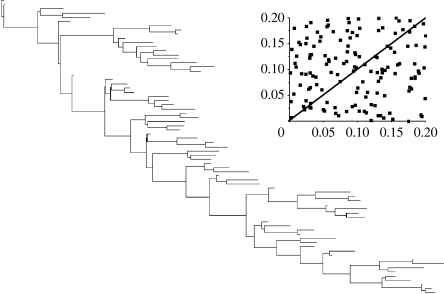
We generated a random topology of 70 taxa, and then drew two random sets of branch lengths from a uniform distribution ranging between 0 and 0.2 (figure 2, inset shows the two sets of branch lengths are uncorrelated). We generated 1500 simulated nucleotides for each branch-length set, using Seqgen (Rambaut & Grassly 1997) and a common general-time-reversible (GTR) model of sequence evolution (Swofford et al. 1996). We combined the alignments from the two randomly varying branch-length sets into a single alignment of 3000 simulated nucleotides.
Figure 2.
Random topology of 70 taxa with branch lengths corresponding to branch-length set 1 drawn on the uniform interval 0–0.2. Inset: scatterplot of branch lengths from set 1 versus branch lengths from set 2.
We analysed the simulated alignment using a conventional GTR likelihood model with one set of branches, and also with the heterotachy mixture model fitting two full branch-length sets, and finally using the reversible-jump algorithm.
5. Application to published data
We applied the model to the five published datasets described in table 1, including algae, plants, nematode worms and salamanders. The alignments include nuclear and mitochondrial genes of both ribosomal and protein coding functions. Each alignment contains at least two genes, and no fewer than 3600 nucleotides, but the number of taxa ranges from just 14 to 119 (all datasets available from Treebase, Piel et al. 2002). In our previous study of heterotachy (Meade & Pagel 2008), we found that each of these datasets could be described by two branch-length sets, save for the Plethodontids for which the AIC supported three branch-length sets, but a BIC supports two. Accordingly, we study the behaviour of the reversible-jump mixture model for two branch-length sets for each alignment.
Table 1.
Details of the five published datasets.
| taxa | no. of taxa | genes | no. of sites |
|---|---|---|---|
| Chlorophyceae; Buchheim et al. (2001) | 38 | 18S and 26S | 3684 |
| Gnetales; Rydin et al. (2002) | 119 | 26s, 18s, rbcL, atpB | 5923 |
| Caenorhabditis; Kiontke et al. (2004) | 14 | 18s, 28s, RNAP2, Par6, pkc3 | 7652 |
| Plethodontids; Mueller et al. (2004) | 27 | complete mitochondrion | 14 040 |
| Costaceae (Zingiberales); Specht (2006) | 66 | ITS, trml-F, trnK, matK | 5898 |
6. Results
(a) Simulated data
Table 2 shows the mean log-likelihoods for the conventional model with one branch-length set, and the improvement in likelihood from the two branch-length set mixture models and the reversible-jump mixture model. The addition of a second branch-length set, as expected, accounts for a large improvement in the log likelihood (significant by both AIC and BIC). The mixture model is able accurately to estimate the additional branches: the correlation between the simulated branch lengths and the mean of the posterior sample of the estimated branch lengths is 0.997 for set 1 and 0.995 for set 2. This is almost identical to what Zhou et al. (2007) reported for a similar set of simulations. The second branch-length set requires 138 additional parameters (2×70−3 branch parameters plus one weight parameter). The reversible-jump model accounts for 92.4 per cent of the likelihood improvement over the conventional model but with a mean of just 73 additional branches or roughly half the number of parameters.
Table 2.
Results for 70-taxa simulated data. (BLS, branch-length set. RJ, reversible jump.)
| log-L±s.da | log-improvement | proportion RJ/2 BLS | no. of branches | RJ branches | proportion RJ branches | |
|---|---|---|---|---|---|---|
| 2 BLS | RJ | |||||
| 131 951±9 | 923 | 853 | 0.92 | 138 | 73±2.5 | 0.53 |
harmonic mean of likelihoods from converged chain.
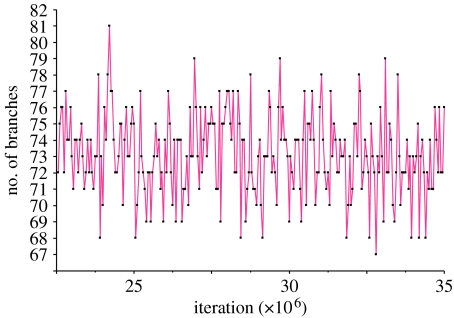
Figure 3 plots the posterior distribution of additional branches in the reversible-jump model as a time series, showing that the chain is stable at 73±2.5 branches and that it mixes well, sometimes including more branches and sometimes fewer. From the inset to figure 2, a guess can be made about why the reversible-jump model uses 65 fewer parameters than the full branch-length set mixture model. Despite being uncorrelated, just by chance some of the randomly drawn branches in the first set are similar in length to their matching branch in the second set—those falling near to the 1: 1 line shown there. Figure 4 confirms this view, plotting the posterior probability of an edge in the tree having two distinct branches as a function of the difference in length of the two randomly generated branches. The posterior distribution for each edge is found from the proportion of times in the posterior sample a particular edge had one versus two branches.
Figure 3.
Time-series plot taken from a segment of the converged Markov chain of the posterior distribution of extra branches derived from the reversible-jump algorithm. Mean=73±2.5.
Figure 4.
The posterior probability of the reversible-jump algorithm identifying two branches as a function of the absolute difference in the true lengths of the two branches. P50 denotes the point on the x-axis (0.040, red circle) where there is a 50% chance of detecting a second branch. S-shaped curve is a two-parameter logistic model.
Figure 4 shows that for a difference of just 0.04 branch-length units there is a 50 per cent chance of a second branch being acquired in the reversible-jump posterior sample. Branches that differ by an amount less than that by definition display little or no heterotachy and so a second branch is not required for them, or at least is not detectable even with an alignment of 3000 independent sites. We think that this 0.04 figure is not just a function of our branches varying on the interval 0–0.2. Even for very short or relatively long branches (near 0 or near to 0.2, respectively) in our simulations, we find that a second branch is required if it is estimated to differ by more than this 0.04 amount from the first.
(b) Real datasets
In real datasets, we might expect a range of degrees of heterotachy, from it being confined to a small number of branches up to including nearly every branch in the tree. Especially when it is confined to a small number of branches, the reversible-jump mixture model should be more sensitive to detecting these effects.
Table 3 reports the mean log likelihoods for the conventional one branch-length set model as applied to the five published datasets, and the improvement in likelihood from the two branch-length sets and reversible-jump mixture models. The results from the reversible-jump model explain why the AIC often supports two full branch-length sets in the conventional mixture model but the BIC does not. The last column of table 3 shows that the proportion of branches showing evidence of heterotachy ranges from just 5 to approximately 60%. For the Chlorophyceae the full branch-length sets mixture model is forced to fit 74 branches as opposed to just 4±3 for the reversible-jump model. The message here is that real datasets can be expected to vary greatly in the amount of heterotachy they show and in how many branches in the tree are affected. It is not necessary to test the reversible-jump results further: the mean of the posterior distributions of extra branches directly estimates how much heterotachy is in the tree. Nevertheless, each of the reversible-jump results would be judged significant using the more stringent BIC as applied to their likelihood improvement and numbers of additional branches required.
Table 3.
Results for real datasets. (BLS, branch-length set; RJ, reversible jump.)
| dataset | log-L 1 BLSa | log improvement | proportion RJ/2 BLS | no. of branches | RJ branches mean±s.d. | proportion RJ branches | |
|---|---|---|---|---|---|---|---|
| 2 BLS | RJ | ||||||
| Chlorophyceae | −26 260 | 112 | 23 | 0.21 | 74 | 4.0±3.0 | 0.05 |
| Gnetales | −77 650 | 517 | 424 | 0.82 | 236 | 44.5±3.1 | 0.19 |
| Caenorhabditis | −43 622 | 106 | 95 | 0.90 | 26 | 15.4±1.4 | 0.60 |
| Plethodontids | −185 521 | 190 | 111 | 0.58 | 52 | 13.1±1.6 | 0.25 |
| Costaceae | −28 411 | 285 | 149 | 0.52 | 130 | 13.5±1.6 | 0.10 |
The reversible-jump algorithm usefully pinpoints the positions in the tree most affected by heterotachy. Figure 5 shows the consensus tree derived from the posterior distribution of trees for the 119 species in the Gnetales alignment (table 1), pruned to make it easier to view. The Gnetales are a small group of seed plants that may form a sister group to the angiosperms. The branch lengths of the consensus tree are, in the case of edges with more than one length, the weighted averages of the two branches found for that edge. The reversible-jump algorithm settles on an average of 44.5±3.1 extra branches (table 3) and those with a posterior probability greater than 0.5 of having more than one length are coloured in red. This reveals a concentration of heterotachy in the clade in the lower portion of the tree, corresponding to lycopods, ferns and equisetum. The inset shows the two very different sets of branch lengths the reversible-jump algorithm finds for this clade. Meade & Pagel (2008) show that this heterotachy is associated principally with acceleration in the evolution of the rbcl and atpB protein coding genes in this clade (lower sub-clade), genes associated with ATP synthesis and photosynthesis. By comparison, the two ribosomal genes evolve slowly in this clade (upper sub-clade) and in general have a more uniform rate throughout the tree.
Figure 5.
The phylogeny of the Gnetales (table 1). The tree is derived as a consensus of the posterior sample of trees produced by the reversible-jump mixture model. For edges with more than one branch length, the consensus length is the weighted mean of the lengths for that edge. Colours identify edges with a greater than 50% chance of having two distinct lengths in the posterior sample. Inset shows the two sets of branch lengths for the clade with pronounced heterotachy.
7. Discussion
Our results show that a reversible-jump mixture model can identify heterotachy in gene-sequence alignments and often with far fewer parameters than conventional mixture-models that rely on additional complete branch-length sets (Zhou et al. 2007; Meade & Pagel 2008; Kolaczkowski & Thornton 2008). It is worth bearing in mind that our results with real datasets record the amount of heterotachy over and above any improvements in likelihood that could be attributed to pattern heterogeneity (Pagel & Meade 2004) or rate variation among sites. To our knowledge, ours is the first demonstration of the effects of heterotachy independently of these other sources of variation. The results in table 3, therefore, reinforce earlier suggestions that heterotachy is an important source of variation in real data.
The strength of the mixture-model approach to heterotachy, of using multiple branch-length sets, is that it makes no a priori assumptions about the distribution or occurrence of heterotachy throughout the tree. Rather, the mixture model acts like a non-parametric covarion, the individual branch-length sets each providing a unique and potentially idiosyncratic description of the heterotachy in the tree. But as the reversible-jump algorithm exposes, potentially many of the parameters in the additional branch-length sets are redundant: they may correspond to edges in the tree for which no heterotachy is present. The reversible-jump algorithm solves the problem of redundancy by assigning multiple lengths only to those edges for which the data justify more than one length. In just the few datasets we investigated, the proportion of edges requiring more than one length fell as low as 5 per cent and was never more than 60 per cent.
The figure of 60 per cent, if representative of the upper values we might routinely expect of real datasets, makes the clear statement that, on average, the majority of the parameters that the conventional multiple branch-length sets mixture models estimate are not needed! Zhou et al. (2007), for example, reported correlations of approximately 0.60 between the branch lengths in the two branch-length sets they fitted to one of their datasets, indicative of much redundant information between the two sets. By comparison for the Gnetales data of figure 5, the correlation between the edges that the reversible-jump algorithm identifies as requiring two distinct lengths is −0.11. In the datasets, we examined the average number of extra branches required was just 24 per cent of those used in a complete additional set of branches, a reduction of approximately 75 per cent in the amount of estimation required. We do not think this low figure arises from a lack of power to detect additional branches, but rather points up just how much redundancy there is in the full branch-length set models. Even in our simulated data, we were able to account for 92 per cent of the likelihood supplied by an additional branch-length set, but using roughly half the parameters that the additional set required. These data further suggested that the reversible-jump algorithm was able to assign two lengths to an edge when the true simulated lengths differed by approximately 0.04 units of expected nucleotide substitutions.
The reversible-jump approach may help to manage some of the awkward problems of inference that arise when quasi-frequentist ideas of hypothesis testing are applied to Bayesian problems. Zhou et al. (2007) highlighted the very different outcomes that can arise when the relatively lenient Akaike and more stringent Bayesian information criterion tests are used to select among various mixture models of heterotachy. The reversible-jump algorithm produces a direct estimate of the posterior distribution of extra branches required in the mixture model. The mean of that distribution is a measure of one's posterior confidence in the existence of heterotachy in the data: if the mean is zero one can, other things being equal, be reasonably confident that heterotachy is not affecting the data and as it moves away from zero this provides evidence of more and more heterotachy. In this light, it is interesting that in our real data examples, the AIC always supported a complete additional branch-length set but the BIC does only for one of them (the Caenorhabditis). With no good way to choose between these two criteria, it is difficult to draw any conclusions. However, the BIC always supports the request for the smaller number of extra branches derived from the reversible-jump algorithm. The message is not that one should apply the BIC to test the results derived from the reversible-jump algorithm, rather, that the reversible-jump algorithm on its own is doing its job.
Apart from methodological considerations, an attractive feature of the reversible-jump approach is that it directly highlights the regions of the tree—and thus the taxa and their ancestors—whose evolution has been characterized by changes to rates of evolution. It does this by providing an estimate for each edge of the tree of the posterior belief in how many distinct lengths are needed to describe the evolution along those edges. Along with identifying the taxa involved, it is also straightforward to query the alignment itself to discover which sites are most directly implicated in the heterotachy. For the Gnetales data, this combination of information allows us to say directly that two protein coding genes involved in energy transfer (rbcl and atpB) have greatly accelerated their rate of evolution in the lycopods, ferns and equisetum. Put together, what the model is discovering are taxa–gene interactions in the rate of evolution. This kind of information should prove valuable for understanding the selective forces acting on genes and for reconstructing the history of protein evolution in particular groups.
Acknowledgements
This work was supported by grant NE/C51992X/1 from the Natural Environment Research Council, United Kingdom to M.P. Earlier versions of the model reported here were presented at the annual New Zealand phylogenetics conference (Whitianga, 2005) and the Newton Institute (Cambridge, UK, 2007). We thank the Centre for Advanced Computing and Emerging Technologies (ACET) at the University of Reading for computational support, and Nick Goldman and Ziheng Yang for inviting this paper.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘Statistical and computational challenges in molecular phylogenetics and evolution’.
Supplementary Material
Supporting Materials to accompany Modelling Heterotachy in Phylogenetic Inference
References
- Akaike H. A new look at the statistical model identification. Automatic Control, IEEE Transactions on. 1974;19:716–723. doi:10.1109/TAC.1974.1100705 [Google Scholar]
- Ane C, Burleigh J.G, McMahon M.M, Sanderson M.J. Covarion structure in plastid genome evolution: a new statistical test. Mol. Biol. Evol. 2005;22:914–924. doi: 10.1093/molbev/msi076. doi:10.1093/molbev/msi076 [DOI] [PubMed] [Google Scholar]
- Blackburne, B. P., Hay, A. J. & Goldstein, R. A. 2008 Changing selective pressures during antigenic changes in Human influenza H3. PLoS Pathog 4, e1000058. (doi:10.1371/journal.ppat.1000058) [DOI] [PMC free article] [PubMed]
- Boys R.J, Henderson D.A. A comparison of reversible jump MCMC algorithms for DNA sequence segmentation using hidden Markov models. Comput. Sci. Stat. 2001;33:35–49. [Google Scholar]
- Buchheim M.A, Michalopulos E.A, Buchheim J.A. Phylogeny of the Chlorophyceae with special reference to the Sphaeropleales: a study of 18S and 26S rDNA data. J. Phycol. 2001;37:819–835. doi:10.1046/j.1529-8817.2001.00162.x [Google Scholar]
- Felsenstein J. Sinauer Associates; Sunderland, MA: 2004. Inferring phylogenies. [Google Scholar]
- Fitch W.M. Rate of change of concomitantly variable codons. J. Mol. Evol. 1971;1:84–96. doi: 10.1007/BF01659396. doi:10.1007/BF01659396 [DOI] [PubMed] [Google Scholar]
- Fitch W.M, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem. Genet. 1970;4:579–593. doi: 10.1007/BF00486096. doi:10.1007/BF00486096 [DOI] [PubMed] [Google Scholar]
- Galtier N. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 2001;18:866–873. doi: 10.1093/oxfordjournals.molbev.a003868. [DOI] [PubMed] [Google Scholar]
- Gascuel O. In: Mathematics of evolution and phylogeny. Gascuel O, editor. Oxford University Press; Oxford, UK: 2005. [Google Scholar]
- Gelman A. CRC Press; Boca Raton, FL: 2003. Bayesian data analysis. [Google Scholar]
- Gilks W.R, Richardson S, Spiegelhalter D.J. Introducing Markov chain Monte Carlo. In: Gilks W.R, Richardson S, Spiegelhalter D.J, editors. Markov chain Monte Carlo in practice. Chapman and Hall; Boca Raton, FL: 1996. pp. 1–19. [Google Scholar]
- Green P.J. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–732. doi:10.1093/biomet/82.4.711 [Google Scholar]
- Hastings W. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. doi:10.1093/biomet/57.1.97 [Google Scholar]
- Huelsenbeck J.P. Testing a covariotide model of DNA substitution. Mol. Biol. Evol. 2002;19:698–707. doi: 10.1093/oxfordjournals.molbev.a004128. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Nielsen R. Variation in the pattern of nucleotide substitution across sites. J. Mol. Evol. 1999;48:86–93. doi: 10.1007/pl00006448. doi:10.1007/PL00006448 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Larget B, Alfaro M.E. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 2004;21:1123–1133. doi: 10.1093/molbev/msh123. doi:10.1093/molbev/msh123 [DOI] [PubMed] [Google Scholar]
- Kiontke K, Gavin N.P, Raynes Y, Roehrig C, Piano F, Fitch D.H.A. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl Acad. Sci. USA. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. doi:10.1073/pnas.0403094101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Thornton J. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. doi:10.1038/nature02917 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski, B. & Thornton, J. W. 2008 A mixed branch length model of heterotachy improves phylogenetic accuracy. Mol. Biol. Evol 25, 1054–1066. (doi:10.1093/molbev/msh042). [DOI] [PMC free article] [PubMed]
- Koshi J.M, Goldstein R.A. Models of natural mutations including site heterogeneity. Proteins Struct. Funct. Genet. 1998;32:289–295. doi:10.1002/(SICI)1097-0134(19980815)32:3<289::AID-PROT4>3.0.CO;2-D [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1105. doi: 10.1093/molbev/msh112. doi:10.1093/molbev/msh112 [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Meade A, Pagel M. A phylogenetic mixture model for heterotachy. In: Pontarotti P, editor. Evolutionary biology from concept to application. Springer Verlag; Heidelberg, Germany: 2008. pp. 29–41. [Google Scholar]
- Metropolis N, Rosenbluth A.W, Rosenbluth M.N, Teller A.H, Teller E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1092. doi:10.1063/1.1699114 [Google Scholar]
- Mueller R.L, Macey J.R, Jaekel M, Wake D.B, Boore J.L. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc. Natl Acad. Sci. USA. 2004;101:13 820–13 825. doi: 10.1073/pnas.0405785101. doi:10.1073/pnas.0405785101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. doi:10.1038/44766 [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst. Biol. 2004;53:571–581. doi: 10.1080/10635150490468675. doi:10.1080/10635150490522232 [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. Mixture models in phylogenetic inference. In: Gascuel O, editor. Mathematics of evolution and phylogeny. Clarendon Press; Oxford, UK: 2005. pp. 121–142. [Google Scholar]
- Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 2006;167:808–825. doi: 10.1086/503444. doi:10.1086/503444 [DOI] [PubMed] [Google Scholar]
- Penny D, McComish B.J, Charleston M.A, Hendy M.D. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J. Mol. Evol. 2001;53:711–723. doi: 10.1007/s002390010258. doi:10.1007/s002390010258 [DOI] [PubMed] [Google Scholar]
- Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F. Heterotachy and long-branch attraction in phylogenetics. BMC Evol. Biol. 2005;5:50. doi: 10.1186/1471-2148-5-50. doi:10.1186/1471-2148-5-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, W. H., Donoghue, M., Sanderson, M. & Netherlands, L. U. T. 2002 Treebase: a database of phylogenetic information. Research report from the National Institute for Environmental Studies, pp. 41–47.
- Rambaut A, Grassly N.C. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 1997:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Rydin C, Kallersjo M, Friist E.M. Seed plant relationships and the systematic position of Gnetales based on nuclear and chloroplast DNA: conflicting data, rooting problems, and the monophyly of conifers. Int. J. Plant Sci. 2002;163:197–214. doi:10.1086/338321 [Google Scholar]
- Schwarz G. Estimating the dimensions of a model. Ann. Stat. 1978;6:461–464. [Google Scholar]
- Specht C.D. Systematics and evolution of the tropical monocot family Costaceae (Zingiberales): a multiple dataset approach. Syst. Bot. 2006;31:89–106. doi:10.1600/036364406775971840 [Google Scholar]
- Spencer M, Susko E, Roger A.J. Likelihood, parsimony, and heterogeneous evolution. Mol. Biol. Evol. SMBE. 2005;22:1161–1164. doi: 10.1093/molbev/msi123. doi:10.1093/molbev/msi123 [DOI] [PubMed] [Google Scholar]
- Suchard M.A, Weiss R.E, Sinsheimer J.S. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- Swofford D.L, Olsen G.J, Waddell P.J, Hillis D.M, Moritz C, Mable B.K. Molecular Systematics. Sinauer Associates; Sunderland, MA: 1996. Phylogeny reconstruction; pp. 407–514. [Google Scholar]
- Taylor M.S, Kai C, Kawai J, Carninci P, Hayashizaki Y, Semple C.A. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2:e30. doi: 10.1371/journal.pgen.0020030. doi:10.1371/journal.pgen.0020030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffley C, Steel M. Modeling the covarion hypothesis of nucleotide substitution. Math. Biosci. 1998;147:63–91. doi: 10.1016/s0025-5564(97)00081-3. doi:10.1016/S0025-5564(97)00081-3 [DOI] [PubMed] [Google Scholar]
- Venditti C, Meade A, Pagel M. Phylogenetic mixture models can reduce the node-density artifact. Syst. Biol. 2008;58:286–293. doi: 10.1080/10635150802044045. doi:10.1080/10635150802044045 [DOI] [PubMed] [Google Scholar]
- Wang H.C, Spencer M, Susko E, Roger A.J. Testing for covarion-like evolution in protein sequences. Mol. Biol. Evol. 2007;24:294–305. doi: 10.1093/molbev/msl155. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 1994;39:306–314. doi: 10.1007/BF00160154. doi:10.1007/BF00160154 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rodrigue N, Lartillot N, Philippe H. Evaluation of models handling heterotachy in phylogenetic inference. BMC Evol. Biol. 2007;7:206. doi: 10.1186/1471-2148-7-206. 10.1186/1471-2148-7-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Materials to accompany Modelling Heterotachy in Phylogenetic Inference