Abstract
Cubic membranes are soft three-dimensional crystals found within cell organelles in a variety of living systems, despite the aphorism of Fedorov: ‘crystallization is death’. They consist of multi-bilayer lipid–protein stacks, folded onto anticlastic surfaces that resemble triply periodic minimal surfaces, forming highly swollen crystalline sponges. Although cubic membranes have been observed in numerous cell types and under different pathophysiological conditions, knowledge about the formation and potential function(s) of non-lamellar, cubic structures in biological systems is scarce. We report that mitochondria with this cubic membrane organization isolated from starved amoeba Chaos carolinense interact sufficiently with short segments of phosphorothioate oligonucleotides (PS-ODNs) to give significant ODNs uptake. ODNs condensed within the convoluted channels of cubic membrane by an unknown passive targeting mechanism. Moreover, the interaction between ODNs and cubic membrane is sufficient to retard electrophoretic mobility of the ODN component in the gel matrix. These ODN–cubic membrane complexes are readily internalized within the cytoplasm of cultured mammalian cells. Transmission electron microscopic analysis confirms ODNs uptake by cubic membranes and internalization of ODN–cubic membrane complexes into the culture cells. Cubic membranes thus may offer a new, potentially benign medium for gene transfection.
Keywords: cubic membranes, cubic phases, DNA delivery, fluorescence microscopy, cubosomes, transmission electron microscopy
1. Introduction
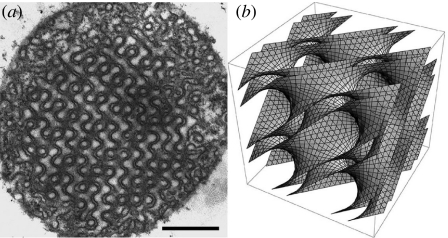
A simple process based on mixing, dispersion and homogenization of lipids in water leads to a variety of mesoscopic phases (Luzzati 1968; Larsson et al. 2006). Besides the parallel leaflet characteristics of lamellar phases, many lipids can also adopt other morphologies, such as the hexagonal phase or various cubic phases. In this respect, the behaviour of these lipid mixtures appears to be closely related to biomolecular assemblies in vivo (Almsherqi et al. 2006). For example, bicontinuous cubic mesophases are geometrically identical to the so-called cubic membranes (figure 1). In both cases, the lipid bilayers are draped on sponge-like triply periodic minimal surfaces of cubic symmetry, also observed in a variety of self-assembled soft molecular mesophases (Hyde et al. 1997; Sagalowicz et al. 2007). Cubic membranes are characterized by very large lattice parameters, typically of the order of 100 nm. These structures are formed reversibly; for example, in amoeba Chaos carolinense, the inner mitochondrial membranes transform reversibly from the usual random tubular arrangement to ordered cubic membrane organization (figure 1) with 50–80 nm intracristal channels in the absence of their natural food organisms (Paramecium multimicronucleatum). The membrane organization inside the starved amoeba mitochondria has been analysed in detail previously (Deng & Mieczkowski 1998; Deng et al. 1999). Cubic membrane organization is not confined to amoeba Chaos but is a common naturally occurring cell organelle membrane structure. Non-lamellar cubic membrane structures have been observed in numerous cell types from all kingdoms of life and in virtually any membrane-bound cell organelles, especially smooth endoplasmic reticulum (Almsherqi et al. 2006), plasma membrane, inner nuclear membrane, mitochondrial inner membrane and chloroplast thylakoid membranes (Landh 1995). However, knowledge about the formation and function of non-lamellar, cubic structures in biological systems is scarce, and research so far is restricted to the descriptive level.
Figure 1.
(a) Two-dimensional transmission electron microscopic image and (b) three-dimensional mathematical model of the same cubic membrane organization observed in 10-day starved amoeba Chaos mitochondrion. Scale bar, 500 nm.
Steps towards understanding the functional roles of non-lamellar structure of cell membranes in biological systems have been paralleled by the efforts in exploiting the polymorphic phase behaviour of lipids for the rational design of lipid-based intracellular drug and gene delivery systems. Lipid polymorphism is an essential aspect to consider in designing liposomes for biologically active agent delivery (Landy et al. 2007). For example, a lamellar phase comprising parallel strands of DNA sandwiched between fluid lipid bilayers is a prevailing system for DNA packaging (Schmutz et al. 1999), and the subsequent induction of a phase transition from the multi-lamellar ‘sandwich’ structure to an inverted hexagonal or cubic phase is associated with the efficient release of DNA molecules (Koynova et al. 2005). More recently, the potential application of dispersed liquid crystalline phases such as bicontinuous cubic phase dispersions (known as cubosomes) for drug delivery has received increasing interest (Larsson & Tiberg 2005). However, their potential as gene delivery systems is likely to be limited: the ability of cubic phase to interact with DNA is largely dependent on the charge interaction between negatively charged DNA molecules and cationic surfactants forming the cubic phases (McLoughlin et al. 2004). In this respect, cubosomes and liposomes share the same mechanism of interaction with DNA molecules. Although the biological significance of cubic membrane organization is still under investigation, the potential applications of cubic membranes deserve special attention. Given the promising capability of cubosomes in drug delivery, it is reasonable to speculate that their biological analogue (cubic membranes) might have similar functional applications. Here, we explore the ability of cubic membranes isolated from amoeba Chaos to encapsulate and then deliver DNA oligonucleotides (ODNs) to the target cells, with a view to both understand the possible function of cubic membranes in biological systems and develop a potential bio-compatible gene delivery vehicle.
2. Material and methods
2.1 Amoeba mass culture and cell harvest
A protocol for the continuous simple mass culture of amoeba, C. carolinense, feeding on only one food organism, P. multimicronucleatum, has been described previously (Tan et al. 2005). Prior to harvesting Chaos cells, the cultures were gently washed several times with amoeba medium to remove the food organism. Chaos cells were dislodged from the bottom of culture dishes by squirting with a washing bottle and placed in tall (500 ml) beakers of growth medium, in which they were allowed to settle to the bottom by gravity. The supernatant was siphoned off and the clean Chaos cells were ready for further processing. No food organisms were added to amoeba Chaos culture for 10 days in starvation treatment to obtain the mitochondria with cubic membrane organization (figure 1a).
2.2 Isolation of amoeba Chaos mitochondria
Samples of fed and starved Chaos mitochondria were isolated in the medium containing 250 mM sucrose, 10 mM EDTA, 100 mM KCl, 50 mM Tris–HCl (pH 7.4), 1 mM K2HPO4 and 2% bovine serum albumin. The harvested Chaos cells were hand-homogenated gently at 4°C. The homogenates were spun down at 1500 r.p.m. for 4 min, and the first pellet (containing many well-preserved nuclei, few mitochondria and some broken fragments of plasmalemma) was discarded. The supernatant was transferred to a new tube and centrifuged at 4500 r.p.m. for 10 min at 4°C. The second pellet, containing mainly the isolated mitochondria with the cubic membrane structure, was used for further experiments. Bradford's method was used to quantify the total mitochondrial proteins in the isolated sample (Bradford 1976).
2.3 Co-localization of ‘cubic’ mitochondria and tagged ODNs
Green fluorescent-tagged phosphorothioate oligonucleotides18 mer (PS-ODNs; 5′-tct ccc agc gtg cgc cat-3′; Sigma-Prooligo) were used in this study. Amoeba Chaos was first incubated with 10 nM MitoTracker (Red) CMXRos (Molecular Probes) for 15 min at room temperature. Mitochondria with cubic membranes (26 μg μl−1) were isolated from Chaos cells and incubated with 1.2 μg μl−1 fluorescent-tagged ODN at 1 : 1 volume ratio for 1 hour at room temperature. The sample was then purified (centrifugation in an Eppendorf centrifuge 5412C at 4500 r.p.m. for 10 min) and washed three times for further fluorescence microscopic examination. The same amount of mitochondrial proteins and ODNs was used for mice (C57BL/6J) liver mitochondria in the comparison study. The samples were viewed under a fluorescence microscope (Zeiss Axioplan2 imaging, Metasystem imaging software) using two different wavelengths (green EX: 520 nm, EM: 495 nm; red EX: 579 nm, EM: 599 nm) for the co-localization study.
2.4 Transmission electron microscopic ultrastructural study and image analysis
Mitochondria pellets (from fed, 10-day starved Chaos cells and mice liver) and treated MCF-7 (human breast adenocarcinoma cell line) cell pellets were examined by the conventional transmission electron microscopic (TEM) method. Mitochondria and MCF-7 cell pellets were first fixed in 2.5% glutaraldehyde, second fixated by 1% osmium tetroxide followed by a series of ethanol dehydration and finally embedded in Epon–Araldite. The ultra-thin sections (70–90 nm) were cut and viewed under a Jeol 1010 TEM.
2.5 Gel retardation (band shift) assay
This procedure is based on the observation that the formation of ODN and protein/or lipid complexes usually reduces the electrophoretic mobility of the ODN component in the gel matrix. The assays were carried out in a final volume of 10 μl. The same amount of mitochondrial proteins from amoeba Chaos cubic mitochondria and mice liver mitochondria (26 μg μl−1) were incubated separately with 1.2 μg μl−1 of ODN molecules at 1 : 1 ratio for 1 hour at room temperature. After incubation, the samples were loaded in the wells of 1% agarose (stained with 0.5 μg ml−1 of ethidium bromide) and electrophoresed in 1× Tris–borate–EDTA buffer for 1 hour at 80 mV at room temperature. The gel was then fixed, dried and photographed under UV light.
2.6 In vitro transfection with ODNs–cubic membrane complexes
MCF-7 (human breast adenocarcinoma) cell line was obtained from American Type Culture Collection (ATCC cat. no.: HTB-22). MCF-7 cells (1×105) were plated in six-well tissue culture plates in Dulbecco's modification of Eagle's medium (DMEM) supplemented with 10% foetal bovine serum and 1% ampicillin. After 24 hours, the medium was removed and replaced with fresh DMEM (1 ml). As cubic mitochondria are large (1–5 μm in diameter) particles (Deng & Mieczkowski 1998), ultrasound sonication of cubic mitochondria containing ODNs leads to fragmentation, forming stable submicrometre-sized particles containing ODNs. After incubation of cubic mitochondria with ODNs, 1 ml of the ODN–cubic membrane complex suspension was sonicated (Soniclean Ultrasonic, Model 250HT 6lw, Australia) for 5 min at the high amplitude setting. The submicrometre-sized complexes (fragmented cubic mitochondria+fluorescent-tagged ODNs) were added to MCF-7 cells and incubated for 5 hours at 37°C. The mixture was then removed and washed three times and replaced with fresh DMEM medium. The same protocol was used for the isolated mice liver mitochondria and tagged ODNs alone in separated wells for the comparison study. The transfected MCF-7 cells were examined under the fluorescence microscope first and further fixed in 2.5% glutaraldehyde for the TEM ultrastructural study.
3. Results
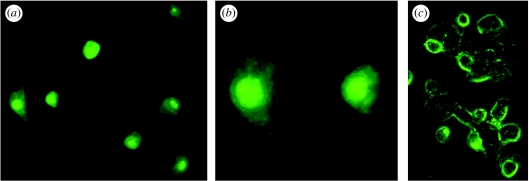
Chaos cells were starved for 10 days to ensure complete transformation of the inner mitochondrial membrane into the cubic membrane structure (Deng & Mieczkowski 1998; figure 1a). After 10 days of starvation, Chaos cells were incubated with a mitochondrial-selective fluorescent probe, MitoTracker Red (Molecular Probe), followed by extraction of the mitochondria via standard isolation techniques. These isolated and highly purified mitochondria containing cubic membranes (figure 3a) were then incubated with single-strand ODNs tagged with green fluorescent markers. An equivalent amount of ‘non-cubic’ mitochondria (isolated either from fed amoeba or mice (C57BL/6J) liver) and ODNs were incubated for comparison (figure 3c,d). Wavelength-tuned fluorescence microscopy allowed us to detect the locations of both mitochondria and ODNs. In the usual (non-cubic) mitochondria, the locations of ODNs and mitochondria were distinct. By contrast, cubic mitochondrial membranes revealed coincidence in the locations of ODNs (figure 2a) and mitochondria (figure 2b); for example, simultaneous excitation of both red and green fluorescent probes gave a visible yellow signal (figure 2c), indicative of ODN location within the mitochondria.
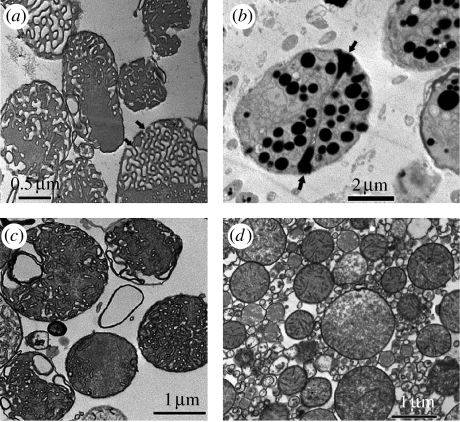
Figure 3.
TEM images of mitochondria containing cubic membrane structure isolated from 10-day starved Chaos cells (a) before and (b) after incubation with ODNs. ODN molecules interact significantly with the mitochondria exhibiting cubic membrane organization (b) but not with the same amount of ‘non-cubic’ mitochondria isolated from (c) well-fed amoeba or (d) mice liver. Multiple electron-dense intra-mitochondrial inclusions may represent cubic membrane-mediated ODN interactions. The electron-dense core may consist of ODNs–membrane lipid complexes encapsulated within distinct vesicles. A significant increase in the mitochondrial size and volume has been observed post ODN treatment, which could be attributed to the (b) ‘adsorption’ of substantial amount of ODNs. The multiple pores (arrows in (a,b)) at the surface of cubic mitochondria may play an important role in facilitating passive uptake of ODNs. Data are from a representative independent experiment performed at least five other times with similar findings.
Figure 2.
Two-colour fluorescence images analysis. (a) Green fluorescent-tagged ODNs were incubated with (b) amoeba Chaos cubic mitochondria labelled with MitoTracker Red for an hour at room temperature. (c) Overlay of the two images shows co-localization of the fluorescent-tagged ODNs and Chaos cubic mitochondria, suggesting mitochondrial uptake of the ODNs. The results illustrate a representative experiment of at least three other times with similar findings.
TEM images substantiate the apparent co-location of ODNs and cubic mitochondria. Cubic mitochondria samples (figure 3a) incubated with ODNs for 1 hour at room temperature show multiple intra-mitochondrial inclusions (figure 3b), most likely due to the association between ODNs and internal membrane lipids in cubic mitochondria. Moreover, as a result of this association, the characteristic morphological signature of the cubic membranes within the mitochondria is lost and no organized membrane morphology could be detected (figure 3b). None of these effects were observed in non-cubic mitochondria isolated from either fed amoeba (figure 3c) or mice liver (figure 3d). The surface pores, which are both abundant and very uniform in diameter in cubic mitochondria, seem to play an important role in facilitating the movement of ODNs into cubic mitochondria (figure 3a,b, arrows). Morphometric calculations suggest that 20–40% of the total volume of the mitochondria is occupied by the inclusion bodies and approximately 90% of all the cubic mitochondria show an association with ODNs.
To further examine the ability of cubic mitochondria to retain ODN molecules, gel retardation tests were performed using mice liver mitochondria and amoeba mitochondria subjected to starvation for 10 days, both incubated with the same amount of ODNs. Since the formation of ODNs–cubic membrane complexes is expected to retard electrophoretic mobility of ODN component in the gel matrix, this assay tests the degree of association between ODNs and mitochondria. As expected from the fluorescence microscopic and TEM studies, the gel tests revealed that the amount of mobile ODNs (i.e. dissociated from the starved amoeba cubic mitochondria) detected by applying an electrical field was much less than that found in the mice liver mitochondria (figure 4).
Figure 4.
Gel retardation study. The same amounts of mitochondrial proteins (26 μg μl−1) from amoeba cubic mitochondria and mice liver mitochondria were incubated with the same amount of ODN (0.1 μg μl−1) molecules. The mitochondria with cubic membrane organization are able to retard ODN mobilization (lane 2) towards the positive pole when compared with the same amount of pure ODNs (lane 1) and ODNs incubated with isolated mice liver mitochondria (lane 3). The results illustrate a representative experiment of at least three other times with similar findings, showing one of the duplicate samples.
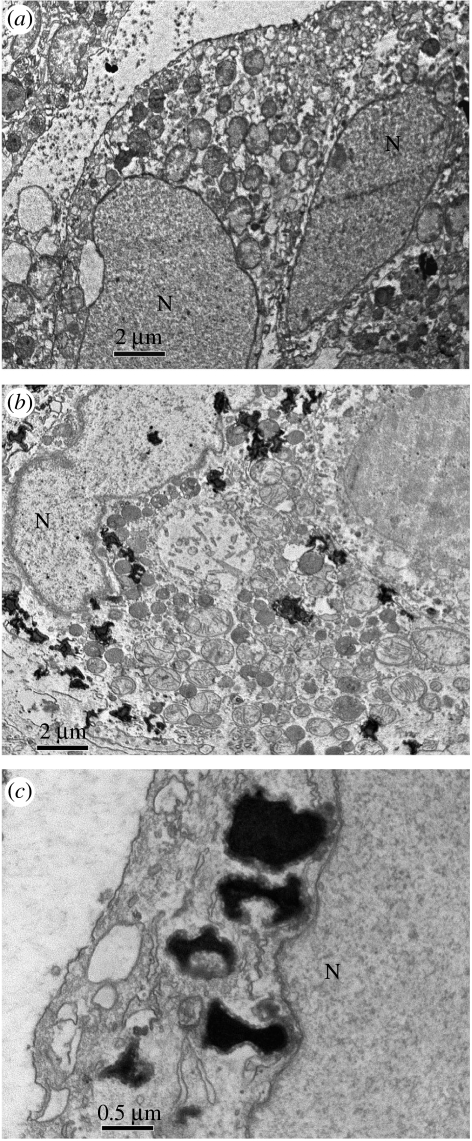
Finally, we have tested targeted cells for the uptake of fragmented ODN–cubic membrane complexes. Cubic mitochondria are large (1–5 μm in diameter); however, ultrasound sonication of starved mitochondria containing ODNs leads to the fragmentation, forming stable submicrometre-sized particles containing ODNs. MCF-7 cells were incubated with these fragmented ODNs–membrane complexes and further examined under the fluorescence microscope (figure 5a,b) and phase-contrast microscope (figure 5c). In MCF-7 cells incubated with ODNs–membrane complexes, the tagged ODNs were found in both cytoplasm and nucleus (figure 5a,b). The ODNs–membrane complexes' internalization procedure was further examined under TEM at different time points, and data showed several cytoplasmic inclusions (figure 6b,c), including some adjacent to the target nucleus (figure 6c). There are no inclusion bodies observed in MCF-7 cells without ODN treatment (figure 6a) or MCF-7 cells incubated with naked ODNs (not shown). Indeed, similar inclusion bodies have been observed in vitro and in vivo (Zabner et al. 1995) in TEM micrograph analysis of culture cells treated with lipoplex, a cationic lipid–DNA complex. Evidently, the mitochondrial fragments offer robust delivery and release vehicles for at least short strands of DNA.
Figure 5.
In vitro study of ODN–cubic membrane complex internalization into MCF-7 cells. The MCF-7 cells were incubated with stable submicrometre-sized particles of cubic membranes containing ODNs. After 5 hours of treatment, highly fluorescent MCF-7 cells were observed under (a) low, (b) high magnification of fluorescence and (c) phase-contrast microscope. The green fluorescent-tagged ODNs are observed in the cytoplasm and the nucleus ((a,c) 64×, (b) 200×). The results illustrate a representative experiment of at least three to five other times with similar findings.
Figure 6.
TEM images of the MCF-7 cells, (a) before and 5 hours post treatment with (b) ODNs–membranes complexes. Multiple electron-dense inclusion bodies (ODN–cubic membrane complexes) are frequently observed within the cytoplasm and some targeted the nucleus (N) of the treated MCF-7 cells (b) while there are no such inclusion bodies observed in MCF-7 cells before ODN treatment (a). At higher magnification, membrane association was frequently observed between electron-dense inclusion bodies and nuclear membranes (c). Data are from a representative experiment performed at least three to five other times with similar findings.
4. Discussion
The morphology of mitochondrial cristae can be modified under a range of physiological and pathological conditions, particularly stressed conditions (Munn 1974). TEM and electron microscopy tomography studies reveal that mitochondrial cristae in amoeba Chaos undergo conformational changes under starvation–stress conditions, leading to the formation of unambiguous cubic membrane morphology (Deng & Mieczkowski 1998; Deng et al. 1999). On the other hand, alternative functions of mitochondria besides ATP synthesis have been proposed, which include the participation in apoptosis (programmed cell death), heat production, control of second messenger levels, steroid and haem syntheses, cell proliferation, cellular redox state and detoxification (Zorov et al. 1997). Usually, mitochondrial membrane organizations support the functional features of the mitochondria, implicating a structure–function relationship. Thus, the recognition of the cubic membrane structure in mitochondria, with a capacity for passive uptake and retention of ODNs suggests a potential role of these membrane organizations in the intracellular macromolecule transportation and/or communication between the mitochondria and other intracellular compartments. Other proposed functions of the cubic membranes in biological systems include cellular and organelle volume regulation, cell space compartmentalization, attenuation of intracellular oxidative stress and light manipulation (Landh 1996; Almsherqi et al. 2006). Furthermore, we demonstrate that this unique property of cubic membranes could be employed to transfer short segments of DNA to targeted mammalian cells.
In spite of their geometrical analogy, there are significant differences between cubic membranes and cubosomes. In particular, the lattice sizes of cell membrane systems are usually larger than 100 nm (Landh 1996). On the other hand, the lattice dimension in lipid systems tends to be smaller than 20 nm (Luzzati 1997). In addition, the size of channels or pores (50–80 nm) in biological membrane networks is approximately 10-fold that observed in cubosomes (Angelov et al. 2003). As a result, cubic membranes could provide a practical and stable vehicle for accommodating ‘rigid’ macromolecules (such as DNA) with the provision of their curved labyrinth in three dimensions.
At this stage, it is difficult to offer a definitive explanation for the affinity of ODNs for cubic membranes formed within the starved amoeba Chaos mitochondria. However, we can assert that the adsorption is not an electrostatic effect induced by cationic lipids. Detailed lipid profile analysis of starved amoeba with cubic mitochondria morphology has been performed using a mass spectra technique (Z. Almsherqi et al. 2006, 2007, unpublished data). The results show that the phospholipids composition of amoeba Chaos is similar to that of mammalian cell membranes. Although unsaturated fatty acids are predominant in Chaos cells, no significant cationic lipids are detectable. The affinity of ODNs for cubic membranes is possibly due to the distinct chemical nature of the exposed curved membranes, induced by the membrane conformation in the vicinity of the pores. For example, heterogeneity of molecular species in these regions may give rise to local charging around the pore opening of the cubic membranes, allowing the absorption of ODNs via electrostatic interactions. An alternative explanation involves the lipid species found in these cubic membranes.
Three possible modes of intracellular trafficking have been proposed: clathrin-mediated endocytosis; caveolae-mediated endocytosis; and macropinocytosis (Conner & Schmid 2003; Kirkham & Parton 2005). No endosomal or lysosomal vesicles have been observed in our TEM images of MCF-7 cells 5 hours post treatment. Thus, cubic membranes as a gene vector would probably avoid endosomal/lysosomal degradation. By contrast, it is well known that most cationic complexes are internalized through endocytosis with only a fraction of DNA molecules escaping lysosomal degradation (Zabner et al. 1995; Labat-Moleur et al. 1996; Howell et al. 2003).
A significant obstacle to gene therapy is the potential toxicity of current delivery vehicles. Non-viral vectors such as cationic lipids have become a prevalent method due to the potential fatality of viral vectors. Although cationic lipids are relatively efficient in packaging DNA, high cellular toxicity remains a concern (Conwell & Huang 2005). It is therefore of interest to explore an alternative delivery vehicle with the features of efficient DNA packaging, DNA release into the cytoplasm and nucleus targeting in addition to the low toxicity. This study is the first attempt in the exploration of the ability of the non-lamellar cubic membranes to absorb, retain and deliver short segments of DNA molecules. However, these results should be regarded as preliminary observations: several parameters such as the efficiency, cytotoxicity and immunogenicity remain to be evaluated and compared with the currently available methods and techniques.
Our results demonstrate that mitochondria containing ordered cubic sponges with hyperbolically curved membranes readily adsorb DNA ODNs, with significant molecular uptake. By contrast, insignificant uptake occurs where the inner mitochondrial membranes display tubular or lamellar organizations. Thus, cubic membranes—prepared from, for example, amoeba Chaos mitochondria—may afford a new transfection vector for short segments of DNA or other agents that are clinically valuable for disease diagnosis and/or treatment, such as small interfering RNA and/or short hairpin RNA.
Acknowledgments
The authors are grateful to Kåre Larsson for initiating this work, Barry Ninham for his insightful comments, Yuhong Xu, Shali Shen, Ratha Mahendran for their fruitful suggestions to set up the experiments during this work and Ms Chwee Wah Low for her technical support in TEM sample preparations. This work is supported by research grant NMRC (R-185-000-058-213) from Singapore to Y.D. and an Australian Research Council Federation Fellowship to S.H.
References
- Almsherqi Z.A, Kohlwein S.D, Deng Y. Cubic membranes: a legend beyond the Flatland* of cell membrane organization. J. Cell Biol. 2006;173:839–844. doi: 10.1083/jcb.200603055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov B, Angelova A, Ollivon M, Bourgaux C, Campitelli A. Diamond-type lipid cubic phase with large water channels. J. Am. Chem. Soc. 2003;125:7188–7189. doi: 10.1021/ja034578v. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Conner S.D, Schmid S.L. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Conwell C.C, Huang L. Recent advances in non-viral gene delivery. Adv. Genet. 2005;53:1–18. doi: 10.1016/S0065-2660(05)53001-3. [DOI] [PubMed] [Google Scholar]
- Deng Y, Mieczkowski M. Three-dimensional periodic cubic membrane structure in the mitochondria of amoebae Chaos carolinensis. Protoplasma. 1998;203:16–25. doi: 10.1007/BF01280583. [DOI] [Google Scholar]
- Deng Y, Marko M, Buttle K.F, Leith A, Mieczkowski M, Mannella C.A. Cubic membrane structure in amoeba (Chaos carolinensis) mitochondria determined by electron microscopic tomography. J. Struct. Biol. 1999;127:231–239. doi: 10.1006/jsbi.1999.4147. [DOI] [PubMed] [Google Scholar]
- Howell D.P, Krieser R.J, Eastman A, Barry M.A. Deoxyribonuclease II is a lysosomal barrier to transfection. Mol. Ther. 2003;8:957–963. doi: 10.1016/j.ymthe.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Hyde S, Andersson S, Larsson K, Blum Z, Landh T, Lidin S, Ninham B.W. Elsevier; Amsterdam, The Netherlands: 1997. The language of shape. The role of curvature in condensed matter: physics, chemistry, and biology. [Google Scholar]
- Kirkham M, Parton R.G. Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim. Biophys. Acta. 2005;1746:349–363. doi: 10.1016/j.bbamcr.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Koynova R, Wang L, Tarahovsky Y, MacDonald R.C. Lipid phase control of DNA delivery. Bioconjug. Chem. 2005;16:1335–1339. doi: 10.1021/bc050226x. [DOI] [PubMed] [Google Scholar]
- Labat-Moleur F, Steffan A.M, Brisson C, Perron H, Feugeas O, Furstenberger P, Oberling F, Brambilla E, Behr J.P. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;11:1010–1017. [PubMed] [Google Scholar]
- Landh T. From entangled membranes to eclectic morphologies: cubic membranes as subcellular space organizers. FEBS Lett. 1995;369:13–17. doi: 10.1016/0014-5793(95)00660-2. [DOI] [PubMed] [Google Scholar]
- Landh, T. 1996 Cubic cell membrane architectures. Taking another look at membrane bound cell spaces. PhD thesis, Lund University, Sweden.
- Landy P, Pollien P, Rytz A, Leser M.E, Sagalowicz L, Blank I, Spadone J.C. Model studies on the release of aroma compounds from structured and nonstructured oil systems using proton-transfer reaction mass spectrometry. J. Agric. Food Chem. 2007;55:1915–1922. doi: 10.1021/jf062643p. [DOI] [PubMed] [Google Scholar]
- Larsson K, Tiberg F. Periodic minimal surface structures in bicontinuous lipid–water phases and nanoparticles. Curr. Opin. Colloid Interface Sci. 2005;9:365–369. doi: 10.1016/j.cocis.2004.12.002. [DOI] [Google Scholar]
- Larsson K, Quinn P, Sato K, Tiberg F. In: Lipids: structure, physical properties and functionality. Barnes P.J, editor. The Oily Press; Bridgwater, UK: 2006. [Google Scholar]
- Luzzati V. X-ray diffraction studies of lipid–water systems. In: Chapman D, editor. Biological membranes. Academic Press; New York, NY: 1968. pp. 71–123. [Google Scholar]
- Luzzati V. Biological significance of lipid polymorphism: the cubic phases. Curr. Opin. Struct. Biol. 1997;7:661–668. doi: 10.1016/S0959-440X(97)80075-9. [DOI] [PubMed] [Google Scholar]
- McLoughlin D, Imperor-Clerc M, Langevin D. A new cubic phase containing DNA and a surfactant. ChemPhysChem. 2004;5:1619–1623. doi: 10.1002/cphc.200400163. [DOI] [PubMed] [Google Scholar]
- Munn E.A. Academic Press; London, UK: 1974. The structure of mitochondria. [Google Scholar]
- Sagalowicz L, Acquistapace S, Watzke H.J, Michel M. Study of liquid crystal space groups using controlled tilting with cryogenic transmission electron microscopy. Langmuir. 2007;23:12 003–12 009. doi: 10.1021/la701410n. [DOI] [PubMed] [Google Scholar]
- Schmutz M, Durand D, Debin A, Palvadeau Y, Etienne A, Thierry A.R. DNA packing in stable lipid complexes designed for gene transfer imitates DNA compaction in bacteriophage. Proc. Natl Acad. Sci. USA. 1999;96:12 293–12 298. doi: 10.1073/pnas.96.22.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan O.L.-L, Almsherqi Z.A, Deng Y. A simple mass culture of the amoeba Chaos carolinense: revisit. Protistology. 2005;4:185–190. [Google Scholar]
- Zabner J, Fasbender A.J, Moninger T, Poellinger K.A, Welsh M.J. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18 997–19 007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- Zorov D.B, Krasnikov B.F, Kuzminova A.E, Vysokikh M.Y, Zorova L.D. Mitochondria revisited. Alternative functions of mitochondria. Biosci. Rep. 1997;17:507–520. doi: 10.1023/A:1027304122259. [DOI] [PubMed] [Google Scholar]