Abstract
The objective of this study was to examine the suitability of multiplex ligation-dependent probe amplification (MLPA) in chorionic villus samples as a replacement for traditional karyotyping for the detection of (an)euploidies of chromosomes 21, 18, 13, X, and Y. Chorionic villus samples were diagnosed by traditional karyotyping using short-term cultures (STC) and long-term cultures (LTC), and by MLPA using kit P095. DNA was extracted after digestion of whole villi with proteinase K and/or trypsin and collagenase. Different cell-dissociation procedures were tested to obtain MLPA results representative of the cytotrophoblast layer and the mesenchymal core. Over 95% of the MLPA results were in concordance with the traditional karyotyping of STC and LTC. Traditional karyotyping revealed seven mosaics. After digestion of whole villi with proteinase K, only abnormal cell lines confined to the STC gave rise to abnormal MLPA results. In one sample, the complete discrepancy between STC and LTC was resolved after enzymatic dissociation of cells from the cytotrophoblast layer and the mesenchymal core. MLPA in chorionic villus samples was found to be a reliable test for the detection of (an)euploidies of chromosomes 21, 18, 13, X, and Y. Whole villi digestion with proteinase K resulted in the over-representation of cytotrophoblasts in the DNA pool. To obtain MLPA results representative for STC and LTC, enzymatic dissociation of cells from the cytotrophoblast layer and mesenchymal core is required.
Chorionic villus sampling has been widely accepted as a technique for first trimester prenatal diagnosis and is performed from 11 weeks of gestation. Until recently, prenatal diagnosis of chorionic villus samples (CVS) was accomplished through tissue culture and subsequent cytogenetic analysis. This procedure is labor intensive and time-consuming. Therefore, more rapid and comprehensive methods for the prenatal diagnosis of CVS are currently being developed and implemented. In a number of prenatal centers in Europe, quantitative fluorescent PCR (QF-PCR) analysis is already being offered to women undergoing invasive testing by chorionic villus sampling.1 In parallel, we have implemented multiplex ligation-dependent probe amplification (MLPA) for the rapid detection of (an)euploidies of chromosomes 21, 18, 13, X, and Y in amniotic fluid cells.2,3,4 A general disadvantage of the use of CVS in comparison with amnion fluid is the extra-embryonic nature of this tissue. Although fetus and placenta originate from the same zygote, a discrepancy between the chromosomal constitution of cells in the placenta and cells in the fetus, known as chromosomal mosaicism, can occur. Such mosaicisms are well documented in the literature and are detected in 1% to 2% of the CVS.5,6 Abnormal mosaic cells can be found in both fetal and placental tissues, or may be confined to either the placenta (confined placental mosaicism, cpm) or the fetus.7 Karyotypes of CVS represent cells from chorionic ectoderm (cytotrophoblasts) in short-term cultures (STC) and chorionic mesoderm (mesenchymal core) in long-term cultures (LTC). In molecular testing of CVS it is, therefore, of obvious importance to establish that both cell lineages are adequately represented by the pool of cells from which the DNA is extracted.8 In this study we investigated the suitability of the MLPA test for the detection of (an)euploidies in CVS and assayed to what extent this test compares to traditional karyotyping (TK) of STC, LTC, or both.
Materials and Methods
Clinical Samples
CVS with a weight of >30 mg (N = 152), were collected at the outpatient clinics located in Nijmegen, Arnhem, Tilburg, ‘s-Hertogenbosch and Enschede (Network Prenatal Diagnostics Nijmegen, the Netherlands) from pregnant women at 11 to 21 weeks of gestation. From these 152 CVS, 125 were consecutively collected between May 2006 and June 2007. Additionally, twenty CVS with known aneuploidies for one of the target chromosomes and seven CVS diagnosed by TK as mosaic were added. The referral reasons of the pregnant women ranged from low-risk to high-risk. The CVS were washed in PBS and the villi were separated from maternal decidua and blood clots under an inverted microscope.
Karyotyping
Approximately 20 to 30 mg of the villi was used for conventional karyotyping according to standard STC and LTC procedures. Briefly, 10 to 15 mg of the villi was used for STC and, subsequently, incubated for 30 minutes in colcemid, followed by a short hypotonic treatment after which the cells were fixed in methanol/acetic acid (3:1) and rehydrated. Finally, the trophoblast (interphase and metaphase) cells were released from the villus core using 60% acetic acid and spread on microscopic slides. The remaining 10 to 15 mg of the villi were used for LTC, after incubation for one hour in trypsin-EDTA and a 40-minute incubation in collagenase. Metaphases were harvested in situ using standard procedures on Labtek II chamber slides. Cytogenetic investigation of STC and LTC was routinely performed and 4 and 8 metaphases were analyzed, respectively, to exclude discrepancies between STC and LTC.9 Cytogenetic examination of the LTC was expanded to 29 metaphases when an abnormal karyotype was detected in STC or LTC cells. CVS karyotypes with a tetrasomy or triploidy were excluded from this study.
Definition of Mosaic Levels
CVS encompass cells from both the trophoblasts and the mesenchymal core. Discordant findings between STC and LTC and/or fetal tissues have been either referred to as pseudomosaicisms or true mosaicisms.10 Here, three levels to define mosaicism were used: level I, detection of a single abnormal cell (single cell pseudomosaicism), level II, the same abnormality was detected in two or more cells in the same culture vessel (multiple cell pseudomosaicism) or level III, the same abnormality was observed in two or more independent culture vessels (true mosaicism).10 Discrepancies between karyotypes of villus and fetal tissues may occur as a result of a cpm. Three types of cpm can be discerned and categorized by the placental cell lineage exhibiting the abnormal cell line, ie, confined to the cytotrophoblast (type I), the mesenchymal core (type II), or both (type III).5
Fluorescence in Situ Hybridization Analysis
Interphase fluorescence in situ hybridization (I-FISH) analyses were performed on nuclei (N = 100) of STC villi cells in samples exhibiting a mosaicism of any of the target chromosomes using an AneuVysion prenatal detection kit (Vysis Inc, Downers Grove, Ilinois) for an accurate establishment of the distribution of normal and abnormal cells.
DNA Extraction Following Proteinase K Treatment of Whole Villi
This method was used for all CVS. DNA from at least two chorionic villi was extracted using a QIAamp DNA Mini Kit (Qiagen) following the protocol “Isolation of DNA from soft tissues using the TissueLyser and QIAamp DNA Mini Kit” (Qiagen, Westburg bv, The Netherlands) In this procedure, incubation at 56°C with proteinase K results in lysis of the villi before DNA extraction. Proteinase K has a specific activity and degrades tissue to facilitate the purification of the DNA. Finally, DNA was eluted in 50 μl AE buffer (10 mmol/L Tris-HCl, 0.5 mmol/L EDTA, pH 9.0). The DNA was quantified using a Nanodrop spectrophotometer (NanoDrop Technologies Wilmington, Delaware) and varied from 2.5 to 650 ng/μl elution buffer.
DNA Extraction Following Enzymatic Dissociation of Villi
Two methods were designed to obtain cell populations from the cytotrophoblast layer and the mesenchymal core separately. In the first method, a modification of the method described by Mann,8 ie, digestion of cleaned villi with collagenase (800 units/ml; 37°C, 30 minutes), was followed by trypsin digestion (0.5% trypsin/EDTA, 37°C, 30 minutes). After collagenase digestion, the suspension was separated from the remaining villi and transferred to a tube containing 4 ml PBS and 10% fetal calf serum to stop the digestion. After centrifugation (1200 rpm, 5 minutes) the supernatant was removed and the cell pellet was re-suspended in 300 μl PBS (fraction C). After digestion of the remaining villi with trypsin, 4 ml PBS + 10% fetal calf serum were added to stop the reaction. After centrifugation (1200 rpm, 5 minutes) the supernatant was removed and the cell pellet was re-suspended in 300 μl PBS (fraction M). Finally, 100 μl of fraction C and 100 μl of fraction M were mixed (1:1) (fraction T) and used for DNA isolation as described below.
In the second method, digestion of cleaned villi was first performed with trypsin (0.5% trypsin/EDTA, 37°C, 1 hour) followed by collagenase digestion (800 units/ml; 37°C, 40 minutes). After both digestion steps, cell fractions C, M, and T were obtained as in the first method.
DNA extractions were performed by incubation of the cell population with proteinase K at 56°C. DNA was purified using a QIAamp DNA Mini Kit (Qiagen) following the protocol “Blood and body fluid spin protocol.” Finally, DNA was eluted in 50 μl AE buffer (10 mmol/L Tris-HCl, 0.5 mmol/L EDTA, pH 9.0).
Both methods were performed on CVS case 7 with discrepant results in STC and LTC, ie, 46,XY and 47,XY,+18, respectively.
MLPA Test
The MLPA test was performed with SALSA MLPA kit P095 for aneuploidy detection as described by Schouten (MRC Holland, Amsterdam, The Netherlands).11 The MLPA analyses were performed blinded to the cytogenetic analyses until completion. Data analyses were performed on the transfer of electropherogram-based GeneMapper results to a modified spreadsheet for normalization and ratio computation of the peak areas (Kooper et al, submitted). First an intrasample normalization of each probe peak area was performed, followed by an intersample normalization with tray-specific references. Normalization is essential because variations in experimental conditions may lead to quantitative differences.
As a consequence of our normalization procedure, mean probe ratios skew in case of a trisomy, since the probe ratios of the targets with two copies scale down to 80% when a trisomy for one of the targets is present. This decrease in mean probe ratios is not corrected in the data analysis. The mean and SD of each target chromosome were determined, followed by a 95% confidence interval (CI). In cases of disomy (two copies of each target chromosome), these calculations will result in a theoretical (expected) value of 1.0, representing two copies of the target sequence in the sample. One or three copies of the target sequence in a sample will result in theoretical values of 0.5 or 1.5, respectively. A result is considered abnormal when the theoretical value (= expected value in a normal case) is not included in the confidence interval and the mean probe ratio is more or less than 10% of the expected value. Follow-up of these samples is performed by karyotyping and/or amniocentesis.
MLPA does not detect female triploidies. As a consequence of the normalization procedure ie, relative peak areas do not differ in intrasample normalizations as the copy numbers of all of the targets is the same. A male triploidy will yield normal mean probe ratios for 21, 18, and 13, an increased mean probe ratio for X and a decreased mean probe ratio for Y. In addition, there is limited potential to discriminate maternal contamination in a normal male sample from 69,XXY and, therefore, careful CVS cleaning before sample preparation is of critical importance.
The Levene's test was used to determine equality of variances of the pooled standard deviations of the mean probe ratios for the target chromosomes 13, 18, 21, and X between CVS and amniotic fluid samples.
Results
Proteinase K Treatment of Whole Villi
In all 152 CVS, the MLPA test was performed successfully. The results were compared with cytogenetic analyses of both STC villi and LTC villi. There was a correct assignment of sex in all 152 samples. No evidence for the presence of maternal contamination was noted in male samples, ie, the expected normalization effect of a decreased mean probe ratio for the Y chromosome combined with an increased mean probe ratio for the X chromosome was observed in none of the male samples.
A summary of the test results on all 152 samples is displayed in Figure 1, showing a boxplot with the mean ratios for the chromosome 21, 18, 13, X, and Y probes in euploid (N = 124: normal male N = 58 and normal female N = 66), aneuploid (N = 21), and (pseudo)mosaic samples (N = 7). In all normal samples, the mean ratios for the autosomes were close to the expected value of 1.0. The mean probe ratios for samples with a trisomy 21, 18, or 13 were all significantly increased (mean ratios of >1.0 with 95% CI not including 1.0) and close to the expected value of 1.5. In all normal male and female samples the mean probe ratios for X and Y were close to the expected values: 1.0 for X in female samples and 0.5 for X and Y in male samples, and for X in samples with a monosomy X. The variation in mean probe ratios was slightly higher for Y, which may be due to the limited number of targets included in the P095 kit for this chromosome.
Figure 1.
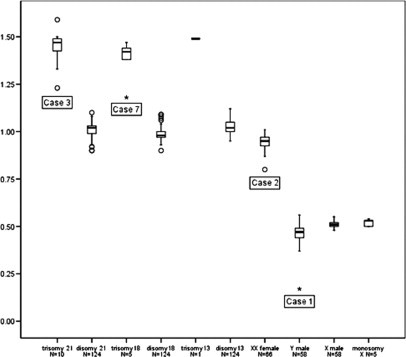
Quantitative MLPA analysis with mean probe ratios of the 21, 18, 13, X, and Y targets with 50% of the samples within the box. The outliers are cases ‘o’ with values between 1.5 and 3 box-lengths from the boundaries of the box. Case 3 shows an increased mean probe ratio for chromosome 21 of 1.24 (95% CI, 1.02−1.46). Inspection of individual probe ratios of this chromosome showed an increase for all individual probes, indicative for a mosaic of trisomy 21. Case 7 is illustrated as an extreme (*) in the box plot with an increased mean probe ratio for chromosome 18 of 1.18 (95% CI, 1.08−1.28), ie, this ratio is increased for the expected value of 1.0 for a disomy 18 and decreased for the expected ratio of 1.5 for a trisomy and, therefore, indicative for a mosaic trisomy 18. Case 1 is illustrated as an extreme (*) in the box plot with a mean probe ratio for the Y chromosome of 0.17 (95% CI, 0.14−0.21). Case 2 is illustrated as an outlier ‘o’. The mean probe ratio for X is 0.80 (95% CI, 0.70−0.90), ie, this ratio is increased for the expected value of 0.5 for a monosomy X and decreased for the expected ratio of 1.0 for a disomy and, therefore, indicative for a mosaic 45,X/46,XX. The other outliers in the box plots result in increased or decreased mean probe ratios, all related to broad confidence intervals. The expected values of 0.5, 1.0, or 1.5 are within the confidence intervals in all outliers.
Normal Karyotypes
In 124 of the CVS the normal results of MLPA and TK were in concordance for all five target chromosomes (58 and 66 male and female samples, respectively).
Aneuploidies in both STC and LTC
In total, 21 aneuploidies for one of the target chromosomes were detected; one with trisomy 13, five with trisomy 18, ten with trisomy 21 and five with monosomy X. There was complete concordancy between the MLPA and TK results. In all 145 non-mosaic samples, the unexplained SD of the mean probe ratio for the target chromosomes 13, 18, 21 and X turned out to be 0.041. The Y chromosome was excluded in this calculation because of the presence of only four probes (see above). This residual variation was significantly lower (P < 0.001, using F test to compare two variances) than the corresponding value of 0.068 determined in a set of amniotic fluid samples (Kooper et al, submitted).
(Pseudo)mosaicisms
Seven samples displayed discordancies between TK (STC and/or LTC) and MLPA. The results of TK (STC and LTC), I-FISH, referral reason, gestational age and follow-up of these cases are summarized in Table 1. MLPA results of these mosaic cases were compared with the TK results (golden standard) and are summarized in Table 2.
Table 1.
Results of Traditional Karyotyping of STC, LTC, and Interphase FISH for One of the Target Chromosomes in Seven CVS, Including Mosaic and Pseudomosaic Cases
| Case no | Referral reason | Gestational weeks | STC karyotype | STC I-FISH (% in 100 nuclei) | LTC karyotype | Diagnosis amniocentesis | Mosaicism level (STC/LTC) | CPM type |
|---|---|---|---|---|---|---|---|---|
| 1 | US-abn | 21 | 45,X[2]/46,XY[1] | Test failure | 46,XY[29] | n.a. | II/− | |
| 2 | US-abn | 11 | 45,X[5]/47,XXX[1] | Monosomy X (88) | 47,XXX[29] | n.a. | II/− | |
| Disomy X (3) | ||||||||
| Trisomy X (9) | ||||||||
| 3 | US-abn | 13 | 47,XY,+21[2]/ 46,XY[5] | Disomy 21 (76) Trisomy 21 (24) | 47,XY,+21[29] | n.a. | II/− | |
| 4 | MA | 11 | 46,XX[4] | Monosomy X (5) Disomy X (95) | 45,X[3]/46,XX[27] | 46,XX | −/II | II |
| 5 | RIS | 13 | 46,XY[4] | Test failure | 47,XY,+13[2]/ 46,XY[27] | 46,XY | −/II | II |
| 6 | DNA | 11 | 45,X[3]/46,XX[1] | Monosomy X (26) Disomy X (74 | 45,X[1]/46,XX[28] | 46,XX | II/I | III |
| 7 | US-abn | 11 | 46,XY[12] | Disomy 18 (100) | 47,XY,+18[29] | n.a. | −/− |
MA, maternal age >35 years; CPM II, confined placental mosaicism type II; CPM III, confined placental mosaicism type III; US-abn, ultrasound abnormality; RIS, at risk for Down after first trimester screening; n.a., not available (not performed); STC, short-term culture; LTC, long-term culture; DNA, primary reason for invasive testing was the presence of a familial DNA mutation.
Table 2.
Overview of the Chromosome Mean Probe Ratios in Seven Mosaic Cases after Preparation of Whole Villi with Proteinase K Treatment
| Case | STC karyotype | LTC karyotype | Mean probe ratios target chromosomes (95% CI) |
|---|---|---|---|
| 1 | 45,X[2]/46,XY[1] | 46,XY[29] | #13: 1.00 (0.90−1.09) |
| #18: 0.97 (0.94−1.01) | |||
| #21: 1.04 (0.95−1.13) | |||
| #X: 0.52 (0.48−0.56) | |||
| #Y: 0.17 (0.14−0.21) | |||
| 2 | 45,X[5]/47,XXX[1] | 47,XXX[29] | #13: 1.04 (0.98−1.10) |
| #18: 0.93 (0.82−1.04) | |||
| #21: 1.05 (0.91−1.19) | |||
| #X: 0.80 (0.70−0.90) | |||
| #Y: − | |||
| 3 | 47,XY,+21[2]/46,XY[5] | 47,XY,+21[29] | #13: 0.89 (0.62−1.16) |
| #18: 0.93 (0.75−1.10) | |||
| #21: 1.24 (1.02−1.46) | |||
| #X: 0.55 (0.45−0.65) | |||
| #Y: 0.43 (0.34−0.52) | |||
| 4 | 46,XX[4] | 45,X[3]/46,XX[27] | #13: 1.00 (0.84−1.15) |
| #18: 0.97 (0.83−1.10) | |||
| #21: 1.07 (0.93−1.21) | |||
| #X: 0.89 (0.80−0.99) | |||
| #Y: − | |||
| 5 | 46,XY[4] | 47,XY,+13[2]/46,XY[27] | #13: 1.02 (0.91−1.13) |
| #18: 0.97 (0.89−1.05) | |||
| #21: 1.02 (0.94−1.11) | |||
| #X: 0.49 (0.45−0.52) | |||
| #Y: 0.48 (0.46−0.50) | |||
| 6 | 45,X[3]/46,XX[1] | 45,X[1]/46,XX[28] | #13: 1.05 (0.96−1.13) |
| #18: 0.96 (0.90−1.03) | |||
| #21: 1.01 (0.92−1.10) | |||
| #X: 0.90 (0.83−0.96) | |||
| #Y: − | |||
| 7 | 46,XY[12] | 47,XY,+18[29] | #13: 0.91 (0.83−0.99) |
| #18: 1.18 (1.08−1.28) | |||
| #21: 0.92 (0.85−0.99) | |||
| #X: 0.51 (0.47−0.56) | |||
| #Y: 0.45 (0.35−0.54) |
, chromosome.
Case 1: STC villi showed a mosaic karyotype 45,X[2]/46,XY[1] and a normal karyotype in LTC. I-FISH of STC nuclei resulted in a test failure. The MLPA results showed a decreased mean probe ratio for Y of 0.17 (95% CI, 0.14−0.21) and a normal mean probe ratio for X. A normal mean probe ratio for X indicates that the decrease of Y is not related to the presence of maternal contamination in this male sample. Case 1 is illustrated as an extreme (*) in the box plot in Figure 1 (with a mean probe ratio of more than three box-lengths from the boundaries of the box) and indicative for a mosaic 45,X/46,XY.
Case 2: STC villi showed a mosaic karyotype 45,X[5]/47,XXX[1] and LTC villi showed a non-mosaic triple X karyotype. I-FISH of STC nuclei resulted in a mosaic monosomy X/disomy X/trisomy X in 68%, 3%, and 9% of the cells, respectively. The MLPA results showed a decreased mean probe ratio for X of 0.80 (95% CI, 0.70−0.90). The ratio is increased for the expected value of 0.5 for a monosomy X and decreased for the expected ratio of 1.0 for a disomy and, therefore, indicative for a mosaic 45,X/46,XX. Case 2 is illustrated as an outlier ‘o’ in the box plot in Figure 1 with a mean probe ratio between 1.5 and 3 box-lengths from the boundaries of the box.
Case 3: STC villi showed a mosaic karyotype 47,XY,+21[2]/46,XY[5] and LTC villi showed a non-mosaic trisomy 21 karyotype. I-FISH of STC nuclei resulted in a mosaic trisomy 21/disomy 21 in 76% and 24% of the cells, respectively. The mean probe ratio for chromosome 21 was increased to 1.24 (95% CI, 1.02−1.46). Inspection of individual probe ratios of this chromosome showed an increase for all individual probes indicative for a mosaic trisomy 21. There was discordance between the MLPA results and the results of TK of the LTC villi: the expected value of 1.5 was not within the 95% CI.
Case 4: LTC villi showed a mosaic karyotype 45,X[3]/46,XX[27] and STC villi showed a normal female karyotype. I-FISH of STC nuclei showed disomy X in 95% of the nuclei. The MLPA resulted in normal mean probe ratios of all target chromosomes (mean probe ratio × 0.89, 95% CI 0.80−0.99, this value is in the gray zone and interpreted as normal). Follow-up cytogenetic analysis of amniotic fluid cells was normal, indicating that the abnormal cell line was probably confined to the placenta and the LTC (cpm type II).
Case 5: LTC villi showed a mosaic karyotype of a low-grade mosaicism 47,XY,+13[2]/46,XY[27] and STC villi showed a normal male karyotype. I-FISH of STC nuclei resulted in a test failure. The MLPA resulted in normal mean probe ratios of all target chromosomes. Follow-up cytogenetic analysis of amniotic fluid cells was normal, indicating that the abnormal cell line was confined to the placenta and the LTC (cpm type II).
Case 6: both STC and LTC villi showed a mosaic karyotype 45,X/46,XX. I-FISH of STC nuclei confirmed a mosaic monosomy X/disomy X in 26% and 74% of the cells, respectively. The MLPA resulted in a slight decrease of all probe ratios for the X chromosome of 0.90 (95% CI, 0.83−0.96). Follow-up cytogenetic analysis of amniotic fluid cells was normal, indicating that the abnormal cell line was confined to the placenta, both the STC and LTC (cpm type III).
Case 7 showed a complete discrepancy between STC and LTC: the STC showed a 46,XY and the LTC a 47,XY,+18 karyotype. The MLPA resulted in a slight increase of the mean probe ratio for chromosome 18 of 1.18 (95% CI, 1.08−1.28). The MLPA results of DNA from whole villi were compared to the results of the enzymatically dissociated cell pools (Table 3). The mean probe ratios for chromosome 18 in fraction C of both methods showed a 95% CI including the value of 1.0, indicating that the cell pool isolated in fraction C represented a pool of cytotrophoblastic cells. The mean probe ratios for chromosome 18 in fraction M of both methods showed a 95% CI including the value of 1.5, indicating that the cell pool isolated in fraction M represented cells from the mesenchymal core. Fraction T, a mixture of fraction C and M (1:1), yielded results indicative for a mosaic trisomy 18, using both methods.
Table 3.
MLPA Results of CVS (Case 7) with STC 46,XY and LTC 47,XY,+18 Using Three Different Sample Preparation Methods
| Sample preparation method | Mean probe ratios (95% CI) fraction C | Mean probe ratios (95% CI) fraction M | Mean probe ratios (95% CI) fraction T |
|---|---|---|---|
| Whole villi proteinase K treatment | - | - | #13: 0.91 (0.83−0.99) |
| #18: 1.18 (1.08−1.28) | |||
| #21: 0.92 (0.85−0.99) | |||
| #X: 0.51 (0.47−0.56) | |||
| #Y: 0.45 (0.35−0.54) | |||
| Enzymatic dissociation | #13: 1.00 (0.90−1.10) | #13: 0.82 (0.77−0.87) | #13: 0.96 (0.86−1.06) |
| (coll, tryp/EDTA) | #18: 1.00 (0.86−1.14) | #18: 1.44 (1.35−1.54) | #18: 1.16 (1.01−1.30) |
| #21: 1.02 (0.89−1.14) | #21: 0.80 (0.75−0.86) | #21: 0.90 (0.81−1.00) | |
| #X: 0.49 (0.41−0.58) | #X: 0.45 (0.43−0.47) | #X: 0.49 (0.42−0.57) | |
| #Y: 0.51 (0.45−0.58) | #Y: 0.42 (0.33−0.50) | #Y: 0.47 (0.37−0.57) | |
| Enzymatic dissociation | #13: 0.94 (0.74−1.13) | #13: 0.84 (0.75−0.93) | #13: 0.86 (0.81−0.91) |
| (tryp/EDTA, coll) | #18: 0.92 (0.80−1.05) | #18: 1.41 (1.26−1.57) | #18: 1.37 (1.30−1.44) |
| #21: 1.17 (0.99−1.36) | #21: 0.81 (0.75−0.88) | #21: 0.82 (0.78−0.86) | |
| #X: 0.59 (0.39−0.78) | #X: 0.44 (0.39−0.49) | #X: 0.46 (0.41−0.51) | |
| #Y: 0.60 (0.31−0.90) | #Y: 0.43 (0.33−0.53) | #Y: 0.45 (0.36−0.54) |
, chromosome; tryp, trypsin; coll, collagenase.
We conclude that these modified protocols for CVS preparation have resulted in an accurate representation of the cytotrophoblast layer and the mesenchymal core, which is in full concordance with the results obtained by karyotyping of STC and LTC.
Abnormalities Undetectable by MLPA
In six samples abnormalities were revealed by TK that remained undetected by MLPA. These included three samples with a familial chromosomal rearrangement, two samples with a mosaicism with an extra marker chromosome with no clinical relevance (reason for referral nuchal translucency and advanced maternal age) and one sample with a mosaic trisomy 10 (reason for referral advanced maternal age). Follow-up by ultrasound in this latter pregnancy revealed an oligohydramnion.
Discussion
The suitability of the MLPA test on CVS as a replacement for TK in the detection of prevalent aneuploidies of chromosomes 21, 18, 13, X, and Y was assayed in 152 samples. By doing so, we found a complete concordance between the MLPA and TK results for the diagnosis of all euploidies and non-mosaic aneuploidies, respectively, yielding an absolute specificity and sensitivity of the MLPA test of 100%. However, a reliable prenatal diagnosis based on CVS may be complicated by several factors related to the cellular composition of CVS tissue, which at the cytogenetic level may be expressed as (pseudo)mosaicisms and/or maternal contaminations. Between 1 and 2 percentage of the CVS karyotypes is mosaic, and in more than 80% of the cases the mosaicism is confined to the placenta.12,13 Mosaicism for trisomies 21, 18 and 13 have been reported to occur in 0.26% of CVS.14 It is important to bear in mind that mosaicisms for one of these target chromosomes may also extend into the fetal cell lineages and, as such, they must be considered as risk factors for fetal abnormalities. Therefore, it is relevant to establish the detection level of the MLPA test in mosaic cases. We found that all mosaics tested exhibited a mean probe ratio outside the 95% CI, and that all individual probes of the target chromosomes involved yielded abnormal MLPA results.
The mosaics in our study were only detected by MLPA if the abnormal cell line was present in the STC. This result suggests that proteinase K digestion of whole villi results in an over-representation of DNA from cytotrophoblastic cells and, as such, closely resembles STC cell DNA. This notion is in line with previous studies demonstrating the cellular complexity of CVS tissues and the relevance of CVS preparation methods for molecular testing to minimize the risk of false-positive or -negative results.15,16 As such, MLPA analysis of CVS on DNA isolation of whole villi digested with proteinase K as presented here, is only appropriate as a replacement for TK of STC villi. In case 7, in which the abnormal cells were confined to the LTC, the MLPA result for chromosome 18 was indicative for a mosaic trisomy 18, with a 95% CI of 1.08−1.28. Neither 1.0 or 1.5 was included in the 95% CI. This indicated that only a small proportion of the cells digested with proteinase K was of mesenchymal origin. In the other two cases, with an abnormality confined to the LTC (case 4 and 5), this phenomenon was not observed probably because the abnormalities in the LTC were present as low-grade mosaicisms and, therefore, were not observed in the MLPA results.
In case 7, we tested two different methods for enzymatic dissociation of the villi to obtain cells from both the cytotrophoblast and the mesenchyme cell lineages. From our data, we conclude that these two methods resulted in proper representations of the cytotrophoblast layer and the mesenchymal core, and that the MLPA results obtained are in concordance with the results obtained by karyotyping of STC and LTC, respectively. Enzymatic dissociation with trypsin/EDTA followed by collagenase showed that the total cell population (fraction T) consisted predominantly of cells from the mesenchymal core. This notion was confirmed by the MLPA electropherogram from fraction C. Digestion with collagenase followed by trypsin/EDTA gave the best results, representing a total cell population (fraction T) with a distribution of 60% cytotrophoblastic layer and 40% mesenchymal core cells. QF-PCR results obtained with this CVS preparation method were consistent with the presence of mesenchyme and cytotrophoblast cells in almost equal proportions.8 Recently, it was proposed by Mori et al that the cytotrophoblast layer of chorion villi becomes thinner during gestation, resulting in a gradual over-representation of the mesenchymal lineage.17 Since in case 1 we found that the CVS in the 21st week of gestation displayed MLPA results representative of the cytotrophoblast lineage, we could not confirm this latter notion.
The potential presence of maternal cell contamination (MCC) poses a serious risk for prenatal misdiagnosis.18 We have taken into account that MCC in female samples cannot be detected by MLPA (see material and methods). In the male samples tested by MLPA, however, no evidence was found for the presence of maternal contamination. Additionally, we tested two of the male DNA samples for the presence of MCC using another PCR-based system (AmpFLSTR Identifiler Kit, Applied Biosystems). Again, we failed to obtain any evidence for MCC. It appears that the MCC problem can be minimized if maternal deciduas are carefully removed from the villi and/or if sample (DNA) preparation procedures are thoroughly optimized. Nevertheless, we suggest that validation studies are required to determine a further assessment of MCC as potential cause of misdiagnosis in prenatal testing of CVS. Enzymatic dissociation of CVS yields cell pools representative for the cytotrophoblast layer and mesenchymal core separately. Therefore, rapid aneuploidy testing with MLPA on these cell pools is comparable to karyotyping of STC and LTC.
Although QF-PCR has the same inherent limitations as MLPA in that it will not detect structural chromosome aberrations1,19 it is sensitive for the detection of MCC and triploidy (69,XXX). In reverse, MLPA has the advantage of being able to assess the copy number of up to 50 loci in a single assay. As such, MLPA can easily be extended to other genomic regions of known clinical relevance20 and can also be used as a highly efficient technique for the detection of subtelomeric imbalances.21 The MLPA technology involves ligation of probes corresponding to a chromosome-specific sequence that is unique within the genome. In contrast to polymorphic loci used for QF-PCR, these chromosome-specific sequences show little or no variation, which avoids non-informativeness of the targeted sequences. Therefore, the MLPA technology may be well-suited for combining speed and targeted testing of specific chromosomal inter- and/or intragenic regions. Until recently, rapid aneuploidy detection was mostly performed by I-FISH. MLPA and QF-PCR have some advantages over I-FISH, ie, the tests are less labor-intensive, more cost-effective and/or better suited for high throughput analyses. QF-PCR and MLPA are considered to be valid alternatives to karyotyping for specific referral reasons, although some clinically significant abnormalities will remain undetected. In a retrospective study of 3700 CVS of women with referral reason elevated maternal age, we determined that for every 1000 CVS performed up to 25 chromosomal aberrations, of which six with potential clinical significance, would remain undetected if MLPA were to replace TK (unpublished data). As far as the clinical relevance of these anomalies is concerned, it should be borne in mind that 70% of the (structurally) unbalanced chromosomal anomalies lead to intrauterine death or miscarriage before birth, and/or are associated with abnormalities detectable by ultrasound.22
The development and implementation of additional novel molecular-cytogenetic techniques, such as array CGH, is continuously increasing the resolution of the detection of chromosome abnormalities.23 In prenatal diagnostics array CGH may be applied to pregnancies with ultrasound abnormalities and a normal karyotype. However, retrospective validation studies have indicated that more insight into normal versus abnormal copy number variation within the human genome24,25 and a comprehensive detection of mosaicisms is required before such a technology can be applied into routine prenatal diagnostic care.
We conclude that MLPA is a powerful technique for the detection of aneuploidies of chromosomes 21, 18, 13, X, and Y in CVS. In addition, we conclude that DNA extraction methods for CVS have a major impact on the genetic make-up of the DNA pool and affect the reliability of the molecular diagnosis of aneuploidies. In anticipation of molecular targeted testing for prevalent aneuploidies in CVS, we have designed a laboratory flowchart (Figure 2). In this flowchart, the implementation of rapid aneuploidy detection is based on DNA extracted from the cytotrophoblasts and the mesenchymal core, respectively. In contrast to Gerdes et al26 concordant abnormal results in these duplicate measurements are considered as final test results, irrespective of whether an abnormality is detected by ultrasound or serum screening.
Figure 2.
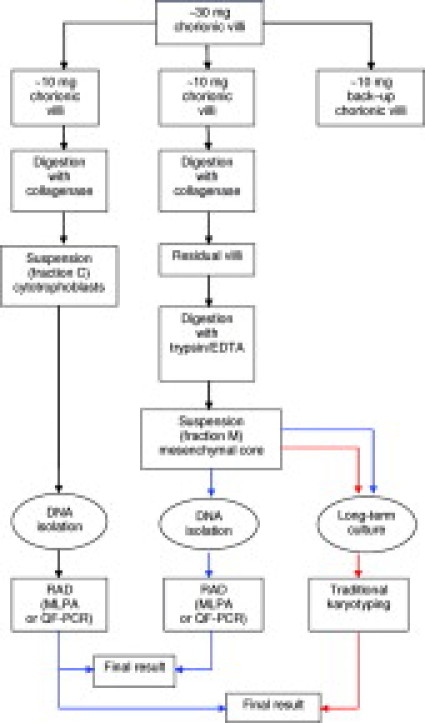
Laboratory flowchart for integrating rapid aneuploidy detection (RAD) in CVS into the cytogenetic diagnostic service. Approximately 30 mg of cleaned villi is split into three fractions (∼10 mg each). Two independent cell preparation procedures based on enzymatic digestion with collagenase and/or trypsin/EDTA are performed on fractions I and II to obtain suspensions from cytotrophoblasts (fraction C) and the mesenchymal core (fraction M), separately. A small amount of fraction M is used for LTC. Fraction III is stored for back-up. DNA is extracted from fractions C and M and RAD by MLPA or QF-PCR are assayed independently. The blue and red arrows indicate the routing for RAD as stand-alone test and the routing for TK, respectively. In this flow chart, TK of the STC is replaced by RAD on DNA from the cytotrophoblast fraction. Discordant results with RAD between fractions C and M or test failures is indicative for TK of the LTC or a repeat experiment using back-up fraction III. When the results of RAD are abnormal, follow-up karyotype analysis is performed to confirm the nature of the aneuploidy.
Taken together, we conclude that MLPA as targeted stand-alone CVS test in pregnancies with an increased risk for Down syndrome is a fast and reliable alternative for traditional karyotyping on STC and LTC.
Acknowledgements
We thank Judith Derks-Willemen for technical support and database development. We are grateful to Dr. Schouten from MRC Holland, who supplied the MLPA kits.
Footnotes
MRC Holland supplied the MLPA kits.
References
- 1.Cirigliano V, Voglino G, Cañadas MP, Marongiu A, Ejarque M, Ordoñez E, Plaja A, Massobrio M, Todros T, Fuster C, Campogrande M, Egozcue J, Adinolfi M. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR. Assessment on 18,000 consecutive clinical samples. Mol Hum Reprod. 2004;10:839–846. doi: 10.1093/molehr/gah108. [DOI] [PubMed] [Google Scholar]
- 2.Kooper AJA, Faas BHW, Kater-Baats E, Feuth T, Janssen JCJA, van der Burgt I, Lotgering FK, Geurts van Kessel A, Smits APT. Multiplex ligation-dependent probe amplification (MLPA) as a stand-alone test for rapid aneuploidy detection in amniotic fluid cells. Prenat Diagn. 2008;28:1004–1010. doi: 10.1002/pd.2111. [DOI] [PubMed] [Google Scholar]
- 3.Hochstenbach R, Meijer J, van de Brug J, Vossebeld-Hoff I, Jansen R, van der Luijt RB, Sinke RJ, Page-Christiaens GC, Ploos van Amstel JK, de Pater JM. Rapid detection of chromosomal aneuploidies in uncultured amniocytes by multiplex ligation-dependent probe amplification (MLPA) Prenat Diagn. 2005;25:1032–1039. doi: 10.1002/pd.1247. [DOI] [PubMed] [Google Scholar]
- 4.Slater HR, Bruno DL, Ren H, Pertile M, Schouten JP, Choo KH. Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA) J Med Genet. 2003;40:907–912. doi: 10.1136/jmg.40.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalousek DK, Vekemans M. Confined placental mosaicism. J Med Genet. 1996;33:529–533. doi: 10.1136/jmg.33.7.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stetten G, Escallon CS, South ST, McMichael JL, Saul DO, Blakemore KJ. Reevaluating confined placental mosaicism. Am J Med Genet A. 2004;15(131):232–239. doi: 10.1002/ajmg.a.30363. [DOI] [PubMed] [Google Scholar]
- 7.Simoni G, Fraccaro M. Does confined placental mosaicism affect the fetus? Hum Reprod. 1992;7:139–140. doi: 10.1093/oxfordjournals.humrep.a137605. [DOI] [PubMed] [Google Scholar]
- 8.Mann K, Kabba M, Donaghue C, Hills A, Ogilvie CM. Analysis of a chromosomally mosaic placenta to assess the cell populations in dissociated chorionic villi: implications for QF-PCR aneuploidy testing. Prenat Diagn. 2007;27:287–289. doi: 10.1002/pd.1663. [DOI] [PubMed] [Google Scholar]
- 9.Mellink CH, Mw. de Pater JM, Poddighe PJ, Smeets DFCM. Kwaliteit van klinisch cytogenetisch onderzoek: voorwaarden, normen en toetsen. Guideliness in Dutch of NAV-VGKN. 2003;9:1–31. [Google Scholar]
- 10.Gardner RJM, Sutherland GR. Chromosome abnormalities and genetic counseling. 3rd ed. Oxford University Press; Oxford, London: 2004. pp. 363–392. [Google Scholar]
- 11.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahnemann JM, Vejerslev LO. Accuracy of cytogenetic findings on chorionic villus sampling (CVS)–diagnostic consequences of CVS mosaicism and non-mosaic discrepancy in centres contributing to EUCROMIC 1986–1992. Prenat Diagn. 1997;17:801–820. doi: 10.1002/(sici)1097-0223(199709)17:9<801::aid-pd153>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 13.Grati FR, Grimi B, Frascoli G, Di Meco AM, Liuti R, Milani S, Trotta A, Dulcetti F, Grosso E, Miozzo M, Maggi F, Simoni G. Confirmation of mosaicism and uniparental disomy in amniocytes, after detection of mosaic chromosome abnormalities in chorionic villi. Eur J Hum Genet. 2006;14:282–288. doi: 10.1038/sj.ejhg.5201564. [DOI] [PubMed] [Google Scholar]
- 14.Smith K, Lowther G, Maher E, Hourihan T, Wilkinson T, Wolstenholme J. The predictive value of findings of the common aneuploidies, trisomies 13, 18 and 21, and numerical sex chromosome abnormalities at CVS: experience from the ACC U.K. Collaborative Study. Association of Clinical Cytogeneticists Prenatal Diagnosis Working Party. Prenat Diagn. 1999;19:817–826. [PubMed] [Google Scholar]
- 15.Waters JJ, Mann K, Grimsley L, Ogilvie CM, Donaghue C, Staples L, Hills A, Adams T, Wilson C. Complete discrepancy between QF-PCR analysis of uncultured villi and karyotyping of cultured cells in the prenatal diagnosis of trisomy 21 in three CVS. Prenat Diagn. 2006;27:332–339. doi: 10.1002/pd.1675. [DOI] [PubMed] [Google Scholar]
- 16.Allen SK, Luharia A, Gould CP, MacDonald F, Larkins S, Davison EV. Rapid prenatal diagnosis of common trisomies: discordant results between QF-PCR analysis and karyotype analysis on long-term culture for a case of trisomy 18 detected in CVS. Prenat Diagn. 2006;26:1160–1167. doi: 10.1002/pd.1582. [DOI] [PubMed] [Google Scholar]
- 17.Mori M, Ishikawa G, Luo SS, Mishima T, Goto T, Robinson JM, Matsubara S, Takeshita T, Kataoka H, Takizawa T. The cytotrophoblast layer of human chorionic villi becomes thinner but maintains its structural integrity during gestation. Biol Reprod. 2007;76:164–172. doi: 10.1095/biolreprod.106.056127. [DOI] [PubMed] [Google Scholar]
- 18.Schrijver I, Cherny SC, Zehnder JL. Testing for maternal cell contamination in prenatal samples. A comprehensive survey of current diagnostic practices in 35 molecular diagnostic laboratories. J Mol Diagn. 2007;9:394–400. doi: 10.2353/jmoldx.2007.070017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer LG, Bui TH. Molecular cytogenetic and rapid aneuploidy detection methods in prenatal diagnosis. Am J Med Genet C Semin Med Genet. 2007;145:87–98. doi: 10.1002/ajmg.c.30114. [DOI] [PubMed] [Google Scholar]
- 20.Faas BHW, Nillesen W, Vermeer S, Olde Weghuis D, de Leeuw N, Smits APT, van Ravenswaaij-Arts CMA. 2008. Detection of cryptic subtelomeric imbalances in fetuses with ultrasound abnormalities. Eur J Med Genet. 2008;51:511–519. doi: 10.1016/j.ejmg.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Ahn JW, Ogilvie CM, Welch A, Thomas H, Madula R, Hills A, Donaghue C, Mann K. Detection of subtelomere imbalance using MLPA: validation, development of an analysis protocol, and application in a diagnostic centre. BMC Med Genet. 2007;8:9. doi: 10.1186/1471-2350-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung WC, Waters JJ, Chitty L. Prenatal diagnosis by rapid aneuploidy detection and karyotyping: a prospective study of the role of ultrasound in 1589 second-trimester amniocenteses. Prenat Diagn. 2004;24:790–795. doi: 10.1002/pd.985. [DOI] [PubMed] [Google Scholar]
- 23.Shaffer LG, Bejjani BA. A cytogeneticist's perspective on genomic microarrays. Hum Reprod Update. 2004;10:221–226. doi: 10.1093/humupd/dmh022. [DOI] [PubMed] [Google Scholar]
- 24.Rickman L, Fiegler H, Shaw-Smith C, Nash R, Cirigliano V, Voglino G, Ng BL, Scott C, Whittaker J, Adinolfi M, Carter NP, Bobrow M. Prenatal detection of unbalanced chromosomal rearrangements by array CGH. J Med Genet. 2006;43:353–361. doi: 10.1136/jmg.2005.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;23:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerdes T, Kirchhoff M, Lind AM, Larsen GV, Schwartz M, Lundsteen C. Computer-assisted prenatal aneuploidy screening for chromosome 13, 18, 21, X, and Y based on multiplex ligation-dependent probe amplification (MLPA) Eur J Hum Genet. 2005;13:171–175. doi: 10.1038/sj.ejhg.5201307. [DOI] [PubMed] [Google Scholar]


