Abstract
The ability of small interfering RNA (siRNA) to regulate gene expression has potential therapeutic applications, but its use is limited by inefficient delivery. Triggered release of adsorbed poly(ethylene glycol) (PEG)-b-polycation polymers from pH-dependent (PD) liposomes enables protection from immune recognition during circulation (pH 7.4) and subsequent intracellular delivery of siRNA within the endosome (pH ~5.5). Polycationic blocks, based on either poly[2-(dimethylamino)ethyl methacrylate] (31 or 62 DMA repeat units) or polylysine (21 K repeat units), act as anchors for a PEG (113 ethylene glycol repeat units) protective block. Incorporation of 1,2-dioleoyl-3-dimethylammonium-propane (DAP), a titratable lipid, increases the liposome’s net cationic character within acidic environments, resulting in polymer desorption and membrane fusion. Liposomes encapsulating siRNA demonstrate green fluorescent protein (GFP) silencing in genetically-modified, GFP-expressing HeLa cells and glyceraldehyde-3-phosphate dehydrogenase (GAPD) knockdown in human umbilical vein endothelial cells (HUVEC). Bare and PD liposomes coated with PEG113-DMA31 exhibit a 0.16±0.2 and 0.32±0.3 fraction of GFP knockdown, respectively. In contrast, direct siRNA administration and Oligofectamine complexed siRNA reduce GFP expression by 0.06±0.02 and 0.14±0.02 fractions, respectively. Our in vitro data indicates that polymer desorption from PD liposomes enhances siRNA-mediated gene knockdown.
Keywords: RNAi, siRNA, Genedelivery, Liposome, pH-dependent, PEG
1. Introduction
The discovery of siRNA in mammalian cells [1] provides a new strategy in which protein expression can be regulated. siRNA is able to induce the destruction of specific mRNA sequences that may lead to disease or may regulate diseased cells. For example, siRNA-induced protein regulation has shown therapeutic benefits in breast cancer [2], colon cancer [3,4], dystonia [5] (a muscular disorder), and diabetes [6].
One of the limiting factors in the clinical use of siRNA technology is its delivery to target cells. Previously investigated avenues of delivery include direct intravenous injection of ‘naked’ or chemically-stabilized siRNA [7,8], insertion of siRNA into DNA plasmid vectors [4,6], transposon vectors (transgenic plasmids) [2,9], plasmid-infected viruses [5], virosomes (reconstituted viral envelopes) [10], lentiviral vectors [11,12], and liposomes [7,13]. The most successful of these in protecting siRNA and supporting in vivo transport are viral and liposomal delivery methods. Viral vectors have serious drawbacks such as immunogenicity and insertional mutations [10], whereas liposomes can be engineered to have a wide range of physical and biological characteristics [14].
Nucleic acid delivery can exhibit high transfection efficiency when complexed with lipid formulations that include cationic, fusogenic, and/or PEG-conjugated lipids [15-17]. Physical challenges for siRNA delivery, as compared to delivery of other nucleic acids, arise from these molecules being highly charged, relatively small, susceptible to nucleases, and the delivery of many siRNA molecules within one cell is required for therapeutic benefit. Efficiently loaded liposomes may protect siRNA from degradation and facilitate therapeutic administration.
Herein, we report a pH-dependent liposomal system for siRNA delivery that has the potential to passively target tumors or sites of inflammation. A PEG-b-polycation block copolymer is electrostatically bound to the liposome exterior for protection from immune system recognition. Endocytosis of the polymer-coated liposomes causes an increase in the cationic character of the liposome; this leads to desorption of the PEG-polycation chains since they are no longer electrostatically anchored to the liposome. The cationic liposome fuses with the anionic endosomal membrane, releasing the siRNA into the cell. Polymer-coated liposomes encapsulating siRNA protect from degradation and immune recognition, allow passive targeting, and provide a mechanism for endosomal release.
2. Materials and methods
2.1. Materials
1,2-Dioleoyl-sn-glycero-3-phosphocholine (PC), 1,2-dioleoyl-3-dimethylammonium-propane (DAP), and 1,2-dioleoyl-sn-glycero-3-phosphoserine (PS) were obtained from Avanti Polar Lipids, Inc (Alabaster, AL). The poly(ethylene glycol)-b-poly[2-(dimethylamino) ethyl methacrylate] (PEG-DMA) polymers, were synthesized at the University of Sheffield (Sheffield, South Yorkshire, UK) as described by Deshpande et al. [18]. The block copolymers are designated as PEG “X”-DMA“Y”, where X is the number of ethylene glycol repeat units and Y is the number of cationic DMA repeat units. The poly(ethylene glycol)-b-poly(lysine) (PEG122-K21, with 122 ethylene glycol repeat units and 21 K repeat units) sample was prepared at the University of California, Santa Barbara (Santa Barbara, CA) as described by Yu et al. [19]. GFP-and GAPD-interfering siRNA were purchased from Dharmacon RNA Technologies (Lafayette, CO). N-(7-Nitro-2,1,3-benzoxadiazol-4-yl)-1,2-dioleoyl-sn-phosphatidyethanolamine (NBD-PE), N-(lissamine rhodamine B sulfonyl)-1,2-dioleoyl-sn-phosphatidylethanolamine (Rh-PE), and Oligofectamine were acquired from Invitrogen (Carlsbad, CA). Quant-IT RiboGreen RNA reagent was procured from Molecular Probes (Eugene, OR). N-Tris(hydroxymethyl)methyl-2-aminoethane-sulfonic acid (TES) was purchased from Sigma-Aldrich (St. Louis, MO). Qiagen RNeasy mini kit and Triton X-100 were obtained from VWR International (Westchester, PA). Primers and reagents for quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) were purchased from Applied Biosystems (Foster City, CA).
2.2. Liposome preparation
PC (100 mol% PC), PD (PC:DAP, 9:1, mol:mol), and RET (PC/PS/NBD-PE/Rh-PE, 58:38:2:2, mol:mol:mol:mol) liposomes were prepared as described previously [20]. Stock solutions of 10 mM lipid were solubilized in chloroform. Lipids were mixed in a glass vial to a final concentration of 5 mM lipid. Evaporation of solvent results in the formation of bilayers. To this, 2 ml of an aqueous solution consisting of 2.5 μM siRNA in a TES Buffer (20 mM TES, 150 mM NaCl, pH 7.4) was added. The samples were vortexed, subjected to 7 freeze/thaw cycles in liquid nitrogen and a room temperature water bath, and then extruded. Samples were extruded through a Lipex extruder (Northern Lipids, Inc., Vancouver, British Columbia, Canada) 10 times through two 0.1 μm polycarbonate filters (Whatman, Florham Park, NJ) with nitrogen gas (200–250 psi).
A phosphate assay was performed to determine lipid composition as described previously [21]. Briefly, samples were ashed with 10% sulfuric acid at 200 °C for 1 h, followed by addition of 30% hydrogen peroxide and further heating at 200 °C for 40 min. After cooling, a color reagent (5% ammonium molybdate, 10% ascorbic acid) was added to the samples and incubated for 20 min. Absorption was measured on a SpectraMax Plus spectrophotometer (Molecular Devices, Sunnyvale, CA) at 820 nm, with appropriate backgrounds.
Samples were characterized for size and zeta potential on a ZetaPALS Zeta Potential Analyzer (Brookhaven Instruments, Corp., Holtsville, NY) in TES buffer (20 mM TES, 150 mM NaCl, pH 7.4). The prepared solutions were dialyzed (Float-A-Lyzer tubes, 1 ml, MWCO 25 kDa, Spectrum Laboratories, Los Angeles, CA) against TES Buffer (20 mM TES, 150 mM NaCl, pH 7.4), and sterile-filtered through a 0.22 μm polycarbonate membrane (Aerodisc Syringe Filters, Pall Life Sciences, Corp., East Hills, NY). For zeta potential measurements, a 1:20 dilution of liposomes into either a TES (20 mM TES, pH 7.4) or sodium citrate (20 mM Na Citrate, pH 5.5) buffer resulted in solutions with 7.5 mM NaCl.
2.3. Encapsulation efficiency
A Ribogreen assay (Molecular Probes, Eugene, OR) was performed to determine the encapsulation efficiency of siRNA within the liposome samples. A calibration curve was performed with appropriate backgrounds. Spectrofluorometry, (excitation 500 nm, emission 525 nm, Photon Technologies International, Birmingham, NJ, USA) was performed on diluted liposome samples. Equal concentrations of lipid were incubated with RiboGreen and measured for fluorescence, before and after administration of Triton X-100 [22]. Triton X-100 is a surfactant that lyses liposomes. Similar methods have been used previously [23]. The encapsulation efficiency is the encapsulated siRNA concentration divided by the initial siRNA concentration times 100.
2.4. Polymer adsorption
Liposomes were prepared at a concentration of 1.0 mM lipid in either 0.5 ml TES buffer (1 mM TES, pH 7.4) or 0.5 ml sodium citrate buffer (1 mM sodium citrate, pH 5.5). Polymer was added to achieve the desired concentration. All samples were vortexed and allowed to equilibrate overnight at room temperature with gentle shaking (American Rotator V, Miami, FL). The samples were added to Nanosep tubes and centrifuged at 1000 ×g for 10 min. The supernatant was then assayed for phosphate and polymer content.
The polymer concentration was quantified by an assay described by Baleux [24], wherein 25 μl of an iodine-potassium iodide solution (0.04 M I2, 0.12 M KI) was added to 1 ml of a diluted supernatant sample. Samples were diluted to an optimal adsorption range (0.1 < AU < 1.0). After 5 min, the optical density (OD) of the solution was determined for λ =500 nm at ambient temperature. Liposomes were retained by the Nanosep membrane and did not influence the Baleux assay. The variation in the calibration curve with different polymer architectures is due to the differences in hindrance to helix formation, which is the origin of the colored complex.
2.5. Membrane fusion assay
Fusion between liposomes containing both NBD-PE and Rh-PE and liposomes devoid of fluorescent lipid was measured by the decrease in resonance energy transfer resulting from probe dilution [25,26]. RET (PC/PS/NBD-PE/Rh-PE, 58:38:2:2, mol:mol:mol:mol) liposomes were prepared in TES buffer (20 mM TES, 150 mM NaCl, pH 7.4). PC and PD liposomes were incubated with polymer at the appropriate concentrations for 2 h. PC and PD liposomes with or without adsorbed polymer were incubated with RET liposomes at a 9:1 molar ratio (at a final 1 mM lipid) in a TES buffer (20 mM TES, 150 mM NaCl, pH 7.4). Fluorescence was monitored with excitation of 450 nm, emission of 535 nm, and an emission cut-off filter at 530 nm. After 15 min, the pH was adjusted to 5.5 with 1 mM HCl. Fluorescence (F) was normalized by subtracting the initial fluorescence (F0) and dividing by the fluorescence achieved by maximal probe dilution, achieved by addition of Triton X-100 (Fmax). The percent change in fluorescence is given by:
2.6. Transfection
HeLa cells (CCL-2, ATCC, Rockville, MD, USA) and green fluorescent protein (GFP) modified HeLa cells (donated from P. Sharp and C. Novina, MIT, Cambridge, MA) were plated at 40 cells/mm2 in 200 μl medium (5% FBS in DMEM) [27]. HUVEC (CC2517, Cambrex, East Rutherford, NJ, USA) were plated at 40 cells/mm2 in 200 μl endothelial growth medium (EGM). Cells were incubated at 37 °C in a CO2 incubator for 24 h. Prior to liposome transfection, cells were at 80% confluency. Cells were transfected with either naked siRNA, liposomes encapsulating siRNA, or polymer-coated liposomes encapsulating siRNA. Polymer-coated liposomes were prepared by incubating liposomes with the appropriate concentration of polymer to achieve 0.5, 1, or 2 times full surface coverage (Γ*).
siRNA concentrations were conserved; the final lipid concentration was within 8% for administration of siRNA encapsulated in PC and PD liposomes. To achieve equivalent siRNA loading, more PC liposomes were delivered than PD liposomes due to encapsulation efficiency, size, and a higher lipid concentration relative to siRNA concentration per volume. Transfection efficiency was determined by flow cytometry and qRT-PCR.
2.7. Flow cytometry
After 48 h incubation, transfected cell samples were prepared for analysis by Fluorescence Acquired Cell Sorting (FACS, FACScan/Cell Quest Pro, Becton Dickinson, Franklin Lakes, NJ, USA). Cells were treated with 0.25% trypsin/2.6 mM ethylenediaminetetraacetic acid (EDTA) solution, transferred to 12×75 mm polystyrene tubes, and pelletized using a centrifuge (Beckman Coulter, Fullerton, CA, USA) at 1000 ×g for 3 min. Cells were resuspended with 5% FBS in phosphate-buffered saline (PBS) and treated with PI (1:20 dilution in 5% FBS in PBS) for determination of cell viability. Integration of the peaks yields the numbers of cells that no longer express GFP and cells that continue to express GFP. The fraction of GFP knockdown can be calculated [(siRNA-treated GFP-HeLa negative cells-HeLa negative cells)/(GFP positive cells-HeLa negative cells)].
2.8. qRT-PCR
Transfected HUVEC were treated with 0.25% trypsin/2.6 mM EDTA solution for 3 min and washed with PBS. RNA extraction and purification was performed using the Qiagen RNeasy mini kit. RNA was detected and quantified with a spectrophotometer (Spectramax Plus 384, Molecular Devices, Sunnyvale, CA, USA) and mixed with 0.2 U/μL μl Ribonuclease Inhibitor. Reverse transcription (RT) was conducted according to the Applied Biosystems Taqman RT protocol using RT Reagents. The RT final concentration included 1× RT Buffer, 5.5 mM Magnesium Chloride, 0.5 mM per dNTP, 2.5 μM oligo d(T)16, 0.4 U/μl RNase Inhibitor, and 1.25 U/μl MultiScribe Reverse Transcriptase. Each 50 μl reaction was incubated for 10 min at 25 °C, 30 min at 48 °C, and 5 min at 95 °C.
Detection and quantification of RNA were performed using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). A 50 μl PCR sample included 5 μl RT product, 25 μl of 2x TaqMan® Universal PCR Master Mix, and 2.5μl TaqMan® Gene Expression Assay (containing the forward and reverse primers and the TaqMan probe) in 17.5 μl RNase free water. The reactions were incubated in a 96-well plate at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. All samples were run in quadruplicate and quantified with threshold cycles (CT) in order to measure knockdown of human GAPD gene expression. All samples were normalized based on a TaqMan® Gene Expression Assay for beta actin (β actin).
3. Results and discussion
Polymer-coated PD liposomes were examined as a strategic method for siRNA delivery. Fig. 1A illustrates a polymer-coated liposome undergoing a physiologically relevant pH change in response to endocytosis of intravenously administered liposomes. Adsorbed PEG-b-polycation block copolymers may protect PD liposomes (PC:DAP, 9:1, mol:mol) from immune recognition during circulation and can be subsequently desorbed via a pH-trigger (Fig. 1B). Uncoated cationic liposomes are then able to fuse with the endosomal membrane and deliver the encapsulated siRNA within the cytoplasm (Fig. 2A, B). PD liposomes have increased cationic character when the environment becomes acidic (from pH 7.4 in the bloodstream to pH ~5–6 within the endosome) (Fig. 3B) [28]. We investigated the ability of siRNA to knockdown GFP from constitutively expressing GFP-HeLa cells by direct administration of siRNA, Oligofectamine associating siRNA, or intracellular delivery of siRNA encapsulating, PEG-b-polycation coated liposomes (Figs. 4A, B, 5). siRNA knockdown of GAPD was also measured in a less endocytotic cell line, HUVEC, to demonstrate system versatility (Fig. 6A, B).
Fig. 1.
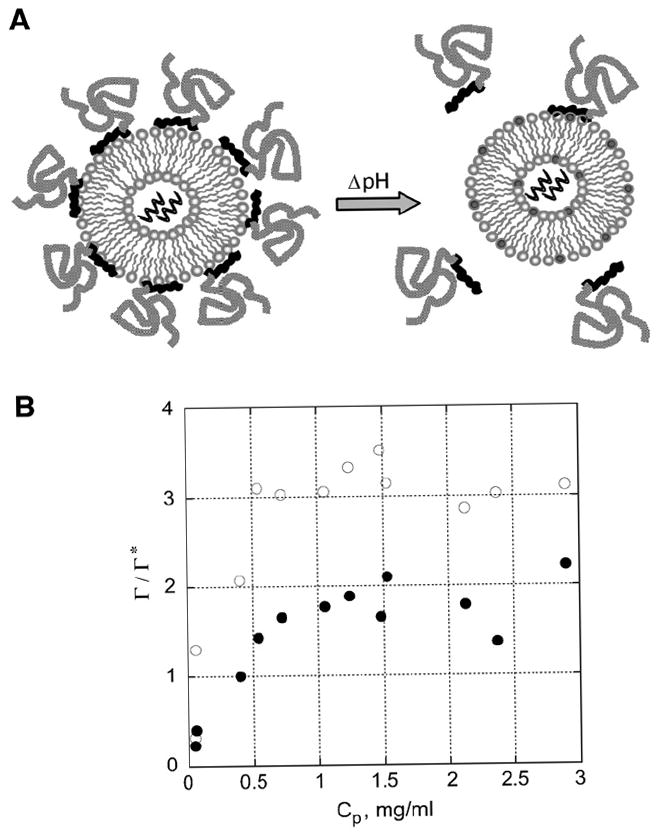
Deprotection of a polymer-protected liposome via a pH shift is depicted in a schematic diagram (A) and demonstrated by adsorption isotherms of PEG113-DMA31 at two pHs (B). (A) Schematic of a PD (PC:DAP, 9:1, mol:mol) liposome with adsorbed PEG-b-polycation conjugates at pH 7.4 undergoing a pH shift which occurs upon transfer from the bloodstream (pH 7.4) to the endosome (pH 5–6) of a cell. (B) Adsorption isotherms for PEG113-DMA31 on PD liposomes in either 1 mM TES buffer (pH 7.4, ○) or 1 mM sodium citrate buffer (pH 5.5, ●). PEG113-DMA31 is a block copolymer with 113 ethylene glycol repeat units and 31 cationic DMA repeat units. The graph depicts the ratio of surface coverage, Γ/Γ*, versus the free polymer in solution, Cp (mg/ml). Each sample contained 1.0 mM lipid and 0.1 to 3.5 mg/ml PEG113-DMA31. Samples were equilibrated for 24 h before separating the polymer-coated liposomes from free polymer in solution.
Fig. 2.
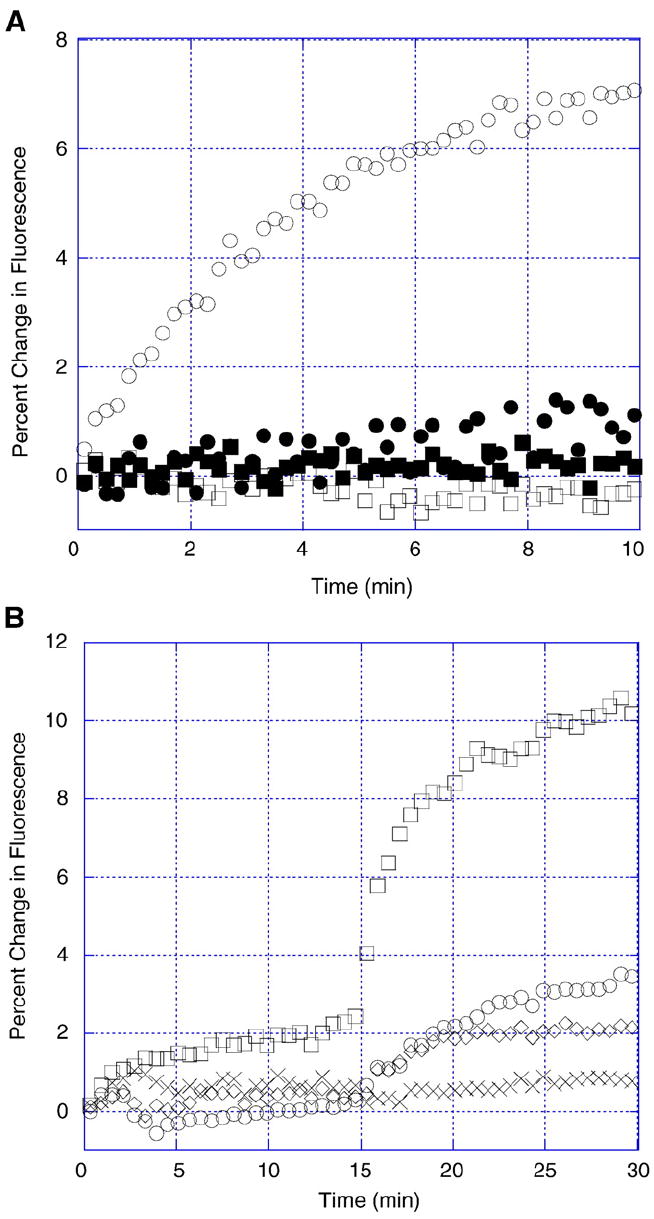
Membrane fusion between (A) bare PC and PD liposomes and (B) polymer-coated PD liposomes at pH 7.4 and 5.5. (A) RET liposomes were incubated for 10 min with bare PC liposomes at pH 7.4 (■) and pH 5.5 (□) and bare PD liposomes at pH 7.4 (●) and pH 5.5 (○). Dilution of NBD-PE and Rh-PE as a result of fusion is monitored by an increase in fluorescence. (B) Bare PD liposomes (□) and PD liposomes coated with PEG113-DMA31 (○), PEG113-DMA62 (◊), and PEG122-K21 (×) were incubated with RET liposomes for 15 min at pH 7.4 and 15 min at pH 5.5. The pH was adjusted by addition of 1 mM HCl. Liposomes were equilibrated with polymer at pH 7.4 for 2 h prior to start of experiment. PC and PD liposomes were incubated with RET liposomes at a 9:1 molar ratio in TES buffer (1 mM TES, pH 7.4).
Fig. 3.
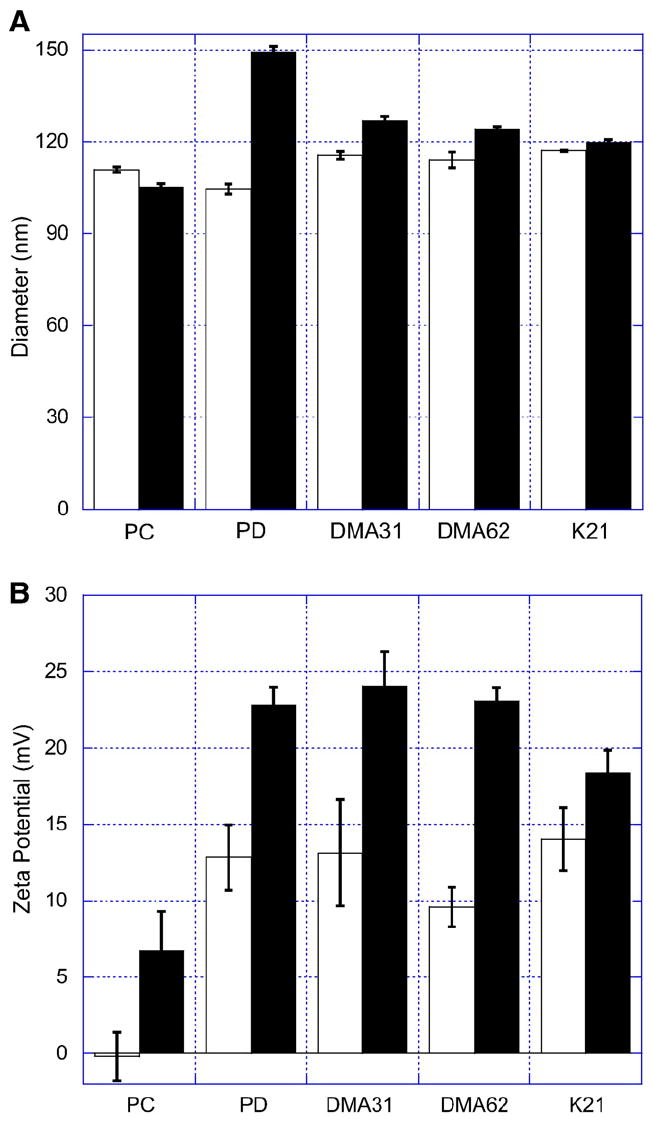
Dynamic light scattering and electrophoresis were used to determine (A) liposome size and (B) zeta potential before (white, pH 7.4) and after fusion (black, pH 5.5). Membrane fusion occurred between RET liposomes and bare PC liposomes (PC), PD liposomes (PD), PEG113-DMA31 coated PD liposomes (DMA31), PEG113-DMA62 coated PD liposomes (DMA62), and PEG122-K21 coated PD liposomes (K21). Liposomes encapsulated siRNA (GAPD). Electrophoresis was conducted using either TES (20 mM TES, 7.5 mM NaCl, pH 7.4) or sodium citrate (20 mM citrate, 7.5 mM NaCl, pH 5.5) buffers. Conductance was maintained at 6.6±0.2 mS. Error is the standard deviation of the mean, n = 3.
Fig. 4.
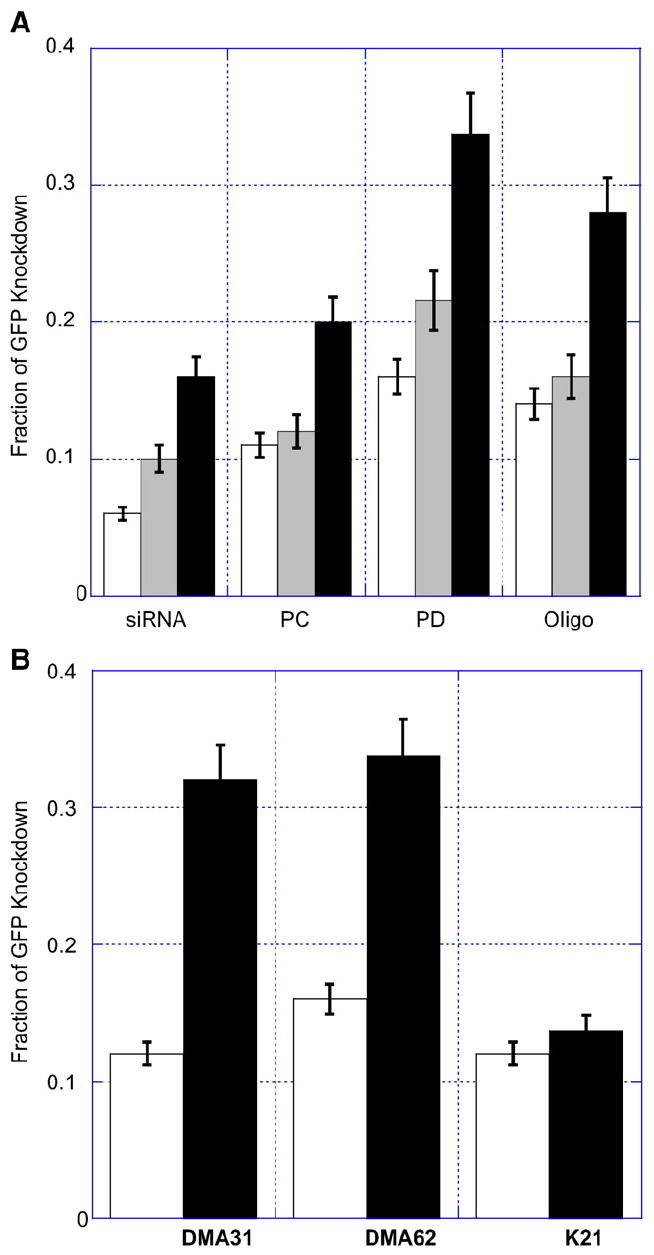
Fraction of GFP knockdown for (A) bare liposomes and (B) polymer-coated liposomes encapsulating siRNA incubated for 48 h with HeLa and GFP-HeLa cells. Flow cytometry was performed to determine shifts in GFP fluorescence of siRNA-treated GFP-HeLa cells relative to autofluorescence of HeLa cells. (A) 40 (white), 80 (grey), or 160 (black) pmol siRNA was administered directly (siRNA), encapsulated within PC liposomes (PC), encapsulated within PD liposomes (PD), or associating with Oligofectamine (Oligo). The sample volume (50 μl) was constant per well. The liposome concentration was diluted to administer the same amount of siRNA to each well and the resulting variation in lipid concentration was within 8%. (B) PC (white) and PD (black) liposomes with full surface coverage of PEG113-DMA31 (DMA31), PEG113-DMA62 (DMA62), and PEG122-K21 (K21) exhibit GFP knockdown from constitutively expressing GFP-HeLa cells. Extruded liposomes encapsulating 40 pmol siRNA were pre-incubated with polymer for 1 h and then added to GFP-HeLa cells. After 48 h, flow cytometry was performed to evaluate changes in cellular GFP expression as a function of liposome formulation and protecting polymer. Error is the standard deviation of the mean, n = 3.
Fig. 5.
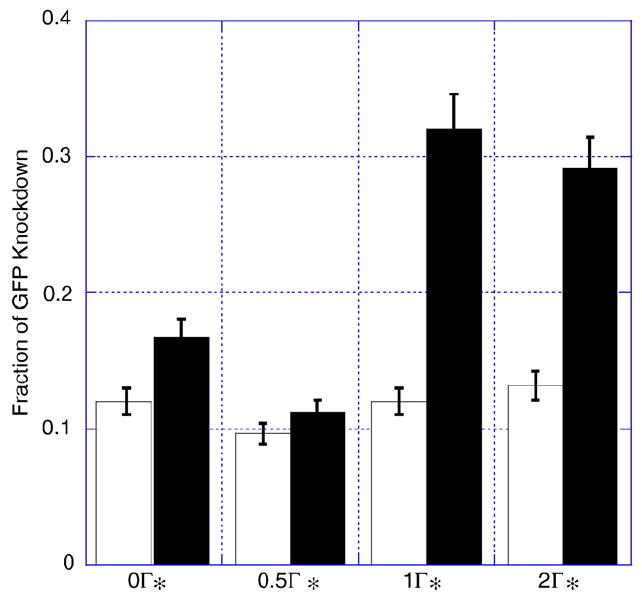
PC (white) and PD (black) liposomes with increasing surface coverage of PEG113-DMA31 polymer exhibit knockdown of GFP from constitutively expressing GFP-HeLa cells. Liposomes encapsulating 40 pmol siRNA were pre-incubated with different concentrations of polymer for 1 h and then added to GFP-HeLa cells. Full surface coverage Γ* for PEG113-DMA31 was 0.7 mg/m2. After 48 h, flow cytometry was performed to evaluate changes in GFP expression as a function of liposome formulation and polymer surface coverage. Error is the standard deviation of the mean, n = 3.
Fig. 6.
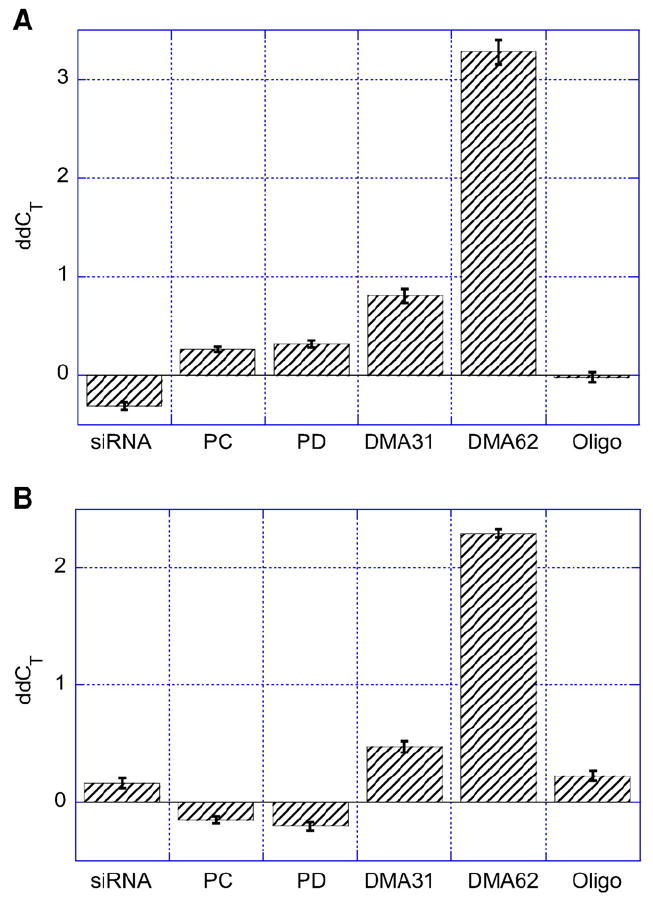
GAPD knockdown after incubation with HUVEC for (A) 24 h and (B) 48 h. 200 pmol siRNA was administered directly (siRNA) or encapsulated within PC liposomes (PC), PD liposomes (PD), PEG113-DMA31 coated PD liposomes at full surface coverage (DMA31), PEG113-DMA62 coated PD liposomes at full surface coverage (DMA62), or associated with Oligofectamine (Oligo). The sample volume (50 μl) was constant per well. The liposome concentration was diluted to administer the same amount of siRNA to each well. Resulting variation of lipid concentration was within 8%. The threshold cycles CT values obtained from qRT-PCR designate the number of amplifications required before entering the exponential phase. The ddCT denotes that the CT values are relative to both β actin and untreated HUVEC. High ddCT values indicate successful GAPD gene expression knockdown. Error is the standard deviation of the mean, n = 4.
3.1. Liposome characterization
Two liposome formulations were prepared to compare siRNA delivery of PD liposomes and net neutral (PC) liposomes and to examine the effectiveness of pH-triggered, PEG-b-polycation desorption. PC and PD liposomes had uniform diameters of 110.5±1.8 nm and 104.2 ±2.5 nm as determined by dynamic light scattering, respectively. The lipid formulation or siRNA encapsulation did not impact the final liposome diameter.
Liposome size and surface charge density can influence in vivo gene expression. Templeton et al. [29] demonstrated a size-dependent biodistribution of gene expression after intravenous administration, showing primary expression in the lung [29]. Optimization of net cationic character and nucleic acid loading was also described; ratios of 5:1 to 3:1 (mol:mol, cationic lipid:cholesterol) and 100 to 150 μg nucleic acid showed increased levels of gene expression [29]. There may be differences in the in vivo delivery of each liposome formulation. However, for the in vitro studies reported here, we did not see a difference in performance with respect to liposome diameter, composition, or loading (data not shown).
The siRNA encapsulation efficiency was quantified using Ribo-Green®, which fluoresces when exposed to siRNA. The quantity of encapsulated siRNA for each liposome formulation was determined by lysing dialyzed liposomes. Thus, the encapsulation efficiency is equal to the amount of siRNA encapsulated divided by the initial siRNA loading times 100. The siRNA encapsulation efficiencies were similar for the PC and PD liposome formulations, 19.4±1.8% and 18.7±2.7%, respectively.
3.2. Polymer adsorption
Polymer adsorption (Γ, mg/m2) describes the mass of polymer adsorbed per square meter lipid. Table 1 provides a description of the three polymers (PEG113-DMA31, PEG113-DMA62, and PEG122-K21) investigated and the mass of polymer required to either coat (Γ*) or neutralize (Γn*) one square meter of liposome surface.
Table 1.
Description of PEG-b-polycation polymers
| Polymer | MW (kg/mol) | PEG: CA | Γ* (mg/m2) | Γn* (mg/m2) | K, pH 7.4 (mg/m2)/(mg/ml) | K, pH 5.5 (mg/m2)/(mg/ml) |
|---|---|---|---|---|---|---|
| PEG113-DMA31 | 9.9 | 4 | 0.7 | 0.8 | 3.1 | 1.7 |
| PEG113-DMA62 | 14.7 | 2 | 1.1 | 0.6 | 6.2 | 3.2 |
| PEG122-K21 | 8.6 | 6 | 0.6 | 1.9 | 1.2 | 0.2 |
Notes: The PEG:CA is the molar ratio of PEG monomers to cationic anchors (CA). Full mushroom coverage (Γ*) is defined as the mass of polymer required to coat an area of lipid, with the PEG chain area defined by the Flory radius of the polymer (A=πr2). Liposome neutralization (Γn*) occurs when each phosphate group binds to one cationic DMA anchoring group. It is defined as the mass of polymer per lipid area required to neutralize the surface charge of the liposome. Equilibrium constants were determined for electrostatic adsorption of PEG-b-cation polymers on PD (PC:DAP, 9:1 mol:mol) liposomes.
To determine the amount of polymer needed to coat the surface, we first calculate the surface area of each polymer. PEG chains follow random-walk statistics and occupy an area at the interface given by a sphere of diameter [30]
| (2) |
where mb is the molecular weight of the chain. From the sphere diameter, we determine the area occupied at the surface using π(ξ/2)2. Thus, PEG113-DMA31 has a diameter of 54 Å, and occupies an unconstrained area of 2270 Å2 per polymer. The number of polymers required for full surface coverage is equal to 1 m2 divided by the unconstrained polymer area. The quantity of polymer to coat one square meter lipid, Γ*, is 73 nmoles, or 0.7 mg, of PEG113-DMA31. The association constant, K, is determined from the initial slope (first 4 data points) of the adsorption profile, such that
| (3) |
where Cp is the concentration of polymer in equilibrium with the adsorbed polymer.
To determine the mass of polymer required to neutralize 1 m2 of the liposome surface, we first calculate the number of polymers needed by dividing the number of phosphate moieties per m2 by the number of cationic anchors of the polymer. The number of polymers is converted to mass by using the molecular weight and Avogadro’s number.
Fig. 1B depicts the adsorption profile of PEG113-DMA31 on PD liposomes in either a TES buffer (1 mM TES, pH 7.4) or sodium citrate buffer (1 mM sodium citrate, pH 5.5). PEG113-DMA31 shows 50% greater adsorption at pH 7.4 than pH 5.5, indicating the potential for desorption which should facilitate liposome and endosomal membrane fusion and siRNA delivery. In addition, plotting r/r* clearly illustrates the regions of mushroom coverage (low polymer density, < 1) and the brush regime (high polymer density, N1). A maximum in surface coverage is observed at 3Γ* for pH 7.4 and at 2Γ* for pH 5.5. Adsorption data for PEG113-DMA62 and PEG122-K21 have been reported previously and show similar trends in desorption; their adsorption parameters are described in Table 1 [30].
Polymer adsorption is negatively affected by increasing salt concentration, as described previously [30]. We maximized the electrostatic interactions in the adsorption profiles in Fig. 1B. Electrostatically-anchored polymers remain bound to PC liposomes but desorb from PD liposomes when the pH is lowered from 7.4 to 5.5 (adsorption data not shown). PC and PD liposomes were assessed for pH-triggered polymer desorption (at physiological salt concentrations) and subsequent fusion by measuring the ability of polymer-coated liposomes to fuse with a resonance energy transfer (RET) liposome at pH 7.4 and 5.5.
3.3. Membrane fusion
RET liposomes contain a quencher and a fluorescer pair, which result in fluorescence upon dilution. We monitored the fluorescence of RET liposomes upon fusion with PC and PD liposomes. Fig. 2A showed no fusion occurring for PC liposomes at pH 7.4 and pH 5.5 and PD liposomes at pH 7.4. We observed fusion for PD liposomes at pH 5.5. This provided evidence that pH-induced cationicity of PD liposomes facilitates fusion with other lipid membranes.
In Fig. 2B, we expanded the membrane fusion study to include bare PD liposomes and PD liposomes protected with either PEG113-DMA31, PEG113-DMA62, or PEG122-K21. Fusion was monitored for 15 min at pH 7.4 and for 15 min at pH 5.5. Bare PD liposomes were effective in fusing with RET liposomes. Polymer adsorption on PD liposomes inhibited fusion at pH 7.4; desorption of polymer at pH 5.5 revealed recovery of PD liposome fusion. PEG113-DMA31, PEG113-DMA62, and PEG122-K21 coated liposomes exhibited an increase in fusion at pH 5.5 relative to bare PD liposomes of 35%, 21%, and 9%, respectively. Thus, we confirm that the electrostatically-anchored polymers were able to desorb from the pH-dependent liposomes within minutes, resulting in membrane fusion.
We measured changes in liposome size (Fig. 3A) and zeta potential (Fig. 3B) before (pH 7.4) and after fusion (pH 5.5). Liposome size remained approximately constant for PC liposomes, whereas it significantly increased by 35% for PD liposomes. PEG113-DMA62 and PEG113-DMA31 coated liposomes experience a 7% or 8% increase in liposome diameter after fusion, respectively. PEG122-K21 coated liposomes exhibited no change in size, within error.
The zeta potential of bare PC and PD liposomes and polymer-coated PD liposomes at pH 7.4 and pH 5.5 are reported in Fig. 3B. PC liposomes were slightly negative at pH 7.4 and carried an effective positive charge of 6.2±2.8 mV at pH 5.5. The zeta potential of PD liposomes increased from 12.5±2.5 mV at pH 7.4 to 22.8±1.4 mV at pH 5.5. Polymer-coated liposomes followed a similar trend to bare PD liposomes. Liposomes coated with the different polymers have similar zeta potentials at pH 7.4 and 5.5. For PEG113-DMA31 coated PD liposomes, the zeta potential increased from 13.4±3.8 mV at pH 7.4 to 22.4±2.6 mV at pH 5.5. The overall net increase in liposome cationic surface charge, which may facilitate siRNA delivery, was not hindered by polymer adsorption.
3.4. siRNA (GFP) delivery to HeLa and constitutively expressing GFP-HeLa cells
HeLa and constitutively expressing GFP-HeLa cells were incubated with PC or PD liposomes encapsulating either 40, 80, or 160 pmol siRNA. For comparison, naked siRNA and Oligofectamine (a popular transfection agent) complexed siRNA were added to cells. After 48 h, flow cytometry was performed to determine the fraction of GFP-HeLa cells no longer expressing GFP. Flow cytometry revealed a distinct population of fluorescent GFP-HeLa cells (positive control) and autofluorescent HeLa cells (negative control). The shift in fluorescence from positive to negative was used to calculate the fraction of GFP knockdown.
Fig. 4A demonstrates the performance of naked siRNA, siRNA encapsulated within PC and PD liposomes, and siRNA complexed with Oligofectamine. Naked siRNA showed the lowest GFP knockdown. A 0.06±0.005 fraction of GFP knockdown was observed when 40 pmol of siRNA was directly administered to cells. This was approximately half the GFP knockdown observed when 40 pmol siRNA is encapsulated within PC liposomes and approximately one-third the GFP knockdown when encapsulated within PD liposomes. GFP knockdown of 80 pmol siRNA alone yielded similar results to administering 40 pmol siRNA encapsulated within PC liposomes. Four times the siRNA dose (160 pmol siRNA) yielded GFP knockdown equivalent to bare PD liposomes encapsulating 40 pmol siRNA, a 0.16±0.2 fraction of cells exhibited GFP knockdown. The highly anionic character of siRNA may reduce its uptake into cells, whereas encapsulation within neutral (PC) or cationic (PD) liposomes increased siRNA delivery and GFP knockdown.
Increasing the amount of encapsulated siRNA that is delivered improves GFP knockdown, due to more siRNA copies being delivered to each cell. At 40 pmol siRNA, Oligofectamine demonstrated similar GFP knockdown to PD liposomes. However, at 80 and 160 pmol siRNA, PD liposomes increased the fraction of GFP knockdown to 0.22±0.02 and 0.33±0.03, respectively. Using Oligofectamine, GFP knockdown was limited to 0.15±0.02 and 0.28±0.02 for 80 and 160 pmol, respectively. We observed that the amount of siRNA delivered and the mode of delivery was important in achieving siRNA-mediated gene knockdown.
Polymer-coated PC and PD liposomes were investigated for their ability to knockdown GFP expression (Fig. 4B). PC and PD liposomes encapsulating 40 pmol siRNA were incubated with either PEG113-DMA31, PEG113-DM62, or PEG122-K21 for 1 h to obtain full surface coverage (Γ*) and then added to cells. A comparison of PC and PD liposomes distinguished the importance of polymer desorption for successful siRNA delivery.
Polymer-coated PC liposomes resulted in 0.11±0.01, 0.16±0.01, and 0.12±0.01 fractions of GFP knockdown at full surface coverage of PEG113-DMA31, PEG113-DMA62, and PEG122-K21, respectively. These results were consistent with 40 pmol siRNA encapsulated PC liposomes without adsorbed polymer (Fig. 4A). PEG113-DMA62 had a slightly improved GFP knockdown relative to PEG113-DMA31 and PEG122-K21, which may result from the long cationic anchor acting as a proton sponge [31].
Polymer-coated PD liposomes showed enhanced GFP knockdown relative to polymer-coated PC liposomes. Delivery of 40 pmol siRNA from polymer coated PD liposomes resulted in 0.30±0.03, 0.34±0.04, and 0.13±0.02 fractions of GFP knockdown at full surface coverage of PEG113-DMA31, PEG113-DMA62, and PEG122-K21, respectively. Bare PD liposomes showed a 0.16±0.02 fraction of GFP knockdown (Fig. 4A), greater membrane fusion (Fig. 2B), and similar zeta potential (Fig. 3B) to polymer-coated PD liposomes. The enhanced GFP knockdown observed from PEG113-DMA31 and PEG113-DMA62 coated PD liposomes may be a result of polymer desorption within the endosome. The additional cationic anchor groups may facilitate a proton sponge like effect (as described with polyethyleneimine [31]). This may increase in vitro membrane fusion or disruption relative to naked PD liposomes.
The three polymers enhanced GFP knockdown in parallel with their increasing association constants. PEG113-DMA62 coated PD liposomes showed a 0.34±0.04 fraction of GFP knockdown and exhibited the strongest adsorption, K = 6.2 (mg/m2)/(mg/ml). With half the cationic anchor and association constant (K = 3.1 (mg/m2)/(mg/ml)), PEG113-DMA31 exhibited a 0.30±0.03 fraction of GFP knockdown. PEG122-K21 adsorbed weakly (K = 1.2 (mg/m2)/(mg/ml)) to PD liposomes and did not show any benefit over PC liposomes for improving GFP knockdown.
We anticipated that PEG113-DMA31 might perform better than PEG113-DMA62 for the following reasons: 1 — the number of administered polymer chains is higher for PEG113-DMA31 (crowding may result in faster desorption), 2 — PEG113-DMA31 binds more weakly and may therefore desorb more quickly, and 3 — fewer cationic charges may displace more readily. In contrast, we find that PEG113-DMA62 performs equal to or better than PEG113-DMA31. This may be a result of the following: 1 — the 62 cationic repeat units act as a proton sponge within the endosome, 2 — the large cationic charge more strongly repels the cationic charge of the liposome in the endosome, and 3 — less polymer is needed so the distribution of PEG on cells and the endosomal membrane is reduced.
3.5. Dependence on polymer surface coverage
Performance of the PEG-b-polycation polymers was dependent on the polymer surface coverage. Surface coverage was attained by equilibrating polymer adsorbed on liposomes with free polymer in solution [28]. Fig. 5 demonstrates the ability of PC and PD liposomes to deliver siRNA with varying amounts of PEG113-DMA31 adsorbed. We compared uncoated liposomes to liposomes with 0.5, 1, or 2 times full surface coverage.
PC liposomes coated with various amounts of PEG113-DMA31 exhibited GFP knockdown similar to bare PC liposomes. Unlike PC liposomes, PD liposomes showed a dependence on polymer adsorption. Below full surface coverage, PD liposomes showed low GFP knockdown, similar to PC liposomes. Above Γ*, we observed an increase in GFP knockdown. Full and a two-fold excess in surface coverage exhibited a 0.33±0.03 and 0.29±0.03 fraction of GFP knockdown, respectively. Surprisingly, two-fold excess coverage (an increase in polymer density) did not hinder GFP knockdown. Polymer desorption may facilitate membrane fusion and disruption.
3.6. siRNA (GAPD) delivery to HUVEC
We also investigated siRNA delivery to HUVEC, a cell line that was reported to undergo endocytosis at a slow rate [32]. Fig. 6 depicts changes in the gene expression levels of GAPD relative to β actin, the endogenous control, for siRNA-treated HUVEC at 24 and 48 h. GAPD was chosen because of its availability and production in HUVEC cells. The threshold cycle (CT) values obtained from qRT-PCR designate the number of amplifications required before entering the exponential phase. Thus, low CT values indicate high mRNA levels. The ddCT values plotted in Fig. 6 were normalized to both β actin and untreated HUVEC. High ddCT values indicate successful GAPD knockdown.
Polymer-coated PD liposomes showed enhanced siRNA delivery relative to naked siRNA, bare PC and PD liposomes, and Oligofectamine complexed siRNA. After 24 h, PEG113-DMA62 coated PD liposomes showed a 10-fold increase in GAPD knockdown relative to bare PD liposomes (Fig. 6A). We observed ddCT values at 24 h of −0.08±0.1, 0.8 ±0.2, and 3.2 ±0.3 for Oligofectamine complexed siRNA, PEG113-DMA31 coated PD liposomes, and PEG113-DMA62 coated PD liposomes, respectively.
After 48 h, PEG113-DMA62 coated PD liposomes continued to offer improved performance over PEG113-DMA31 coated PD liposomes and Oligofectamine complexed siRNA. The ddCT of PEG113-DMA62 coated PD liposomes was 6-fold higher than PEG113-DMA31 coated PD liposomes and 12-fold higher than Oligofectamine complexed siRNA. PEG113-DMA31 coated PD liposomes and Oligofectamine complexed siRNA had ddCT values of 0.40±0.1 and 0.21±0.1 at 48 h, respectively. Naked siRNA and siRNA encapsulated within PC or PD liposomes showed no change in GAPD gene expression.
3.7. Comparison to siRNA delivery strategies
PEG coated liposomes protect from non-specific protein binding which extends the liposome circulation time [20,33]. However, this also makes them less effective in interacting with cells. Delivery of siRNA was accomplished using PEG coated liposomes [23]. PC liposomes incorporating 10 mol% poly(ethylene glycol)-1,2 phosphatidylethanolamine (PEG113-PE) were prepared and tested for siRNA delivery using similar protocols as described herein, with the exception that PEG113-PE was added during formation of the lipid bilayer (data not shown). Its performance, in terms of siRNA knockdown, was similar to PC liposomes, within error. In our previous work, PEG113-PE was shown to bind weakly to liposomes and to disassociate from the liposome surface [28]. This behavior led to liposomes that were predominantly PC, hence the similarity in behavior for siRNA delivery.
Liposomes that encapsulate siRNA offer several advantages over direct administration of siRNA [7,23]. Stable nucleic acid lipid particles (SNALP) encapsulating engineered siRNA molecules have shown an in vivo therapeutic benefit in reducing hepatitis B virus DNA [23]. The SNALP was formulated with cationic, fusogenic, and PEG bound lipids. Biodistribution and inflammatory studies have shown that encapsulation of siRNA within these carriers have reduced liver uptake and plasma levels of type I interferon and inflammatory cytokines [23]. The SNALP formulation was prepared at 42.4 or 48.0 μg siRNA/μmol lipid. Delivery of 40 pmol siRNA (GFP) in the liposome formulations investigated was equivalent to 2 ng siRNA/μmol lipid, which may be optimized for therapeutic use. Delivery vehicles that reduce the amount of siRNA required for delivery while maintaining therapeutic efficacy will be essential in siRNA-mediated therapies.
siRNA encapsulated within PC liposomes showed in vivo therapeutic benefit against tumor growth and Epha2 gene expression [7]. Epha2 over-expression has been correlated with cancer incidence [34]. Reduction in both Epha2 expression and tumor growth was observed [7]. Encapsulation of siRNA within the PEG113-DMA31 and PEG113-DMA62 coated PD liposomes may provide a substantial benefit compared to direct siRNA administration or siRNA encapsulation within PC liposomes.
Lipid formulations that rely on complexing with siRNA have also been studied [16,29,35]. Complexing a cationic liposome with anionic siRNA or DNA may result in neutralization and aggregation. After endocytosis, the electrostatic complex needs to be deconvoluted. Localization of cationic charge may lead to cell toxicity. Encapsulation of siRNA provides a protective barrier, delivery of multiple siRNA to a single cell, and does not rely on other mechanisms for siRNA release. Both systems show downregulation of protein expression in vivo [7,16,23,35].
Determination of whether PEG-b-polycation coated liposomes can improve siRNA delivery requires validation in vivo. Additionally, the effect of polymer desorption during circulationwill be assessed by relation to the charge-dependent biodistribution and circulation time. We believe applications in cancer therapy could be promising, taking advantage of the enhanced permeability and retention effect [36]. Further modification of the polymer-coated PD liposome formulation with an antibody may allow improved delivery to specific disease sites of interest.
4. Conclusions
Regulation of cellular protein expression may provide therapeutic treatment of cancers [2,3,4] and diabetes [6], but is limited by inefficient delivery of siRNA [16]. Our strategy utilizes the shift in pH from the bloodstream to the endosome to trigger the release of PEG-b-polycation polymers from pH-dependent liposomes. Triggered desorption of polymer from PD liposomes is demonstrated (Fig. 1B). In addition, we show the ability of bare and polymer-coated PD liposomes to fuse with RET liposomes (Figs. 2A, B, 3A, and B). siRNA encapsulation within PEG113-DMA31 and PEG113-DMA62 coated PD liposomes results in enhanced (up to 10-fold) siRNA knockdown relative to bare PD and PC liposomes. In comparison to Oligofectamine complexed siRNA and direct siRNA administration, polymer-coated PD liposomes show a significant performance benefit which may be even more substantial based on their ability to evade the immune system and alter the biodistribution. Our data suggest that polymer desorption may contribute to improved siRNA delivery.
Acknowledgments
This work was supported by the National Institute of Health Grant number EB000244. SPA is the recipient of a five-year Royal Society-Wolfson Research Merit Award.
References
- 1.Elbashir S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschi T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultures mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Scherr M, Battmer K, Schultheis B, Ganser A, Elder M. Stable RNA interference as an option for ani-bcr-abl therapy. Gene Ther. 2005;12:12–21. doi: 10.1038/sj.gt.3302328. [DOI] [PubMed] [Google Scholar]
- 3.Charames G, Bapat B. Cyclooxygenase-2 knockdown by RNA interference in colon cancer. Int J Oncol. 2006;28:543–549. [PubMed] [Google Scholar]
- 4.Zhang M, Zhou Y, Xie C, Zhou F, Chen Y, Han G, Zhang W. STAT6 specific shRNA inhibits proliferation and induced apoptosis in colon cancer HT-29 cells. Cancer Letters. :1–9. doi: 10.1016/j.canlet.2005.11.020. in press. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Alegre P, Bode N, Davidson B, Paulson H. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DY1 dystonia. J Neurosci. 2005;25:10502–10509. doi: 10.1523/JNEUROSCI.3016-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Valades A, Vidal-Alabro A, Molas M, Boada J, Bermudez J, Batrons R, Perales J. Overcoming diabetes-induced hyperglycemia through inhibition of hepatic phosphoenolpyruvate carboxykinase (GTP) with RNAi. Mol Ther. 2006;13:401–410. doi: 10.1016/j.ymthe.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Landen C, Chavez-Reyes A, Bucana C, Schmandt R, Deavers M, Lopez-Berestein G, Sood A. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 8.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harbroth J, John M, Kesavan V, Lavine G, Pandey R, Racie T, Rajeev K, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vomlocher H. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman C, Izsvak Z, Katzer A, Ivics Z. Frog Prince transposon-based siRNA vectors mediate efficient gene knockdown in human cells. Journal of siRNA and Gene Silencing. 2005;1:97–104. [PMC free article] [PubMed] [Google Scholar]
- 10.de Jonge J, Holtrop M, Wilschut J, Huckriede A. Reconstituted influenza virus envelopes as an efficient carrier system for cellular delivery of small interfering RNAs. Gene Ther. 2005:1–12. doi: 10.1038/sj.gt.3302673. [DOI] [PubMed] [Google Scholar]
- 11.Taulli R, Accomero P, Follenzi A, Mangano T, Morotti A, Scuoppo C, Forni P, Bersani F, Crepaldi T, Chiarle R, Naldini L, Ponzetto C. siRNA technology and lentiviral delivery as a powerful tool to suppress Tpr-Met-mediated tumorigenesis. Cancer Gene Ther. 2005;12:456–463. doi: 10.1038/sj.cgt.7700815. [DOI] [PubMed] [Google Scholar]
- 12.Rubinson D, Dillon C, Kwiatkowski A, Sievers C, Yang L, Kopinja J, Zhang M, McManus M, Gertler F, Scott M, VanParijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells, and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Tang N, Liu X, Liang W, Xu W, Torchilin V. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J Control Release. 2007;112:229–239. doi: 10.1016/j.jconrel.2006.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasic DD. Novel applications of liposomes. Trends Biotechnol. 1998;16:307–321. doi: 10.1016/s0167-7799(98)01220-7. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release. 2008;126:77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spagnou S, Miller A, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry-US. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 17.Winterhalter M, Lasic D. Liposome stability and formation: experimental parameters and theories on the size distribution. Chem Phys Lipids. 1993;64:35–43. doi: 10.1016/0009-3084(93)90056-9. [DOI] [PubMed] [Google Scholar]
- 18.Deshpande M, Garnett M, Vamvakaki M, Bailey L, Armes S, Stolnik S. Influence of polymer architecture on the structure of complexes formed by PEG-tertiary amine methacrylate copolymers and phosphorothioate oligonucleotide. J Control Release. 2002;81:185–199. doi: 10.1016/s0168-3659(02)00052-4. [DOI] [PubMed] [Google Scholar]
- 19.Yu M, Nowak A, Deming T, Pochan D. Methylated mono- and diethyleneglycol functionalized polylysines: nonionic, alpha-helical, water-soluble polypeptides. J Am Chem Soc. 1999;121:12210–12211. [Google Scholar]
- 20.Blume G, Cevc G. Liposomes for the sustained drug release in vivo. Biochim Biophys Acta. 1990;1029:91–97. doi: 10.1016/0005-2736(90)90440-y. [DOI] [PubMed] [Google Scholar]
- 21.Chen PS, Toribara TY, Warner H. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- 22.Jones L, Yue S, Cheung C, Singer V. RNA quantitation by fluorescence-based solution assay: Ribogreen reagent characterization. Anal Biochem. 1998;265:368–374. doi: 10.1006/abio.1998.2914. [DOI] [PubMed] [Google Scholar]
- 23.Morrissey D, Lockridge J, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer C, Jeffs L, Judge A, MacLachlan I, Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 24.Baleux B. Colorimetric determination of nonionic polyethylene oxide surfactants using an iodine-iodide solution. CR Acad Sci Ser C. 1972;279:1617–1620. [Google Scholar]
- 25.Bailey A, Cullis P. Membrane fusion with cationic liposomes: effects of target membrane lipid composition. Biochemistry. 1997;36:1628–1634. doi: 10.1021/bi961173x. [DOI] [PubMed] [Google Scholar]
- 26.Struck D, Hoekstra D, Pagano R. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 27.Stewart S, Dykxhoorn D, Palliser D, Mizuno H, Yu E, An D, Sabatini D, Chen I, Hahn W, Sharp P, Weinberg R, Novina C. Lentivirus-delivered stable gene silencing by siRNA in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auguste D, Armes S, Brzezinska K, Deming T, Kohn J, Prud’homme RK. pH triggered release of protective poly(ethylene glycol)-b-polycation copolymers from liposomes. Biomaterials. 2006;27:2599–2608. doi: 10.1016/j.biomaterials.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Templeton N, Lasic D, Frederik P, Strey H, Roberts D, Pavlakis G. Improved DNA: liposome complexes for increased systemic delivery and gene expression. Nat Biotechnol. 1997;15:647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 30.Auguste D, Armes S, Brzezinska K, Deming T, Kohn J, Prud’homme R. pH triggered release of protective poly(ethylene glycol)-b-polycation copolymers from liposomes. Biomaterials. 2006;27:2599–2608. doi: 10.1016/j.biomaterials.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 31.Akinc A, Thomas M, Klibanov A, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 32.Wiewrodt R, Thomas A, Cipelletti L, Christofidou-Solomidou M, Weitz D, Feinstein S, Schaffer D, Albelda S, Koval M, Muzykantov V. Size-dependent intracellular immunotargeting of therapeutic cargoes into endothelial cells. Blood. 2002;99:912–922. doi: 10.1182/blood.v99.3.912. [DOI] [PubMed] [Google Scholar]
- 33.Chonn A, Semple SC, Cullis PR. Association of blood proteins with large unilamellar liposomes in vivo. J Biol Chem. 1992;267:18759–18765. [PubMed] [Google Scholar]
- 34.Ogawa K, Pasqualini R, Lindberg R, Kain R, Freeman A, Pasquale E. The ephrin-A1 ligand and its receptor, Epha2, are expressed during tumor neovascularization. Oncogene. 2000;19:6043–6052. doi: 10.1038/sj.onc.1204004. [DOI] [PubMed] [Google Scholar]
- 35.Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, Dames S, Loffler K, Fechtner M, Arnold W, Giese K, Klippel A, Kaufmann J. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 36.Allen TM, Chonn A. Large unilamellar liposomes with low uptake into the reticuloendothelial system. FEBS Lett. 1987;223:42–46. doi: 10.1016/0014-5793(87)80506-9. [DOI] [PubMed] [Google Scholar]


